Copyright
©The Author(s) 2021.
World J Clin Cases. Jul 26, 2021; 9(21): 5873-5888
Published online Jul 26, 2021. doi: 10.12998/wjcc.v9.i21.5873
Published online Jul 26, 2021. doi: 10.12998/wjcc.v9.i21.5873
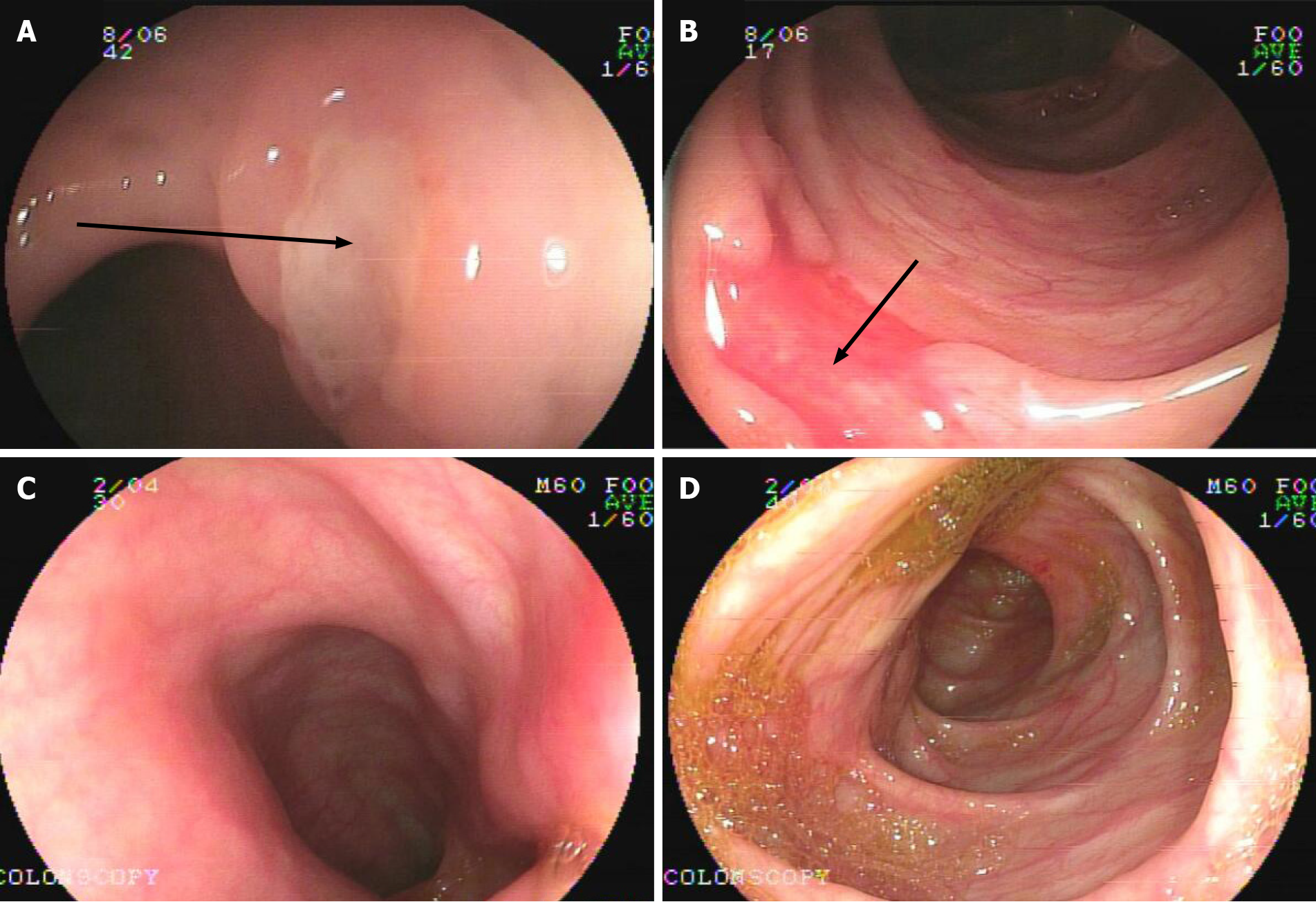
Figure 1 Endoscopy images of patient 2.
A and B: At the onset, ulcers (black arrows) in a gastrointestinal tract resembling Crohn's disease-like colitis; C and D: Three years post-treatment, no mucosal inflammation was found in the gastrointestinal tract.
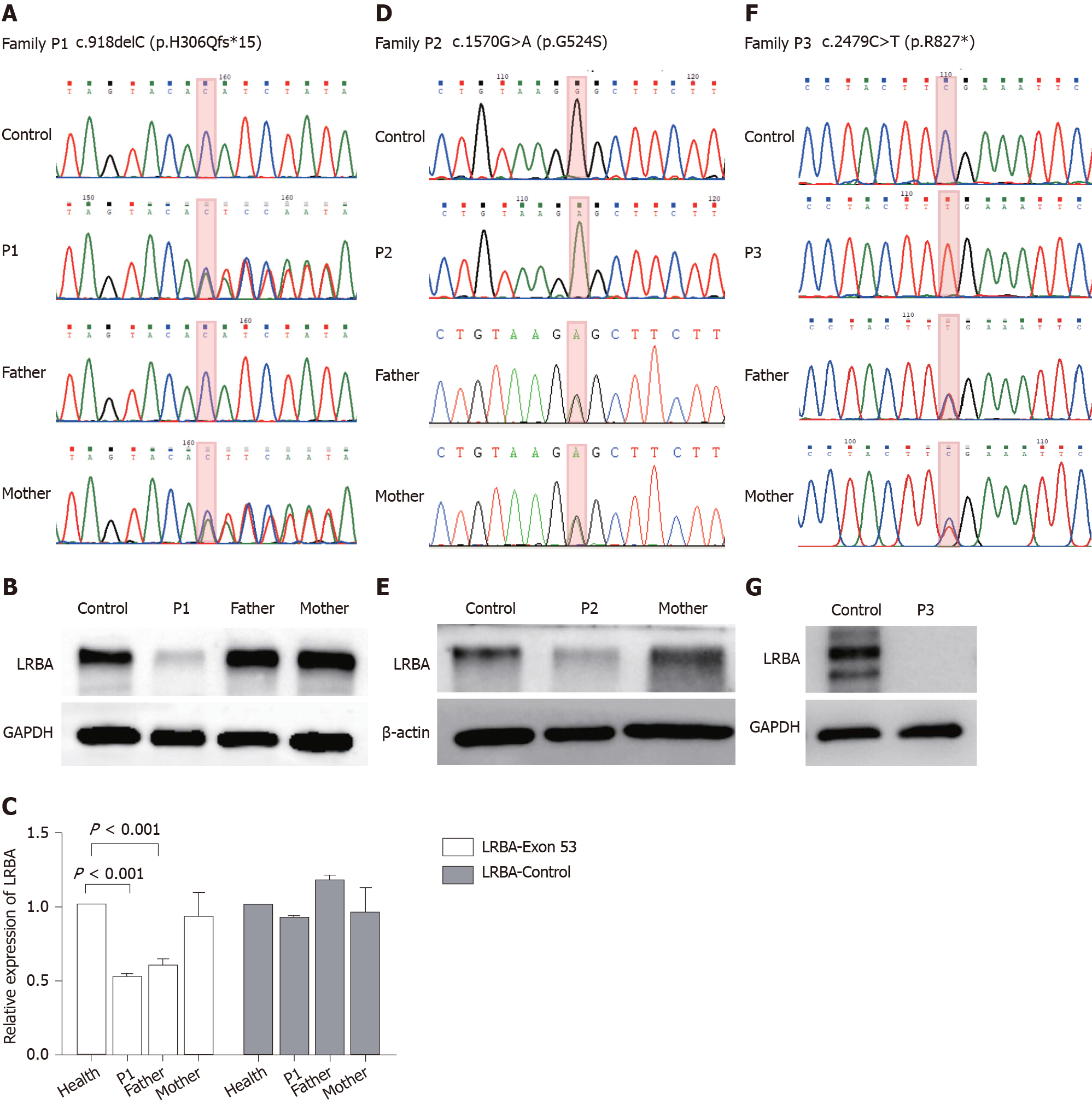
Figure 2 Genetic testing and Western blot analysis results of the three patients.
A: Sanger sequencing confirmed the presence of a mutation in exon 8 (c.918delC, p.H306Qfs*15) of the LABA gene in patient 1. The patient’s mother is a carrier with a heterozygous variant; B: Western blot analysis of the LRBA protein from peripheral blood mononuclear cells (PBMCs) from patient 1, his parents, and a healthy control. GADPH was used as a loading control; C: Quantitative polymerase chain reaction verified a large deletion in exon 53 of the LABA gene in patient 1 and his father; D: Sanger sequencing confirmed the presence of a homozygous mutations in exon 12 (c.1570G>A, p.524G>S) of the LABA gene in patient 2. Her parents are heterozygous variant carriers; E: Western blot of the LPS-responsive beige-like anchor protein (LRBA) protein from PBMCs of patient 2, her mother, and a healthy control. β-actin was used as a loading control; F: Sanger sequencing confirmed the presence of a homozygous mutation in exon 21 (c.2479C>T, p.Arg827*) of the LABA gene in patient 3. His parents are heterozygous variant carriers; G; Western blot of the LRBA protein from PBMCs from patient 3 and a healthy control. GADPH was used as a loading control. LRBA: LPS-responsive beige-like anchor protein.
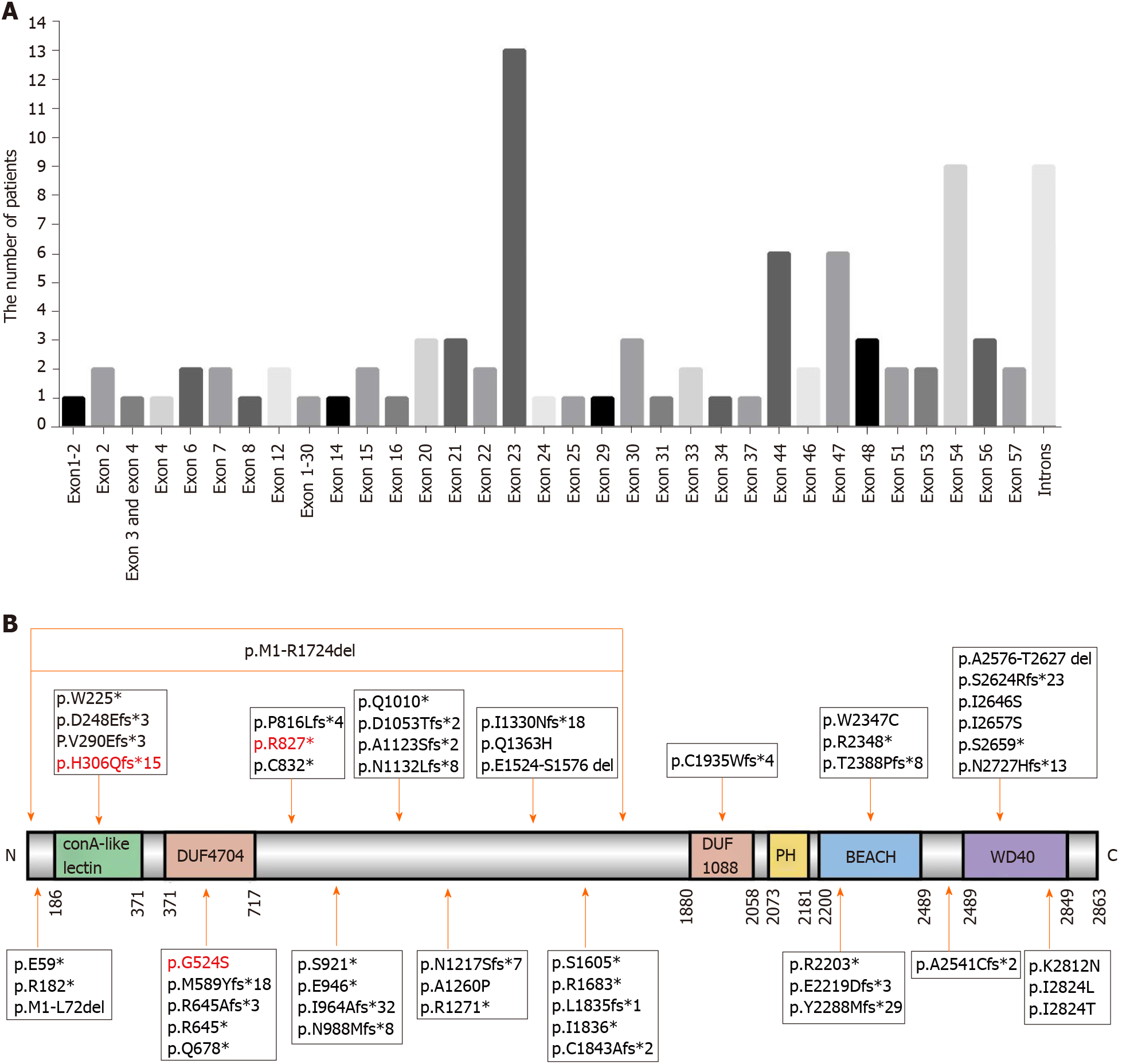
Figure 3 Genetic mutations identified in LABA-deficient patients (83 previously reported patients together with our three patients).
A: The number of patients carrying mutations in different exons/introns; B: LRBA protein map indicating the mutation sites. The location of the present mutations is represented with a orange arrow, and previously reported mutations are represented by black arrows. LRBA: LPS-responsive beige-like anchor protein.
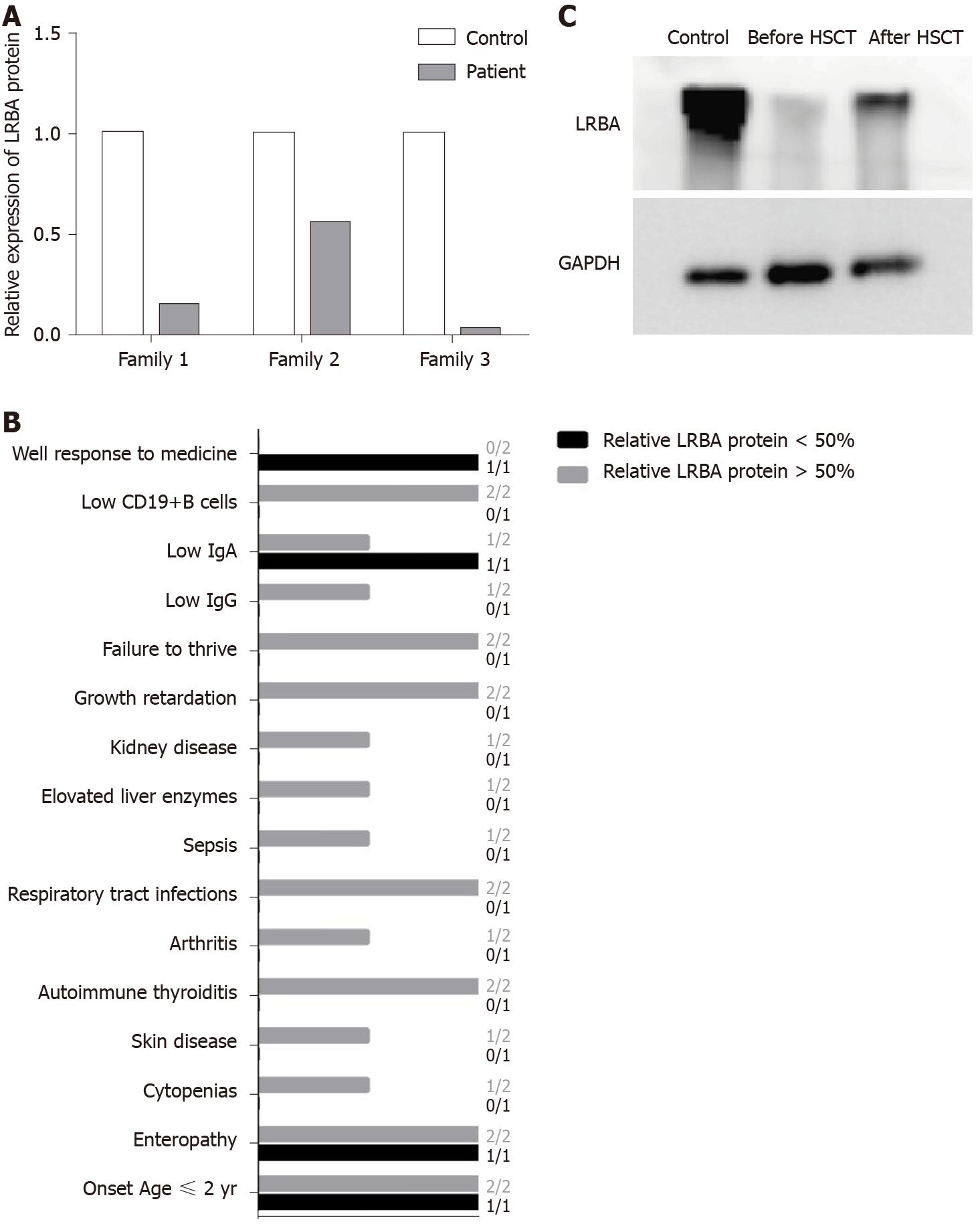
Figure 4 Potential protein–phenotype correlation in the three LPS-responsive beige-like anchor protein deficient patients.
A: Relative residual LPS-responsive beige-like anchor protein (LRBA) protein expression in our three patients was quantified and compared with that in controls; B: The clinical phenotype of the three patients grouped by the LRBA protein level; C: Western blot analysis of the LRBA protein on peripheral blood mononuclear cells from patient 1 before and after hematopoietic stem cell transplantation. LRBA: LPS-responsive beige-like anchor protein; HSCT: Hematopoietic stem cell transplantation.
- Citation: Tang WJ, Hu WH, Huang Y, Wu BB, Peng XM, Zhai XW, Qian XW, Ye ZQ, Xia HJ, Wu J, Shi JR. Potential protein–phenotype correlation in three lipopolysaccharide-responsive beige-like anchor protein-deficient patients. World J Clin Cases 2021; 9(21): 5873-5888
- URL: https://www.wjgnet.com/2307-8960/full/v9/i21/5873.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i21.5873