Published online Oct 28, 2021. doi: 10.35713/aic.v2.i5.60
Peer-review started: October 15, 2021
First decision: October 24, 2021
Revised: October 26, 2021
Accepted: October 27, 2021
Article in press: October 27, 2021
Published online: October 28, 2021
Processing time: 12 Days and 18.2 Hours
Artificial intelligence is a groundbreaking tool to learn and analyse higher features extracted from any dataset at large scale. This ability makes it ideal to facing any complex problem that may generally arise in the biomedical domain or oncology in particular. In this work, we envisage to provide a global vision of this mathematical discipline outgrowth by linking some other related subdomains such as transfer, reinforcement or federated learning. Complementary, we also introduce the recently popular method of topological data analysis that improves the performance of learning models.
Core Tip: In this review, we explore powerful artificial intelligence based models enabling the comprehensive analysis of related problems on oncology. To this end, we described an asserted set of machine learning architectures that goes from the most classical multiple perceptron or neural networks to the novel federated and reinforcement learning designs. Overall, we point out the outgrowth of this mathematical discipline in cancer research and how computational biology and topological features can boost the general performances of these learning models.
- Citation: Morilla I. Repairing the human with artificial intelligence in oncology. Artif Intell Cancer 2021; 2(5): 60-68
- URL: https://www.wjgnet.com/2644-3228/full/v2/i5/60.htm
- DOI: https://dx.doi.org/10.35713/aic.v2.i5.60
The flourishing proliferation of artificial intelligence (AI) worldwide over the last decade has disrupted the way oncologists face cancer. More and more every day, the contribution of AI-based models to different axes of cancer research is not only improving their ability to stratify patients early on or discover new drugs but also influences its fundamentals. By integrating novel structures of data organisation, exploitation, and sharing of clinical data among health institutions, AI is achieving in the short-term to successfully accelerate cancer research. Medical practitioners are becoming familiar with some few mathematical concepts, such as machine learning (ML) or (un/semi) supervised learning. The former is a collection of data-driven techniques with the goal of building predictive models from high-dimensional datasets[1,2], while the latter refers to the grade of human intervention that these models require to make predictions.
These methods are being successfully used in cancer at many levels by simply analysing clinical data, biological indicators, or whole slide images[3-5]. Their application has revealed themselves as an effective way to tackle multiple clinical questions, from diagnosis to prediction of treatment outcomes. For instance, in Morilla et al[3], a minimal signature composed of seven miRNAs and two biological indicators was identified using general linear models trained at the base of a deep learning model to predict treatment outcomes in gastrointestinal cancer. In Schmauch et al[4], 2020, the authors predicted the RNA-Seq expression of tumours from whole slide images using a deep learning model as well.
Indeed, in this particular discipline, ML algorithms have evolved faster. Several approaches have succeeded in the classification of cancer subtypes using medical imaging[6-8]. Mammography and digital breast tomosynthesis have enabled a robust breast cancer detection by means of annotation-efficient deep learning approaches[9]. Epigenetic patterns of chromatin opening across the stem and differentiated cells across the immune system have also been predicted by deep neural networks in ATAC-seq analysis. In Maslova et al[10], solely from the DNA sequence of regulatory regions, the authors discovered ab initio binding motifs for known and unknown master regulators, along with their combinatorial operation.
Another domain where the application of AI-based models has largely been used is single-cell RNA sequencing (sc-RNAseq) analysis. In Lotfollahi et al[11] (2020), a new method based on transfer learning (TL) and parameter optimisation is introduced to enable efficient, decentralised, iterative reference building, and the contextualization of new datasets with existing single-cell references without sharing raw data. In addition, few methods have emerged around genetic perturbations of outcomes at the single-cell level in cancer treatments[12,13].
Finally, some computational topology techniques grouped under the heading of “topological data analysis” (TDA) have also been successfully proven as efficient tools in some cancer subtype classifications[14].
Thus, AI has turned the oncologists and co-workers’ lives around providing them with a new perspective, which was once developed by only a bulk of specialists and is rapidly becoming a reference in the domain. This work revisits, then, most of those techniques and provides a quick overview of their applications in cancer research.
ML or AI models, sometimes a philosophical matter, is a branch of mathematics concerned by numerically mimicking the human brain reasoning as it resolves a given problem. There are many examples of this practice; from those most classic techniques of regression or classification of dataset[15] to the current ground-breaking algorithms as “Deep-Mind, Alpha Fold” for protein-folding prediction[16]. In any case, all of these methods share a common objective: the ML problem. This problem can be mathematically expressed as: $$\hat{C}=\underset{C\in\mathcal{M}}{argmin}\math-bb{E}_{x,y\in\mathcal{X}\times\mathcal{Y}} [\mathcal{B}_{l} (C(x),y)]$$.
For example, if we select the particular loss function binary cross entropy, –Bl–, this equation describes the parameter misapplication of the neural network C by diminishing the expected value of the loss function between the output of this network C(x) and the true label y.
Frequently, the intricate design of models based on any ML technique (i.e., neural networks) makes them more difficult to interpret than simpler traditional models. Hence, if we want to fully exploit the potential of these models, a deeper understanding of their predictions would be advisable in practice. Thus, the predicted efficacy of a personal therapy on a cancer must be well explained, since its decisions directly influence human health. From a methodological point of view, we need to ensure model development with proper interpretations of their partial outputs in order to prevent undesirable effects of the models[17,18]. The two main streams of this discipline are the so-called “feature attribution” and “feature interaction” methods. The former[19-22] individually rewards input features depending on its local causal effect in the model output, whereas the latter examines those features with large second-order derivatives at the input or weight matrices of feed-forward and convolutional architectures[23,24]. However, the robustness of all these approaches may be compromised by the presence of specific types of architecture.
One class of ML models broadly used in current computational cancer research is deep neural networks. Overall, they have succeeded over other non-linear models[25] in the analysis of pathologic image recognition and later patient stratification based on the learned models[26,27]. In brief, deep neural models work in a large number of layers of information that is progressively passing by from one layer to another (i.e., the backpropagation algorithms) to extract relevant features from the original data according to a non-linear model, which is associated with the selected optimisation problem. Their designs can encompass a wide range of algorithms from the classic multiple perceptron networks[28-30] and convolutional neural networks[31-36] to the most recently established long short-term memory (LSTM) recurrent neural networks (RNNs) that are put into the spotlight in the next section[37,38].
RNNs: A different and convenient design other than the more classical neural networks in which the information flows forward are the RNNs. These are computationally more complex models with the skill of capturing hidden behaviours other methods in cancer studies cannot do[39-41]. Recurrent models exhibit an intrinsic representation of the data that allows the exploitation of context information. Specifically, a recurrent network is designed to maintain information about earlier iterations for a period that depends only on the weights and input data at the model’s entrance[42]. In particular, the network’s activation layers take advantage of inputs that come from chains of information provided by previous iterations. This influences the current prediction and enables the gathering of network flops that can retain contextual information on a long-term scale. Thus, by following this reasoning, RNNs can dynamically exploit a contextual interval over the input training history[43].
LSTM: An improvement in of RNNs is the construction of LSTM networks. LSTMs can learn to sort the interexchange between dependencies in the predictive problems addressed by batches. These models have had a major impact on the biomedical domain, particularly in cancer research[44-48]. LSTMs have been successfully proven in analysis where the intrinsic technical drawbacks associated with RNNs have prevented a fair performance of the model[49]. There are two main optimisation problems that must be avoided during the training stage when applying LSTM to solve a problem, namely: (1) vanishing gradients; and (2) exploding gradients[50]. In this sense, LSTM specifically provides an inner structural amelioration concerning the units leveraged in the learning model[51]. However, there is an improvement in the LSTM network calibration that is increasingly used in biomedical research: LSTM bidirectional networks. In these architectures, a bidirectional recurrent neural lattice is applied in order to be able to separately pass by two forward and backward recurrent nets sharing the same output layer during the training task[51].
Recycling is always a significant issue! In ML, we can also reuse a model that was originally envisaged for solving a different task other than the problem that we might be currently facing, but both share a similar structural behaviour. This practice is called TL in ML. Its usage has been progressively increasing in problems whose architecture can consume huge amounts of time and computational resources. In these cases, pre-trained networks are applied as a starting learning point, which largely boosts the performance of new models to approach related problems. Then, TL should ameliorate the current model in another setting if such a model is available for learning features from the first problem in a general way[52,53]. Regarding its benefits in oncology, we can outstand its usage in large datasets of piled images to be recognised for patient stratification, as previously described in the following works[54-61].
Reinforcement learning (RL) is one of the latest ML extensions that ameliorates the global performance of learning models when making decisions. In RL, a model learns a given objective in an a priori fixed uncertainty by means of trial and error computations until a solution is obtained. Then, to guide the model, the AI algorithm associates rewards or penalties with the local performance of the model. The final goal was to maximise the amount of rewards obtained. Remarkably, the ML architecture provides no clues on how to find the final solution, even if it rules the reward conditions. Thus, the model must smooth the optimisation problem from a totally random scenario to a complex universe of possibilities. However, if the learning algorithm is launched into a sufficiently powerful computational environment, the ML model will be able to store thousands of trials to effectively achieve the given goal. Nevertheless, a major inconvenience is that the simulation environment is highly dependent on the problem to be computed.
To sum it up, although RL should not be taken as the definitive algorithm, it promises to blow up the current concept of deep learning in oncology[62-64]. An example with no precedents is the DeepMind algorithm very famous nowadays by performing alpha protein folding[16] predictions at a scale ever done before.
A simple description of federated learning (FL) could be a decentralised approach to ML. Thus, FL boosts and accelerates medical discoveries on partnerships with many contributors while protecting patient privacy. In FL, we only improve and calibrate the results and not the data. Thus, what FL really promises it is a new era in secured AI in oncology: Training, testing, or ensuring privacy that way of learning is an efficient method of using data from a comprehensive network of resources belonging each time to a node of many interconnected hospital institutions[65-68].
Topological ML (TML) is an interaction that has been recently established between TDA and ML. Owing to new advances in computational algorithms, the extraction of complex topological features, such as persistence homology or Betti curves, has become progressively feasible in large datasets. In particular, TDA is commonly referred to as capturing the shape of the data. This method fixes their topological invariants as hotspot to look up relevant structural and categorical information. Indeed, TDA provides ideal completeness in terms of multi-scalability and globalisation missed from the rigidness of their geometric characteristics. In that sense, the use of this tool has been growing in cancer research until it is considered as contextually informative in the analysis of massive biomedical data[69-74]. Multiple studies have exploited the complementary information that emerged from different prisms to gain new insights into the datasets. Its association with ML has enhanced both classical ML methods and deep learning models[75,76].
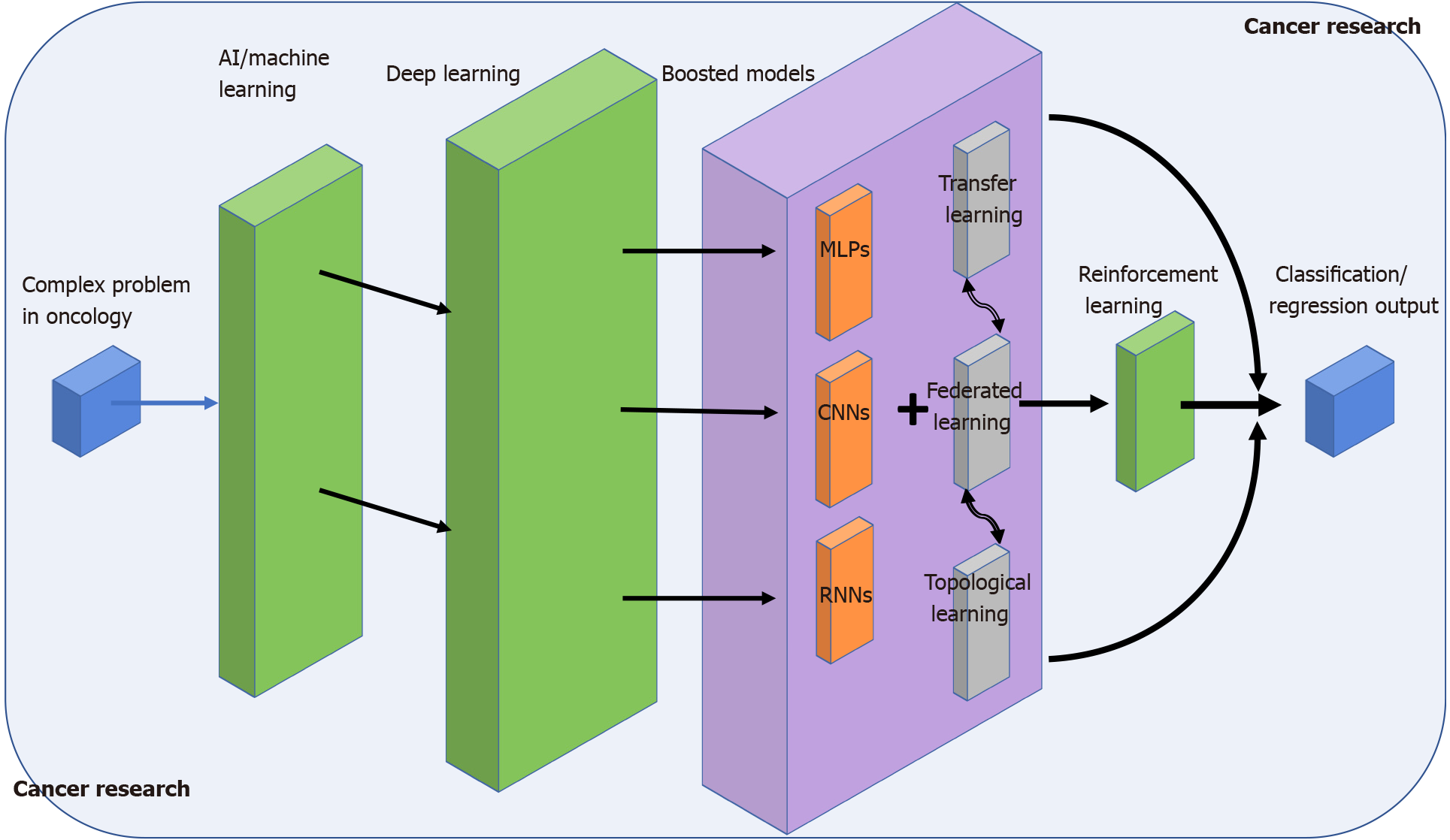
In this work, we summarise the conclusions of some major references of AI in cancer research (Figure 1). Overall, we wanted to point out the rapid AI outgrowth in the biomedical domain and how AI has systematically become familiar to anyone in the domain, expert, or not. This is possibly due to recent advances in learning-oriented algorithms, which have enabled the transformation of data analysis to any scale and complexity provided a suitable environment is available. We have provided many examples of a varied set of learning models (Multi-layer perceptron, convolutional neural networks, RNNs, etc.) that have been successfully proven for related cancer problems such as patient stratification, image-based classification, or recording-device optimisation[77,78]. We have compared different approaches to solve similar questions, and we have introduced novel concepts such as TL, FL, or RL that prevent some of the most classical constraints regarding network architectures or information privacy on high dimensional datasets. Finally, the combination of TDA and ML has also been shown to be a promising discipline where to exploit extra topological features extracted at a higher level. Such tandem promises to contribute to the improvement of the AI algorithm’s performance from a totally different perspective. Although data-driven based AI models have the potential to change the world of unsupervised learning, some limitations could endanger a promising future. The three major issues that hamper a better optimisation and general performance in AI models are related to: (1) the high dependency of the model on the data scale; (2) choice of a proper computational environment, and (3) practical problems of time or computational cost should be assumed. Thus, the future challenges in this discipline begin by smoothing such obstacles as much as possible, which will ultimately end up with AI as the tool of reference in healthcare institutions for a much broader analysis in oncology.
We acknowledge the financial support from the Institut National de la Sant e et de la Recherche Medicale (INSERM), and the Investissements d’Avenir programme ANR-10-LABX-0017, Sorbonne Paris Nord, Laboratoire d’excellence INFLAMEX.
Manuscript source: Invited manuscript
Specialty type: Mathematical and Computational Biology
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Guo XY, Saraiva MM S-Editor: Wang JL L-Editor: A P-Editor: Wang JL
| 1. | Camacho DM, Collins KM, Powers RK, Costello JC, Collins JJ. Next-Generation Machine Learning for Biological Networks. Cell. 2018;173:1581-1592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 525] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 2. | Morilla I, Léger T, Marah A, Pic I, Zaag H, Ogier-Denis E. Singular manifolds of proteomic drivers to model the evolution of inflammatory bowel disease status. Sci Rep. 2020;10:19066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Morilla I, Uzzan M, Laharie D, Cazals-Hatem D, Denost Q, Daniel F, Belleannee G, Bouhnik Y, Wainrib G, Panis Y, Ogier-Denis E, Treton X. Colonic MicroRNA Profiles, Identified by a Deep Learning Algorithm, That Predict Responses to Therapy of Patients With Acute Severe Ulcerative Colitis. Clin Gastroenterol Hepatol. 2019;17:905-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 4. | Schmauch B, Romagnoni A, Pronier E, Saillard C, Maillé P, Calderaro J, Kamoun A, Sefta M, Toldo S, Zaslavskiy M, Clozel T, Moarii M, Courtiol P, Wainrib G. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun. 2020;11:3877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 259] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 5. | Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky JM, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology. 2020;72:2000-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 185] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 6. | Saillard C, Delecourt F, Schmauch B, Moindrot O, Svrcek M, Bardier-Dupas A, Emile JF, Ayadi M, De Mestier L, Hammel P, Neuzillet C, Bachet JB, Iovanna J, Nelson DJ, Paradis V, Zaslavskiy M, Kamoun A, Courtiol P, Nicolle R, Cros J. Identification of pancreatic adenocarcinoma molecular subtypes on histology slides using deep learning models. J Clin Oncol. 2021;39 suppl 15:4141. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 7. | Rhee S, Seo S, Kim S. Hybrid approach of relation network and localized graph convolutional filtering for breast cancer subtype classification. In: Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence. International Joint Conferences on Artificial Intelligence Organization, 2018: 3527–3534. [DOI] [Full Text] |
| 8. | Schutte K, Moindrot O, Hérent P, Schiratti JB, Jégou S. Using stylegan for visual interpretability of deep learning models on medical images. 2021 Preprint. Available from: ArXiv:2101.07563. |
| 9. | Lotter W, Diab AR, Haslam B, Kim JG, Grisot G, Wu E, Wu K, Onieva JO, Boyer Y, Boxerman JL, Wang M, Bandler M, Vijayaraghavan GR, Gregory Sorensen A. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat Med. 2021;27:244-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 185] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 10. | Maslova A, Ramirez RN, Ma K, Schmutz H, Wang C, Fox C, Ng B, Benoist C, Mostafavi S; Immunological Genome Project. Deep learning of immune cell differentiation. Proc Natl Acad Sci U S A. 2020;117:25655-25666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 11. | Lotfollahi M, Naghipourfar M, Luecken MD, Khajavi M, Uttner MB, Avsec Z, Misharin AV, Theis FJ. Query to reference singlecell integration with transfer learning. 2020 Preprint. Available from: bioRxiv:2020.07.16.205997. [DOI] [Full Text] |
| 12. | Hou W, Ji Z, Ji H, Hicks SC. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol. 2020;21:218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 165] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 13. | Zhang Y, Wang D, Peng M, Tang L, Ouyang J, Xiong F, Guo C, Tang Y, Zhou Y, Liao Q, Wu X, Wang H, Yu J, Li Y, Li X, Li G, Zeng Z, Tan Y, Xiong W. Single-cell RNA sequencing in cancer research. J Exp Clin Cancer Res. 2021;40:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 214] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 14. | Dindin M, Umeda Y, Chazal F. Topological data analysis for arrhythmia detection through modular neural networks. In: Goutte C, Zhu X, editors. Advances in Artificial Intelligence. Cham: Springer International Publishing, 2020: 177-188. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Azzaoui T, Santos D, Sheikh H, Lim S. Solving classification and regression problems using machine and deep learning. Technical report, University of Massachusetts Lowell, 2018. [DOI] [Full Text] |
| 16. | Deepmind. Deepmind alphafolding. [Accessed: 2021-10-12] Available from: https://deepmind.com/research. |
| 17. | Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. In: ICML'17: Proceedings of the 34th International Conference on Machine Learning - Volume 70. JMLR.org, 2017: 3319–3328. |
| 18. | Janizek JD, Sturmfels P, Lee SI. Explaining explanations: Axiomatic feature interactions for deep networks. J Mach Learn Res. 2021;22:1-54. |
| 19. | Binder A, Montavon G, Lapuschkin S, Muller KR, Same W. Layer-wise relevance propagation for neural networks with local renormalization layers. In: Villa A, Masulli P, Pons Rivero A, editors. Artificial Neural Networks and Machine Learning – ICANN 2016. ICANN 2016. Lecture Notes in Computer Science, vol 9887. Cham: Springer, 2016: 63-71. [RCA] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 20. | Ribeiro MT, Singh S, Guestrin C. “why should i trust you?” explaining the predictions of any classifier. In: KDD'16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York: Association for Computing Machinery, 2016: 1135–1144. [DOI] [Full Text] |
| 21. | Shirkumar A, Greenside P, Kundaje A. Learning important features through propagating activation diferences. In: Precup D, Teh YW, editors. Proceedings of the 34th International Conference on Machine Learning. JMLR org, 2017: 3145–3153. |
| 22. | Lundeberg SM, Lee SI. A unified approach to interpreting model predictions. In: Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS'17). Red Hook: Curran Associates Inc., 2017: 4768-4777. |
| 23. | Cui T, Marttinen P, Kaski S. Recovering pairwise interactions using neural networks. 2019 Preprint. Available from: arXiv:1901.08361. |
| 24. | Greenside P, Shimko T, Fordyce P, Kundaje A. Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Bioinformatics. 2018;34:i629-i637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 25. | Shavitt I, Segal E. Regularization learning networks: Deep learning for tabular datasets. In: Bengio S, Wallach H, Larochelle H, Grauman K, Cesa-Bianchi N, Garnett R, editors. Advances in Neural Information Processing Systems, volume 31. Red Hook: Curran Associates Inc., 2018. [DOI] [Full Text] |
| 26. | Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). Minneapolis: Association for Computational Linguistics, 2019: 4171-4186. [DOI] [Full Text] |
| 27. | He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2015 Preprint. Available from: arXiv:1512.03385. |
| 28. | Kumar G, Pawar U, O’Reilly R. Arrhythmia detection in ECG signals using a multilayer perceptron network. In: Curry E, Keane MT, Ojo A, Salwala D, editors. AICS, volume 2563 of CEUR Workshop Proceedings. CEUR-WS.org, 2019: 353-364. |
| 29. | Alsmadi MK, Omar KB, Noah SA, Almarashdah I. Performance comparison of multi-layer perceptron (back propagation, delta rule and perceptron) algorithms in neural networks. In: 2009 IEEE International Advance Computing Conference. IEEE, 2009: 296–299. [DOI] [Full Text] |
| 30. | Freund Y, Schapire RE. Large margin classification using the perceptron algorithm. Mach Learn. 1999;37:277-296. [DOI] [Full Text] |
| 31. | Hadush S, Girmay Y, Sinamo A, Hagos G. Breast cancer detection using convolutional neural networks. 2020 Preprint. Available from: arXiv:2003.07911. |
| 32. | Chaturvedi SS, Tembhurne JV, Diwan T. A multi-class skin cancer classification using deep convolutional neural networks. Multim Tools Appl. 2020;28477-27498. [DOI] [Full Text] |
| 33. | Santos C, Afonso L, Pereira C, Papa J. BreastNet: Breast cancer categorization using convolutional neural networks. In: 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS). IEEE, 2020: 463-468. [DOI] [Full Text] |
| 34. | Yoo S, Gujrathi I, Haider MA, Khalvati F. Prostate Cancer Detection using Deep Convolutional Neural Networks. Sci Rep. 2019;9:19518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 35. | Kumar N, Verma R, Arora A, Kumar A, Gupta S, Sethi S, Gann PH. Convolutional neural networks for prostate cancer recurrence prediction. In: Gurcan MN, Tomaszewski JE, editors. Proceedings Volume 10140, Medical Imaging 2017: Digital Pathology. SPIE, 2017: 101400H. [DOI] [Full Text] |
| 36. | Dumoulin V, Francesco V. A guide to convolution arithmetic for deep learning. 2016 Preprint. Available from: arXiv:1603.07285. |
| 37. | Graves A. Long Short-Term Memory. Berlin: Springer Berlin Heidelberg 2012; 37-45. [RCA] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 38. | Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735-1780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61646] [Cited by in RCA: 14010] [Article Influence: 500.4] [Reference Citation Analysis (0)] |
| 39. | Lane N, Kahanda I. DeepACPpred: A Novel Hybrid CNN-RNN Architecture for Predicting Anti-Cancer Peptides. In: Panuccio G, Rocha M, Fdez-Riverola F, Mohamad MS, Casado-Vara R, editors. PACBB, volume 1240 of Advances in Intelligent Systems and Computing. Springer, 2020: 60-69. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Moitra D, Mandal RK. Automated AJCC (7th edition) staging of non-small cell lung cancer (NSCLC) using deep convolutional neural network (CNN) and recurrent neural network (RNN). Health Inf Sci Syst. 2019;7:14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 41. | Chiang JH, Chao SY. Modeling human cancer-related regulatory modules by GA-RNN hybrid algorithms. BMC Bioinformatics. 2007;8:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 42. | Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994;5:157-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4000] [Cited by in RCA: 1492] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 43. | Sak H, Senior AW, Beaufays F. Long short-term memory recurrent neural network architectures for large scale acoustic modeling. In: Li H, Meng HM, Ma B, Chng E, Xie L, editors. Proc. Interspeech 2014. ISCA, 2014: 338–342. [DOI] [Full Text] |
| 44. | Agrawal P, Bhagat D, Mahalwal M, Sharma N, Raghava GPS. AntiCP 2.0: an updated model for predicting anticancer peptides. Brief Bioinform. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 165] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 45. | Jiang L, Sun X, Mercaldo F, Antonella Santone A. Decablstm: Deep contextualized attentional bidirectional lstm for cancer hallmark classification. Knowl Based Syst. 2020;210:106486. [RCA] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 46. | Asyhar AH, Foeady AZ, Thohir M, Arifin AZ, Haq DZ, Novitasari DCR. Implementation LSTM Algorithm for Cervical Cancer using Colposcopy Data. In: 2020 International Conference on Artificial Intelligence in Information and Communication (ICAIIC). IEEE, 2020: 485-489. [DOI] [Full Text] |
| 47. | Bichindaritz I, Liu G, Bartlett CL. Survival prediction of breast cancer patient from gene methylation data with deep LSTM network and ordinal cox model. In: Bartk R, Bell E, editors. FLAIRS Conference. AAAI Press, 2020: 353–356. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 48. | Gao R, Huo Y, Bao S, Tang Y, Antic S, Epstein ES, Balar A, Deppen S, Paulson AB, Sandler KL, Massion PP, Landman BA. Distanced LSTM: Time-distanced gates in long short-term memory models for lung cancer detection. In: Suk HI, Liu M, Yan P, Lian C, editors. Machine Learning in Medical Imaging. MLMI 2019. Lecture Notes in Computer Science, vol 11861. Cham: Springer, 2019: 310-318. [RCA] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 49. | Gers FA, Schmidhuber J, Cummins F. Learning to forget: continual prediction with LSTM. Neural Comput. 2000;12:2451-2471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3498] [Cited by in RCA: 870] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 50. | Graves A, Liwicki M, Fernández S, Bertolami R, Bunke H, Schmidhuber J. A novel connectionist system for unconstrained handwriting recognition. IEEE Trans Pattern Anal Mach Intell. 2009;31:855-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 329] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 51. | Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw. 2005;18:602-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2277] [Cited by in RCA: 927] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 52. | Olivas ES, Guerrero JDM, Sober MM, Benedito JRM, Lopez AJS. Handbook Of Research On Machine Learning Applications and Trends: Algorithms, Methods and Techniques - 2 Volumes. Hershey: IGI Publishing, 2009. [DOI] [Full Text] |
| 53. | Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? In: NIPS'14: Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2. Cambridge: MIT Press, 2014: 3320-3328. |
| 54. | Khamparia A, Bharati S, Podder P, Gupta D, Khanna A, Phung TK, Thanh DNH. Diagnosis of breast cancer based on modern mammography using hybrid transfer learning. Multidimens Syst Signal Process. 2021;1-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 55. | Jayachandran S, Ghosh A. Deep transfer learning for texture classification in colorectal cancer histology. In: Schilling FP, Stadelmann T, editors. Artificial Neural Networks in Pattern Recognition. ANNPR 2020. Lecture Notes in Computer Science, vol 12294. Cham: Springer, 2020: 173-186. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 56. | Shaikh TA, Ali R, Sufyan Beg MM. Transfer learning privileged information fuels CAD diagnosis of breast cancer. Mach Vis Appl. 2020;31:9. [DOI] [Full Text] |
| 57. | de Matos J, de Souza Britto A, Oliveira LES, Koerich AL. Double transfer learning for breast cancer histopathologic image classification. In: 2019 International Joint Conference on Neural Networks (IJCNN). IEEE, 2019: 1-8. [DOI] [Full Text] |
| 58. | Obonyo S, Ruiru D. Multitask learning or transfer learning? application to cancer detection. In: Merelo JJ, Garibaldi JM, Linares-Barranco A, Madani K, Warwick K, editors. Computational Intelligence. ScitePress, 2019: 548–555. |
| 59. | Kassani SH, Kassani PH, Wesolowski MJ, Schneider KA, Deters R. Breast cancer diagnosis with transfer learning and global pooling. In: 2019 International Conference on Information and Communication Technology Convergence (ICTC). IEEE, 2019: 519-524. [DOI] [Full Text] |
| 60. | Dhruba SR, Rahman R, Matlock K, Ghosh S, Pal R. Application of transfer learning for cancer drug sensitivity prediction. BMC Bioinformatics. 2018;19:497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 61. | Vesal S, Ravikumar N, Davari A, Ellmann S, Maier AK. Classification of breast cancer histology images using transfer learning. In: Campilho A, Karray F, ter Haar Romeny B, editors. Image Analysis and Recognition. ICIAR 2018. Lecture Notes in Computer Science, vol 10882. Cham: Springer, 2018: 812–819. [RCA] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 62. | Kalantari J, Nelson H, Chia N. The Unreasonable Effectiveness of Inverse Reinforcement Learning in Advancing Cancer Research. Proc Conf AAAI Artif Intell. 2020;34:437-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 63. | Daoud S, Mdhaffar A, Jmaiel M, Freisleben B. Q-Rank: Reinforcement Learning for Recommending Algorithms to Predict Drug Sensitivity to Cancer Therapy. IEEE J Biomed Health Inform. 2020;24:3154-3161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 64. | Balaprakash P, Egele R, Salim M, Wild S, Vishwanath V, Xia F, Brettin T, Stevens R. Scalable reinforcement-learning-based neural architecture search for cancer deep learning research. In: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. New York: Association for Computing Machinery, 2019: 1-33. [DOI] [Full Text] |
| 65. | Beguier C, Andreux M, Tramel EW. Efficient Sparse Secure Aggregation for Federated Learning. 2020 Preprint. Available from: arXiv:2007.14861. |
| 66. | Andreux M, du Terrail JO, Beguier C, Tramel EW. Siloed federated learning for multi-centric histopathology datasets. 2020 Preprint. Available from: arXiv:2008.07424. |
| 67. | Andreux M, Manoel A, Menuet R, Saillard C, Simpson C. Federated Survival Analysis with Discrete-Time Cox Models. 2020 Preprint. Available from: arXiv:2006.08997. |
| 68. | Rieke N, Hancox J, Li W, Milletarì F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K, Ourselin S, Sheller M, Summers RM, Trask A, Xu D, Baust M, Cardoso MJ. The future of digital health with federated learning. NPJ Digit Med. 2020;3:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 608] [Cited by in RCA: 676] [Article Influence: 135.2] [Reference Citation Analysis (0)] |
| 69. | Hensel F, Moor M, Rieck B. A Survey of Topological Machine Learning Methods. Front Artif Intell. 2021;4:681108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 70. | Groha S, Weis C, Gusev A, Rieck B. Topological data analysis of copy number alterations in cancer. 2020 Preprint. Available from: arXiv:2011.11070. |
| 71. | Loughrey C, Fitzpatrick P, Orr N, Jurek-Loughrey A. The topology of data: Opportunities for cancer research. Bioinformatics. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 72. | Gonzalez G, Ushakova A, Sazdanovic R, Arsuaga J. Prediction in Cancer Genomics Using Topological Signatures and Machine Learning. In: Baas N, Carlsson G, Quick G, Szymik M, Thaule M, editors. Topological Data Analysis. Abel Symposia. Cham: Springer, 2020: 247-276. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 73. | Yu YT, Lin GH, Jiang IHR, Chiang CC. Machine learning-based hotspot detection using topological classification and critical feature extraction. IEEE Trans Comput Aided Des Integr Circuits Syst. 2015;34:460-470. [DOI] [Full Text] |
| 74. | Matsumoto T, Kitazawa M, Kohno Y. Classifying topological charge in SU(3) YangMills theory with machine learning. Prog Theor Exp Phys. 2021;2:023D01. [DOI] [Full Text] |
| 75. | Bukkuri A, Andor N, Darcy IK. Applications of Topological Data Analysis in Oncology. Front Artif Intell. 2021;4:659037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 76. | Huang CH, Chang PM, Hsu CW, Huang CY, Ng KL. Drug repositioning for non-small cell lung cancer by using machine learning algorithms and topological graph theory. BMC Bioinformatics. 2016;17 Suppl 1:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 77. | Morilla I. A deep learning approach to evaluate intestinal fibrosis in magnetic resonance imaging models. Neural Comput Appl. 2020;32:14865-14874. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 78. | Morilla I, Uzzan M, Cazals-Hatem D, Colnot N, Panis Y, Nancey S, Boschetti G, Amiot A, Tréton X, Ogier-Denis E, Daniel F. Computational Learning of microRNA-Based Prediction of Pouchitis Outcome After Restorative Proctocolectomy in Patients With Ulcerative Colitis. Inflamm Bowel Dis. 2021;27:1653-1660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |