Published online Apr 28, 2012. doi: 10.3748/wjg.v18.i16.1903
Revised: September 7, 2011
Accepted: April 5, 2012
Published online: April 28, 2012
AIM: To investigate the effects of small interference RNA (siRNA) targeting of Cdx2 on human gastric cancer MGC-803 cells in vitro and in vivo.
METHODS: The recombinant pSilencer 4.1-Cdx2 siRNA plasmids were constructed and transfected into gastric cancer MGC-803 cells in vitro. The stable transfectants were selected. The effects of Cdx2 siRNA on growth, proliferation, cell cycle, apoptosis, migration and invasiveness of human gastric cancer MGC-803 cells were evaluated and the expression of phosphatase and tensin homolog (PTEN), caspase-9 and caspase-3 was observed in vitro by reverse transcription polymerase chain reaction (RT-PCR) and Western blotting analysis. We also investigated the effect of Cdx2 siRNA on growth of MGC-803 cells in nude mice in vivo.
RESULTS: Cdx2 siRNA led to inhibition of endogenous Cdx2 mRNA and protein expression as determined by RT-PCR and Western blotting analysis. Cdx2 siRNA significantly inhibited cell growth and proliferation, blocked entry into the S-phase of the cell cycle, induced cell apoptosis, and reduced the motility and invasion of MGC-803 cells. Cdx2 siRNA also increased PTEN expression, and activated caspase-9 and caspase-3 in MGC-803 cells in vitro . In addition, siRNA targeting of Cdx2 inhibited the growth of MGC-803 cells and promoted tumor cell apoptosis in vivo in nude mice tumor models.
CONCLUSION: Cdx2 was involved in regulating pro-gression of human gastric cancer cells MGC-803. Manipulation of Cdx2 expression may be a potential therapeutic strategy for gastric cancer.
- Citation: Wang XT, Xie YB, Xiao Q. siRNA targeting of Cdx2 inhibits growth of human gastric cancer MGC-803 cells. World J Gastroenterol 2012; 18(16): 1903-1914
- URL: https://www.wjgnet.com/1007-9327/full/v18/i16/1903.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i16.1903
The transcription factor, Cdx2, is a member of the caudal-related homeobox gene family, and is mainly expressed in the intestine. Cdx2 plays important roles in early differentiation, proliferation, and maintenance of intestinal epithelial cells, and in the transcription of genes such as multidrug resistance 1[1,2]. Overexpression of Cdx2 in the small intestine is associated with reduced postnatal growth, early epithelial maturation, alterations in the development of a differentiated phenotype in crypt base organization, and changes in paneth and goblet cell lineages[3].
Initially, Cdx2 was reported to be a tumor suppressor. Several investigators reported that low levels of Cdx2 is a characteristic feature of human colon and squamous esophageal cancer[4,5], and overexpression of Cdx2 could decrease mobility and antagonize metastasis of colon cancer cells[6]. However, other studies showed that strong and robust expression of Cdx2 was found in > 80% of colorectal cancers and non-small cell lung cancer[7,8]. In addition, Cdx2 was found to enhance proliferation and have tumorigenic potential in the human colon cancer cell lines, LoVo and SW48[9]. These studies suggested that Cdx2 also had oncogenic property. Together, these conflicting findings point to a complex role for Cdx2 in the regulation of cell proliferation.
Gastric cancer is the third most common cancer in China, and is one of the most frequent causes of cancer-related mortality in China, with an incidence of 0.4 million new cases and 0.3 million deaths annually[10]. Intestinal metaplasia has been shown to be a precursor of intestinal-type gastric adenocarcinoma. Since intestinal metaplasia cannot be eradicated, it is important to determine how to reduce the morbidity from intestinal metaplasia to gastric cancer. However, the histogenesis of intestinal metaplasia and factors in the metaplastic epithelium that lead to its development into carcinoma is still in dispute[11,12]. In adult humans, Cdx2 has been reported to be associated with intestinal metaplasia in the stomach in which ectopic expression of Cdx2 is speculated to cause the gastric epithelial cells to trans-differentiate and take the intestinal phenotype[13]. In addition, Cdx2 transgenic mice have been shown to induce intestinal metaplasia and have a high incidence of gastric carcinoma[14,15]. This indicates a direct relationship between Cdx2-induced intestinal metaplasia and gastric carcinogenesis.
In the present study, we constructed small interference RNA (siRNA) sequences targeting of Cdx2, transfected them into the human gastric cancer cell line MGC-803, selected the stable transfectants, and explored changes in growth, proliferation, cell cycle, apoptosis, metastasis and invasiveness. We also observed the effect of Cdx2 siRNA on the expression of phosphatase and tensin homolog (PTEN), caspase-9, and caspase-3. Moreover, we investigated the effects of Cdx2 downregulation on the growth and apoptosis of MGC-803 cells in nude mice.
The human gastric carcinoma cell line, MGC-803, was supplied by the Cell Bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Gaithersburg, MD, United States). All media were supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL). Cells were cultured in an incubator with 5% CO2 at 37 °C with medium changes every 3 d.
Anti-Cdx2, anti-β-actin and secondary antibody were obtained from Santa Cruz Biotechnology Inc., Santa Cruz, CA, United States. Antibodies specific for PTEN, pro-caspase-9, cleaved caspase-9, pro-caspase-3, cleaved caspase-3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were from Cell Signaling Technology, Beverly, MA, United States.
Double strand siRNA oligonucleotides were obtained from Gima Biotechnology Company (China). There were two reversed repeated sequences with 21 inserted sequences (GACAAATATCGAGTGGTGTAC, TAACCCGCGATCTGTTCTGCA) in the complementary sequence, with BamHI and HindIII sites for ligation into the pSilencer 4.1 vector, which contained a neomycin resistance marker for the selection of stable transfectants in the presence of G418. The siRNA targeting site of the transcripted product was nucleotides 115-818 of Cdx2 mRNA (GeneBank No. NM-001265). The negative control was the siRNA sequence with no homology to any human gene sequence.
After ligation, the plasmid was transformed into Escherichia coli TOP10 cells, and then cultured on solid LB medium (LB solid medium containing 50 ng/L ampicillin and 2% agarose gel) at 37 °C for 16 h. Positive clones were identified by DNA sequence analysis (Majorbio Biotech Co., Ltd), and the resulting plasmid was named pSilencer 4.1-Cdx2(+) or pSilencer 4.1-Cdx2(-). Six-well plates were inoculated with MGC-803 cells (1 × 105), and cells were transfected with pSilencer 4.1-Cdx2(+) recombination plasmids. For selection of stable transfectants, G418 (Life Technologies) was added to the cells 48 h after transfection. The concentration of G418 for selection was gradually decreased as follows: 1 mg/mL for 4 d; 750 μg/mL for 4 d; 500 μg/mL for 4 d; and 250 μg/mL as a sustaining dose. At day 20 after transfection, G418-resistant clones were isolated. The selected cell colonies were transferred from 10-mm dishes to 96-well plates, and then from 96-well plates to 24-well plates. The transformants selected by G418 were analyzed by measuring the expression of Cdx2 mRNA and protein. The negative control cells were transfected with vector pSilencer 4.1-Cdx2(-) alone, and maintained under identical conditions. In the case of cells that were selected in medium containing G418, antibiotics were routinely included in their growth medium until 1 to 2 d before experiments were carried out. The cells were divided into 3 groups: MGC-803/Cdx2 siRNA, MGC-803/Cdx2 negative control and MGC-803 group.
Total RNA was extracted from the positive cell clone using TRIzol Reagent (Invitrogen). Neo gene segments were amplified and verified by semi-quantitative reverse transcription polymerase chain reaction (RT-PCR). Complementary deoxyribonucleic acids (cDNAs) were reverse-transcribed from 2 μg of total RNA. Primers used in this study were as follows: Cdx2 forward primer (5’-CGGCAGCCAAGTGAAAAC-3’) and reverse primer (5’-GATGGTGATGTAGCGACTGTAGTG-3’), PCR product: 100 base pairs (bp); β-actin forward primer (5’-AACTCCATCATGAAGTGTGA-3’) and reverse primer (5’-ACTCCTGCTTGCTGATCCAC -3’), PCR product: 247 bp. The PCR products were checked by agarose gel electrophoresis, and the abundance of each mRNA was detected and normalized to that of β-actin mRNA.
Cell lysates were prepared in a buffer containing 100 mmol/L NaCl, 10 mmol/L Tris-Cl (pH 7.6), 1 mmol/L ethylenediaminetetraacetic acid (pH 8.0), 1 μg/mL aprotinin, 100 μg/mL phenylmethylsulfonyl fluoride, and 1% (v/v) NP-40. After protein quantitation using the Lowry protein assay, equal amounts of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and blotted onto nitrocellulose membranes by the semi-dry blotting method using a three buffer system. The membrane was blocked with 5% bovine serum albumin in phosphate buffer solution Tween-20 (PBST) (PBS, pH 7.5, containing 0.1% Tween-20), and incubated with a 1:500 dilution of primary antibody (anti-Cdx2) overnight at 4 °C. The membrane was then washed with PBST and incubated with a peroxidase-conjugated secondary antibody (1:1000) for 1 h. Specific antibody binding was detected using a chemiluminescence detection system (Pierce, Rockford, IL, United States), according to the manufacturer’s recommendations. Western blotting film was scanned, and the net intensities of the bands were quantified using Image-QuanT software (Molecular Dynamics, Sunnyvale, CA, United States). After development, the membrane was stripped and reprobed with antibody against β-actin (1:1000) to confirm equal sample loading.
The growth of MGC-803 cells was determined by an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay using a CellTiter 96 AQueous assay system (Promega, Madison, WI, United States), according to the manufacturer’s instructions. This assay measures dehydrogenase enzyme activity in metabolically active tumor cells, as reflected by the conversion of MTT to formazan, which is soluble in tissue culture medium and is detected by absorbance (A) at 490 nm. The production of formazan is proportional to the number of living cells, with the intensity of the produced color serving as an indicator of cell viability. Briefly, MGC-803 cells were plated at a density of 5 × 103 cells/well in 96-well plates, and cultured for 72 h. The percentage of cell survival was calculated using the background-corrected absorbance: % proliferation rate = 100 × A of experimental well/A of untreated control well. All experiments were performed at least three times.
Cell suspensions from each group were diluted in DMEM with 10% FBS, and immediately re-plated in 6-well plates at a density of 20 cells/cm2. The plates were incubated until cells in control wells formed sufficiently large colonies. After that, the colonies were fixed in 6% glutaraldehyde and stained with 0.5% crystal violet. The plates were photographed and their digital images were analyzed manually to determine colony number.
For cell cycle analysis, MGC-803 cells (1 × 106) were washed twice with ice-cold PBS, treated with trypsin, and then fixed in 70% cold ethanol at 4 °C for 30 min. The cell pellet was incubated in a solution containing 50 ng/mL propidium iodide, 0.2 mg/mL RNase, and 0.1% Triton X-100 at room temperature for 30 min, and then analyzed by flow cytometry using a FACscan (Becton Dickinson, Mountain View, CA, United States). Data were analyzed with the MultiCycle for Windows (Phoenix Flow Systems, San Diego, United States).
Apoptotic cells were determined using the Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (Jingmei Biotech Co., Shenzhen, China) and an EPICS XL-MCL flow cytometer (Becton Dickinson) according to the manufacturer’s instructions. Briefly, 1 × 106 cells were stained with Annexin V/FITC for 30 min at 4 °C in the dark and then stained with propidium iodide for 10 min before flow cytometric analysis.
The cells were cultured to confluence in 6-well plates, and were then treated with mitomycin C to inhibit cell proliferation. A central linear wound was made with a 200 μL sterile pipet tip. Media were changed gently to remove any floating cells. Phase micrographs of the wound cultures were taken at 0 and 36 h. The photographs were analyzed by measuring the distance from the wound edge of the cell sheet to the original wound site. Migratory activity was calculated as the mean distance between edges of three points in 12 fields per well. Relative motility = (mean original distance - mean distance at a time point)/mean original distance × 100%. Each test group was assayed in triplicate.
Cell invasion was assessed using Transwell chambers (6.5 mm; Corning, New York, United States) with 50 μL serum-free DMEM containing 1 μg/mL Matrigel (Department of Biology, Beijing University, China) in the upper chamber. The lower chamber was filled with 50 μL DMEM containing 0.1 μg/mL fibronectin (Beijing University). Cells (1 × 105) were suspended in 100 μL DMEM with 1% fetal calf serum and plated into the upper chamber. PBS (5%) 500 μL was added in the lower chamber. After a 24 h incubation with 5% CO2 at 37 °C, the number of cells with Giemsa staining on the undersurface of the polycarbonate membranes was scored visually in five random fields at a 400 × magnification by light microscopy.
Semi-quantitative RT-PCR was performed as previously described. Primers used in this study were as follows: (1) PTEN forward primer (5’-CTGGAAAGGGACGAACTG-3’) and reverse primer (5’-AGGTAACGGCTGAGGGA-3’), PCR product: 368 bp; (2) Caspase-9 forward primer (5’-GGCTGTCTACGGCACAGATGGA-3’) and reverse primer (5’-CTGGCTCGGGGTTACTGCCAG-3’), PCR product: 200 bp; (3) Caspase-3 forward primer (5’-AAGCGAATCAATGGACTC-3’) and reverse primer (5’-TTCCTGACTTCATATTTCAA-3’), PCR product: 192 bp; (4) GAPDH (a) forward primer (5’-ACAGCAACAGGGTGGTGGAC-3’) and reverse primer (5’-TTTGAGGGTGCAGCGAACTT-3’), PCR product: 252 bp; and (5) GAPDH (b) forward primer (5’-ACCACAGTCCATGCCATCAC-3’) and reverse primer (5’-TCACCACCCTGTTGCTGTA-3’), PCR product: 450 bp. Western blotting analysis was carried out as previously described.
BALB/c male nude mice at 5 wk old were obtained from Guangxi Animal Center, China. All animals were kept under specific pathogen-free conditions and tended to in accordance with institutional guidelines. All experimental studies were approved by the Guangxi Medical University Animal Care and Use Committee. MGC-803/Cdx2 siRNA cells, MGC-803/Cdx2 negative control cells and MGC-803 cells were used for tumor implantation. There were six mice in each group. Approximately 2 × 106 tumor cells were implanted subcutaneously into the flanks of the nude mice. Tumor sizes were measured every 4 d with a caliper, and the diameters were recorded. The tumor volume (TV) was calculated by the formula: TV = W2× L/2, where L was the length and W was the width of the tumor. The relative tumor volume (RTV) was calculated by the formula: RTV = Vt/V0 (V0 is the TV at the day when the chemicals were given, and Vt is the TV of subsequent measurement). After mice were killed, total RNA and protein were extracted from tumor tissues. The expression of Cdx2 mRNA and protein were detected by semi-quantitative RT-PCR and Western blotting analysis, respectively. Tumor cells were assessed for apoptosis using in situ terminal deoxynucleotidyl transferase-mediated 2’-deoxyuridine, 5’-triphosphate nick end labeling assays (TUNEL). Apoptosis was evaluated by counting the positive cells (brown-stained cells) as well as the total number of cells in 10 arbitrarily selected fields at 400 × magnifications in a double-blinded manner. The apoptotic index (per 400 × microscopic field) was calculated as the number of apoptotic cells × 100/total number of cells. Brown-stained nuclei immediately at the edge of a tissue section were excluded from cell counts to minimize false positives.
Data are expressed as mean ± SE. Statistical significance was determined using χ2 test, student’s t test, or one-way analysis of variance. Statistical analysis were carried out using SPSS, version 13.0 (SPSS Inc., Chicago, IL, United States) or Origin 7.5 software programs (OriginLab Co., Northampton, MA, United States). A value of P < 0.05 was considered as statistically significant.
Recombinant pSilencer 4.1-Cdx2(+) and pSilencer 4.1- Cdx2(-) sequences were verified by DNA sequenced analysis (data not shown) which demonstrated that the inserted siRNA coding frames and frame sequences were correct. This confirmed that the construction of Cdx2 siRNA expression plasmid was successful.
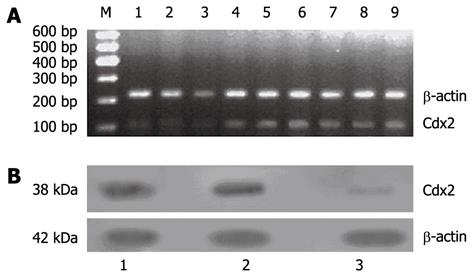
The transfection of pSilencer 4.1-Cdx2(+) plasmid into MGC-803 cells led to remarkable inhibition of Cdx2 mRNA and protein expression. Densitometric analysis showed that Cdx2 mRNA and protein in MGC-803/Cdx2 siRNA cells were about 11- and 7-fold lower, respectively, than those in the two control groups (P < 0.05), while no differences were found between MGC-803/Cdx2 negative control cells and MGC-803 cells (Figure 1).
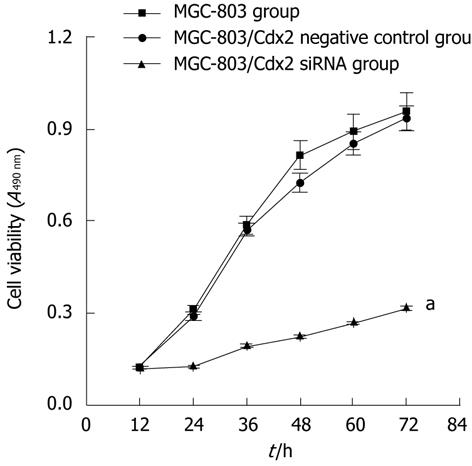
Next, we determined the in vitro survival rates of gastric tumor cells stably transfected with pSilencer 4.1-Cdx2(+) plasmids, using the gastric carcinoma cell line, MGC-803, as a model for gastric cancer. As shown in Figure 2, Cdx2 siRNA significantly reduced cell survival, as assessed by the MTT assay. We observed that MGC-803/Cdx2 siRNA cells obviously grew slower than MGC-803/Cdx2 negative control cells and MGC-803 cells (P < 0.05), which was consistent with the decreased levels of Cdx2 in MGC-803/Cdx2 siRNA cells. Additionally, MGC-803/Cdx2 negative control cells and MGC-803 cells exhibited about 3-fold higher mean proliferation rates than MGC-803/Cdx2 siRNA cells (P < 0.05). These results indicate a suppressive effect of Cdx2 siRNA on MGC-803 cell growth and survival.
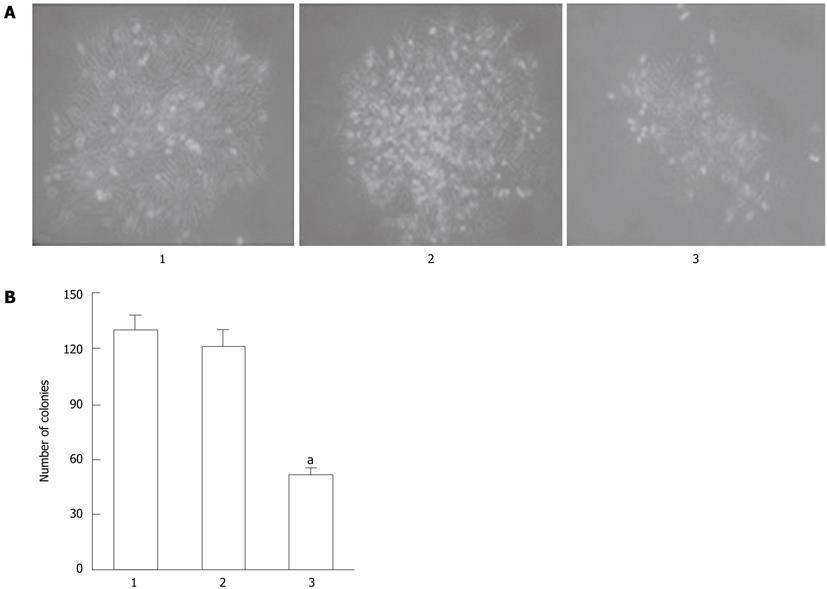
To confirm the inhibitory effect of Cdx2 siRNA on the growth of MGC-803 cells, we performed colony formation assays to measure the capability of the cells to grow in an anchorage-independent environment by culturing the cells in soft agarose. As shown in Figure 3, three cell lines were able to form colonies in soft agarose, but the number of colony formation in MGC-803/Cdx2 siRNA cells after 3 wk was 51.4 ± 3.2, with a 60.1% and 57.6% decrease, compared to the two control groups, respectively (P < 0.05). Together, these data suggest that Cdx2 siRNA inhibits cell growth and proliferation in gastric cancer cells.
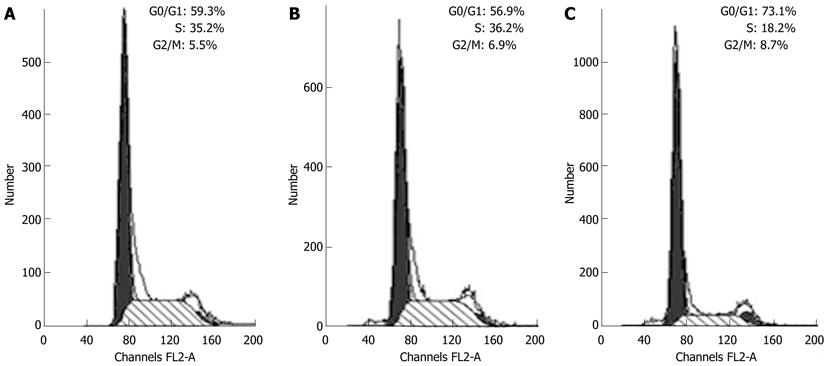
We used flow cytometry to determine whether the inhibitory effect of Cdx2 siRNA on MGC-803 cell proliferation was mediated, at least in part, through affecting cell cycle progression. We found that MGC-803/Cdx2 siRNA cells were 73.1% in G0/G1 phase and 18.2% in S phase, with a 13.8% and 16.2% increase in the G0/G1 phase cell population, and a 17% and 18% decrease in the S phase cell population, compared to MGC-803 cells and MGC-803/Cdx2 negative control cells (P < 0.05) (Figure 4). These data indicate that cell growth inhibition by Cdx2 siRNA is associated with significant cell cycle arrest in G0/G1 phase, and suggest that siRNA directed against the Cdx2 gene suppresses cell proliferation by controlling the G1 and S checkpoints and inducing a specific block in cell cycle progression.
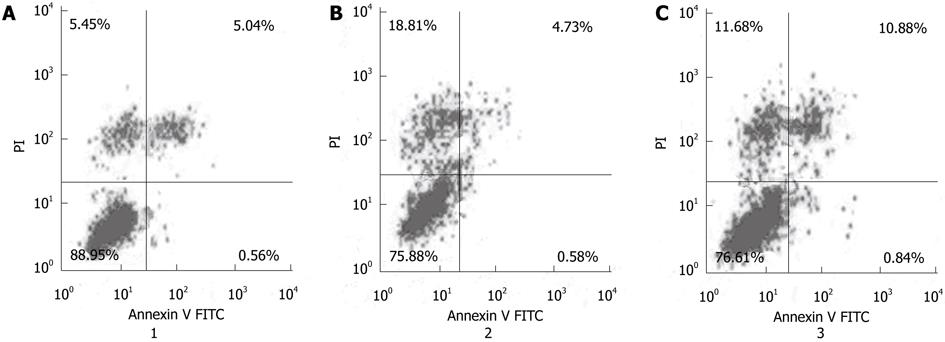
To further study the effect of Cdx2 siRNA on MGC-803 cell apoptosis, cells were stained with Annexin V-FITC and propidium iodide, and then subsequently analyzed by flow cytometry. The dual parameter fluorescent dot plots showed that the viable cells were in the lower left quadrant, and the apoptotic cells were in the right quadrant. As shown in Figure 5, the apoptotic percentage of MGC-803/Cdx2 siRNA cells was 11.7% ± 2.2%, which was significantly higher than that of MGC-803/Cdx2 negative control (5.3% ± 1.3%) and MGC-803 cells (5.6% ± 1.1%) (P < 0.05). This implies that inhibition of Cdx2 is able to induce apoptosis in gastric cancer MGC-803 cells.
We measured the migratory ability of three cell groups using the wound healing assay by scratching the single-layer cells. As shown in Figure 6, the distance between the wound edges was determined at 0 and 36 h and the healing rate was calculated in the three groups. MGC-803/Cdx2 siRNA cells showed a lower migratory ability at 36 h than MGC-803/Cdx2 negative control and MGC-803 cells. The healing rate of MGC-803/Cdx2 siRNA cells after 36 h was 53.7% ± 7.2%, with a 39.9% and 40.8% decrease, as compared to MGC-803/Cdx2 negative control cells and MGC-803 cells (P < 0.05).
Since siRNA targeting of Cdx2 inhibited the expression of Cdx2 gene in gastric cancer cells, we assessed its ability to inhibit cell invasion. After incubation for 24 h in the invasion assay, the numbers of MGC-803/Cdx2 negative control and MGC-803 cells invaded through the membrane of Matrigel chamber were 2.9- and 3.0-fold greater than that of MGC-803/Cdx2 siRNA cells, respectively (P < 0.05) (Figure 7). The results indicate that Cdx2 siRNA reduces the migratory and invasion ability of gastric cancer MGC-803 cells.
To investigate the mechanism by which Cdx2 siRNA induces apoptosis in MGC-803 cells, we detected expression levels of several apoptotic family members including PTEN, caspase-9, and caspase-3 by semi-quantitative RT-PCR and Western blotting analysis. As shown in Figure 8, densitometric analysis showed that PTEN, caspase-9, and caspase-3 mRNA of MGC-803/Cdx2 siRNA cells were higher than that in MGC-803 cells and MGC-803/Cdx2 negative control cells (P < 0.05), while no differences were found between MGC-803/Cdx2 negative control cells and MGC-803 cells. As shown in Figure 9, Cdx2 siRNA led to the cleavage of pro-caspase-9 (47 kDa) and pro-caspase-3 (35 kDa) into other multiple, cleaved, maturation products (data not shown), but only the 37-kDa form of cleaved caspase-9 and the 17-kDa form of cleaved caspase-3 were observed. Densitometric analysis showed that PTEN, p37 cleaved caspase-9, and p17 cleaved caspase-3 protein of MGC-803/Cdx2 siRNA cells were higher, while pro-caspase-9 and pro-caspase-3 were lower than that in MGC-803 cells and MGC-803/Cdx2 negative control cells (P < 0.05). No differences were found between MGC-803/Cdx2 negative control cells and MGC-803 cells.
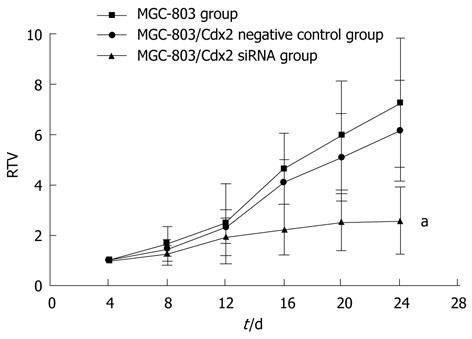
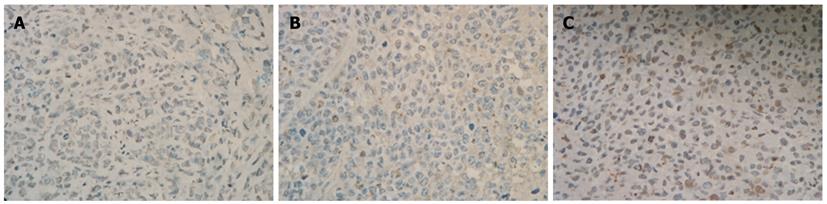
We also examined the effect of Cdx2 siRNA on growth of MGC-803 cells in vivo by implanting MGC-803/Cdx2 siRNA cells subcutaneously into the flanks of BALB/c nude mice. Four weeks after implantation, tumor weight from MGC-803/Cdx2 siRNA cells was 0.773 ± 0.054 g, which was significantly less than 2.334 ± 0.087 g from MGC-803 cells, and 2.356 ± 0.092 g from MGC-803/Cdx2 negative control cells (P < 0.05). As shown in Figure 10, the tumor growth curves indicate the significant growth inhibition in MGC-803/Cdx2 siRNA cells (P < 0.05). Densitometric analysis showed that Cdx2 mRNA expression in MGC-803/Cdx2 siRNA cells (0.305 ± 0.053) was lower than that in MGC-803 cells (1.524 ± 0.323) and MGC-803/Cdx2 negative control cells (1.441 ± 0.269), as determined by semi-quantitative RT-PCR (P < 0.05) (Figure 11A). In addition, the relative protein expression of Cdx2 in MGC-803/Cdx2 siRNA cells (0.134 ± 0.087) also significantly decreased (P < 0.05), when compared to MGC-803 cells (0.634 ± 0.156) and MGC-803/Cdx2 negative control cells (0.569 ± 0.167), as determined by Western blotting analysis (Figure 11B). As shown in Figure 12, the percent of apoptotic tumor cells in MGC-803/Cdx2 siRNA cells was 16.7% ± 5.6%, which was more than 10.5% ± 4.1% in MGC-803/Cdx2 negative control cells and 11.2% ± 4.3% in MGC-803 cells, as determined by the TUNEL method.
The Cdx2 homeobox gene, which is homologous to the Drosophila gene caudal, has an essential role during early development[4]. In adults, Cdx2 expression is restricted to intestinal epithelial cells. Overexpression of Cdx2 in human colon cancer cells induces a less malignant phenotype, inhibiting proliferation, invasion, and migration[16], and Cdx2 expression is progressively reduced in gastric cancer[17]. Moreover, heterozygous-null Cdx2 mice are more sensitive to azoxymethane-induced colonic adenocarcinomas[18], and mice that are compound heterozygotes for Cdx2 and the tumor suppressor Adenomatous Polyposis Coli (Apc) developed more adenomatous polyps in the colon than their heterozygous Apc littermates[19]. These studies suggested that Cdx2 is a putative tumor suppressor.
However, other reports have shown that Cdx2 plays a pivotal role in the development of intestinal metaplasia[20,21]. The implication of Cdx2 in intestinal metaplasia has been demonstrated in intestinal metaplasia of the stomach where Cdx2 was ectopically overexpressed, suggesting that it could play a major role during intestinal metaplasia formation in the stomach[21]. Intestinal metaplasia is a precursor of intestinal-type gastric adenocarcinoma. Long-term intestinal metaplasia induced gastric adenocarcinoma in the Cdx2-transgenic mouse stomach, and no significant changes were noted in wild-type littermates[14]. The tumor incidence was 100% at 100 wk after birth[15]. It can thus be concluded that Cdx2-induced intestinal metaplasia itself is a precancerous lesion leading to gastric carcinoma. Furthermore, Cdx2 is overexpressed in most colorectal tumors compared to matched normal mucosa in adults[7]. Dang et al[22] showed that Cdx2 does not suppress tumorigenicity in the human gastric cancer cell line, MKN45. It can be concluded that, in contrast to the prevailing paradigm, Cdx2 does not serve as a tumor suppressor in the development of most sporadic colorectal tumors. Rather, in the context of earlier observations of its role in promoting the neoplastic phenotype in some cells and tissues, many observations suggest the intriguing possibility that Cdx2 could serve as an oncogene in the gastrointestinal tract[9,23]. This suggests that the level of Cdx2 expression may contribute to its function[9], thereby raising the possibility that intervening with Cdx2 expression in gastric cancer cells with RNA interference may control their growth rate.
Our study indicated that Cdx2 siRNA led to remarkable inhibition of Cdx2 mRNA and protein expression in MGC-803 cells, inhibited cell growth, caused cell cycle arrest in the G0/G1 phase, and induced cell apoptosis. Furthermore, RNAi-directed targeting of Cdx2 in MGC-803 cells reduced the capability of cell motion, invasion, and colony formation. Moreover, a strong anti-tumor effect of Cdx2-siRNA in vivo was observed, as tumor growth was suppressed and tumor apoptosis was increased in nude mice when Cdx2 mRNA and protein was silenced by Cdx2 siRNA. These findings suggest that Cdx2 has tumorigenic potential in the human gastric cancer cell lines MGC-803.
However, our previous study showed that Cdx2 overexpression in human gastric cancer MGC-803 cells produce similar results as Cdx2 siRNA[24]. Moreover, Cdx2 overexpression was associated with cell cycle arrest in the G0/G1 phase which was the same as Cdx2 siRNA. This suggests that Cdx2 plays a double role in the regulation of MGC-803 cell growth and death. Thus, we can only speculate on potential explanations for these observed contrasts. First, appropriate activity and expression levels of Cdx2 are necessary for the normal cell cycle, even in promoting tumor proliferation and regression. Just like E2F-1, both the upregulation and downregulation of E2F-1 can suppress human gastric cancer MGC-803 cell growth in vitro[25,26]. Second, these two conflicting results may involve different mechanisms. Our previous data showed that overexpression of Cdx2 inhibits MGC-803 cell progression via the Wnt signaling pathway (unpublished data). In this result, PTEN, caspase-9 and caspase-3 expression were all increment when Cdx2 was downregulated. The PTEN protein product is a lipid phosphatase that antagonizes PI3K function and consequently inhibits downstream signaling transduction through Akt[27]. Caspase-9, a member of the protease family, is intimately associated with the initiation of apoptosis, and is thought to be activated while Akt is inhibited[28]. Activated caspase-9 is able to cleave caspase-3 in vitro, leading to apoptosis[29]. Therefore, in the present study, inhibition of Cdx2 expression may increase PTEN expression directly or indirectly, leading to activation of caspase-9 and caspase-3 via the PI3K/Akt signaling pathway, which is responsible for inhibition of MGC-803 cell growth in vitro and in vivo. Further studies are needed to confirm our results.
Gastric cancer is a worldwide problem. Besides the undetermined etiological factors, there are also limitations in surgery, chemotherapy and radiotherapy, which to date, are the major therapies for gastric cancer[30]. Many patients lose the chance of surgery because of their systemic condition, while many cannot tolerate the side effects of chemotherapy or radiotherapy. It is important to find a new way to effectively inhibit cancer cell growth and avoid the side effects of drugs or radioactive rays. Gene target therapies have proved to be a promising way to achieve this goal[26]. In this study, we showed that Cdx2 plays a critical role in gastric cancer cell proliferation, invasion, and apoptosis. The downregulation of Cdx2 using RNAi successfully reduced the progression of gastric cancer MGC-803 cells in vitro and in vivo. In conclusion, this study lays the foundation for treatment of gastric cancer through manipulation of Cdx2 expression.
Gastric cancer is a worldwide problem. Besides the undetermined etiological factors, there are also limitations in surgery, chemotherapy and radiotherapy, which to date, are the major therapies for gastric cancer. It is important to find a new way to effectively inhibit cancer cell growth and avoid the side effects of drugs or radioactive rays. Gene target therapies have proved to be a promising way to achieve this goal. The caudal-type homeobox gene, Cdx2, plays an important role in intestinal metaplasia, and is a precursor of intestinal-type gastric carcinoma. However, the effect of Cdx2 in gastric cancer is still not very clear.
Cdx2 plays important roles in early differentiation, proliferation and maintenance of intestinal epithelial cells. The role of Cdx2 as an oncogene or a tumor suppressor gene is still in dispute at the present time. The Cdx2 research hotspot is how it affect the progression of human cancer.
This study for the first time demonstrated that Cdx2 small interference RNA (siRNA) significantly inhibited cell growth and proliferation, blocked entry into the S-phase of the cell cycle, induced cell apoptosis, and reduced the motility and invasion of MGC-803 cells. Cdx2 siRNA also increased phosphatase and tensin homolog expression, and activated caspase-9 and caspase-3 in MGC-803 cells in vitro as determined by reverse transcription polymerase chain reaction and Western blotting analysis. In addition, siRNA targeting of Cdx2 inhibited the growth of MGC-803 cells and promoted tumor cell apoptosis in vivo in nude mice tumor models.
This study lays the foundation for treatment of gastric cancer through manipulation of Cdx2 expression.
The transcription factor, Cdx2, is a member of the caudal-related homeobox gene family, and is mainly expressed in the intestine. It is also known to be a key factor in the development of intestinal metaplasia.
In this study, the authors constructed recombinant pSilencer 4.1-Cdx2 siRNA plasmids and transfected them into human gastric cancer MGC-803 cells in vitro. The authors demonstrated that Cdx2 siRNA led to inhibition of endogenous Cdx2 mRNA and protein expression and Cdx2 siRNA significantly inhibited cell growth and proliferation, blocked entry into the S-phase of the cell cycle, induced cell apoptosis, and reduced the motility and invasion of MGC-803 cells. The authors conclude that Cdx2 is involved in the regulation of tumor growth, proliferation, apoptosis and invasion of gastric cancer cells. Overall this is a well-conducted pilot study.
Peer reviewer: Lin-Feng Chen, Assistant Professor, Department of Biochemistry, COM 190 MSB, MC-714, University of Illinois at Urbana-Champaign, Urbana, IL 61801, United States
S- Editor Cheng JX L- Editor A E- Editor Li JY
| 1. | Boyd M, Hansen M, Jensen TG, Perearnau A, Olsen AK, Bram LL, Bak M, Tommerup N, Olsen J, Troelsen JT. Genome-wide analysis of CDX2 binding in intestinal epithelial cells (Caco-2). J Biol Chem. 2010;285:25115-25125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 2. | Takakura Y, Hinoi T, Oue N, Sasada T, Kawaguchi Y, Okajima M, Akyol A, Fearon ER, Yasui W, Ohdan H. CDX2 regulates multidrug resistance 1 gene expression in malignant intestinal epithelium. Cancer Res. 2010;70:6767-6778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Crissey MA, Guo RJ, Funakoshi S, Kong J, Liu J, Lynch JP. Cdx2 levels modulate intestinal epithelium maturity and Paneth cell development. Gastroenterology. 2011;140:517-528.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 4. | Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3:593-601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 189] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 5. | Guo M, House MG, Suzuki H, Ye Y, Brock MV, Lu F, Liu Z, Rustgi AK, Herman JG. Epigenetic silencing of CDX2 is a feature of squamous esophageal cancer. Int J Cancer. 2007;121:1219-1226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Gross I, Duluc I, Benameur T, Calon A, Martin E, Brabletz T, Kedinger M, Domon-Dell C, Freund JN. The intestine-specific homeobox gene Cdx2 decreases mobility and antagonizes dissemination of colon cancer cells. Oncogene. 2008;27:107-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Witek ME, Nielsen K, Walters R, Hyslop T, Palazzo J, Schulz S, Waldman SA. The putative tumor suppressor Cdx2 is overexpressed by human colorectal adenocarcinomas. Clin Cancer Res. 2005;11:8549-8556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Grimminger P, Ling FC, Neiss S, Vallböhmer D, Lurje G, Schneider PM, Hölscher AH, Metzger R, Brabender J. The role of the homeobox genes BFT and CDX2 in the pathogenesis of non-small cell lung cancer. Anticancer Res. 2009;29:1281-1286. [PubMed] [Cited in This Article: ] |
| 9. | Dang LH, Chen F, Ying C, Chun SY, Knock SA, Appelman HD, Dang DT. CDX2 has tumorigenic potential in the human colon cancer cell lines LOVO and SW48. Oncogene. 2006;25:2264-2272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17-20. [PubMed] [Cited in This Article: ] |
| 11. | Busuttil RA, Boussioutas A. Intestinal metaplasia: a premalignant lesion involved in gastric carcinogenesis. J Gastroenterol Hepatol. 2009;24:193-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Zheng Y, Wang L, Zhang JP, Yang JY, Zhao ZM, Zhang XY. Expression of p53, c-erbB-2 and Ki67 in intestinal metaplasia and gastric carcinoma. World J Gastroenterol. 2010;16:339-344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 28] [Cited by in F6Publishing: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Barros R, da Costa LT, Pinto-de-Sousa J, Duluc I, Freund JN, David L, Almeida R. CDX2 autoregulation in human intestinal metaplasia of the stomach: impact on the stability of the phenotype. Gut. 2011;60:290-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 14. | Almeida R, Silva E, Santos-Silva F, Silberg DG, Wang J, De Bolós C, David L. Expression of intestine-specific transcription factors, CDX1 and CDX2, in intestinal metaplasia and gastric carcinomas. J Pathol. 2003;199:36-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 212] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 15. | Mutoh H, Sakurai S, Satoh K, Tamada K, Kita H, Osawa H, Tomiyama T, Sato Y, Yamamoto H, Isoda N. Development of gastric carcinoma from intestinal metaplasia in Cdx2-transgenic mice. Cancer Res. 2004;64:7740-7747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 135] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 16. | Brabletz T, Spaderna S, Kolb J, Hlubek F, Faller G, Bruns CJ, Jung A, Nentwich J, Duluc I, Domon-Dell C. Down-regulation of the homeodomain factor Cdx2 in colorectal cancer by collagen type I: an active role for the tumor environment in malignant tumor progression. Cancer Res. 2004;64:6973-6977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 111] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 17. | Liu Q, Teh M, Ito K, Shah N, Ito Y, Yeoh KG. CDX2 expression is progressively decreased in human gastric intestinal metaplasia, dysplasia and cancer. Mod Pathol. 2007;20:1286-1297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Bonhomme C, Duluc I, Martin E, Chawengsaksophak K, Chenard MP, Kedinger M, Beck F, Freund JN, Domon-Dell C. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut. 2003;52:1465-1471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 188] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 19. | Aoki K, Tamai Y, Horiike S, Oshima M, Taketo MM. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/- compound mutant mice. Nat Genet. 2003;35:323-330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 184] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 20. | Hinoi T, Lucas PC, Kuick R, Hanash S, Cho KR, Fearon ER. CDX2 regulates liver intestine-cadherin expression in normal and malignant colon epithelium and intestinal metaplasia. Gastroenterology. 2002;123:1565-1577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 168] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 21. | Bai YQ, Yamamoto H, Akiyama Y, Tanaka H, Takizawa T, Koike M, Kenji Yagi O, Saitoh K, Takeshita K, Iwai T. Ectopic expression of homeodomain protein CDX2 in intestinal metaplasia and carcinomas of the stomach. Cancer Lett. 2002;176:47-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 158] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 22. | Dang LH, Chen F, Knock SA, Huang EH, Feng J, Appelman HD, Dang DT. CDX2 does not suppress tumorigenicity in the human gastric cancer cell line MKN45. Oncogene. 2006;25:2048-2059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Kaimaktchiev V, Terracciano L, Tornillo L, Spichtin H, Stoios D, Bundi M, Korcheva V, Mirlacher M, Loda M, Sauter G. The homeobox intestinal differentiation factor CDX2 is selectively expressed in gastrointestinal adenocarcinomas. Mod Pathol. 2004;17:1392-1399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 144] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 24. | Xie Y, Li L, Wang X, Qin Y, Qian Q, Yuan X, Xiao Q. Overexpression of Cdx2 inhibits progression of gastric cancer in vitro. Int J Oncol. 2010;36:509-516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 25. | Xie Y, Wang C, Li L, Ma Y, Yin Y, Xiao Q. Overexpression of E2F-1 inhibits progression of gastric cancer in vitro. Cell Biol Int. 2009;33:640-649. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Xie Y, Yin Y, Li L, Ma Y, Xiao Q. Short interfering RNA directed against the E2F-1 gene suppressing gastric cancer progression in vitro. Oncol Rep. 2009;21:1345-1353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA, Bäsecke J, Stivala F, Donia M, Fagone P. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135-164. [PubMed] [Cited in This Article: ] |
| 28. | Shultz JC, Goehe RW, Wijesinghe DS, Murudkar C, Hawkins AJ, Shay JW, Minna JD, Chalfant CE. Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res. 2010;70:9185-9196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 29. | Yamakawa N, Takahashi A, Mori E, Imai Y, Furusawa Y, Ohnishi K, Kirita T, Ohnishi T. High LET radiation enhances apoptosis in mutated p53 cancer cells through Caspase-9 activation. Cancer Sci. 2008;99:1455-1460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 30. | Cunningham D, Jost LM, Purkalne G, Oliveira J. ESMO Minimum Clinical Recommendations for diagnosis, treatment and follow-up of gastric cancer. Ann Oncol. 2005;16 Suppl 1:i22-i23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |