Copyright
©2012 Baishideng.
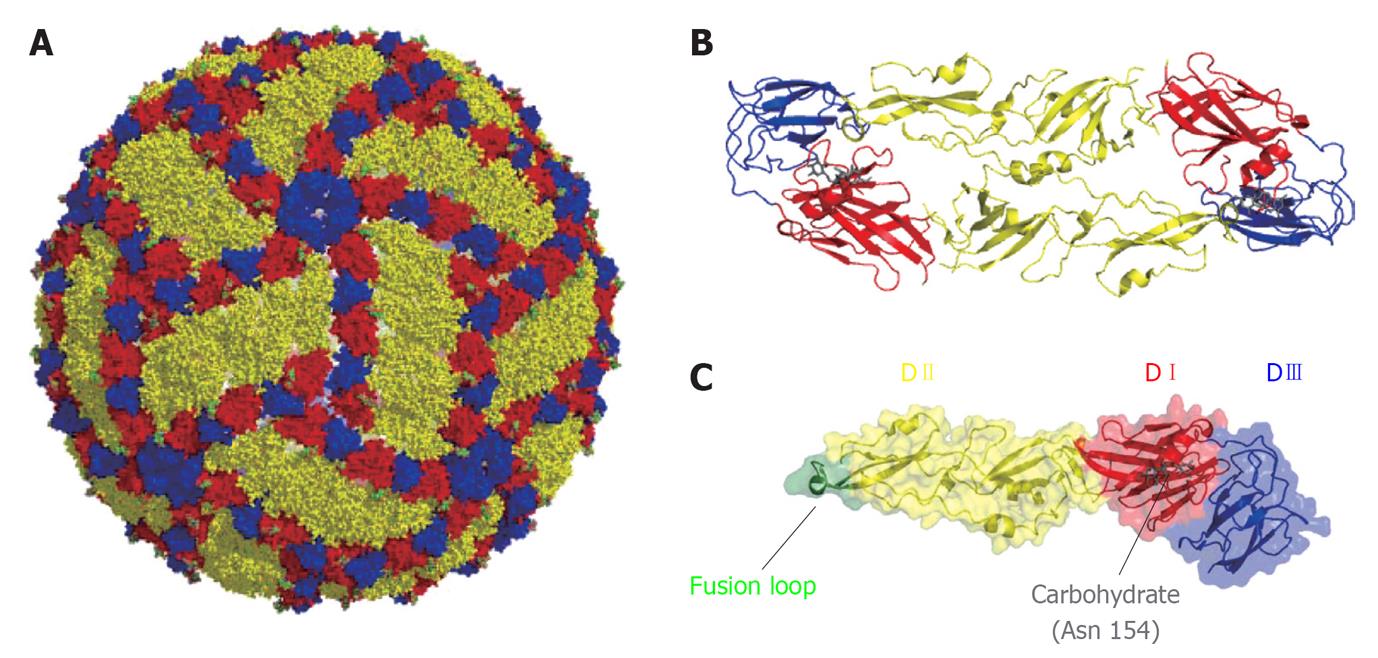
Figure 1 Organization of West Nile virus particle.
A: Representation of West Nile virus particle based on cryoelectron microscopy data[230]. Glycoprotein E is shown on the surface of the particle. Color code: DI (red), DII (yellow) and DIII (blue); B: Ribbon diagram of a dimer of E glycoprotein; C: Structure of a monomer of the soluble ectodomain of E glycoprotein based on the atomic coordinates solved by X-ray crystallography[58]. Fusion loop is highlighted in green. The carbohydrate located at Asn 154 is shown in gray.
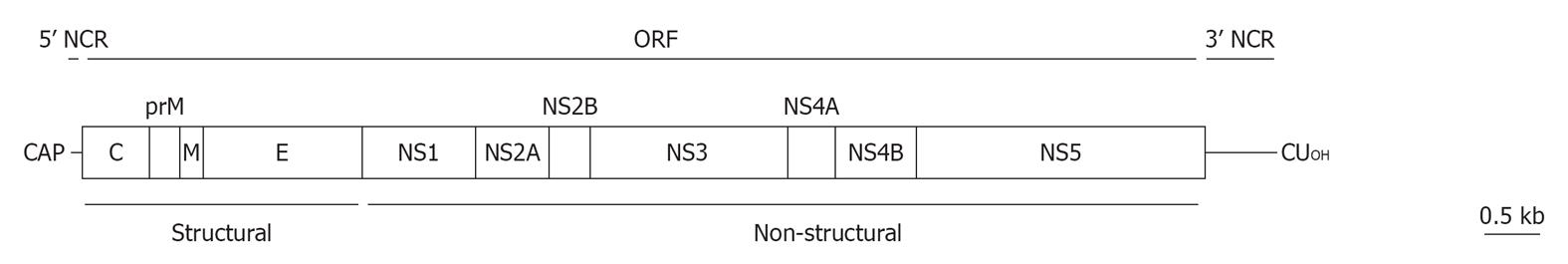
Figure 2 Schematic view of West Nile virus genome organization.
The single ORF (boxes) that encodes both structural and non-structural proteins is flanked by two non-coding regions (NCRs). See text for details.
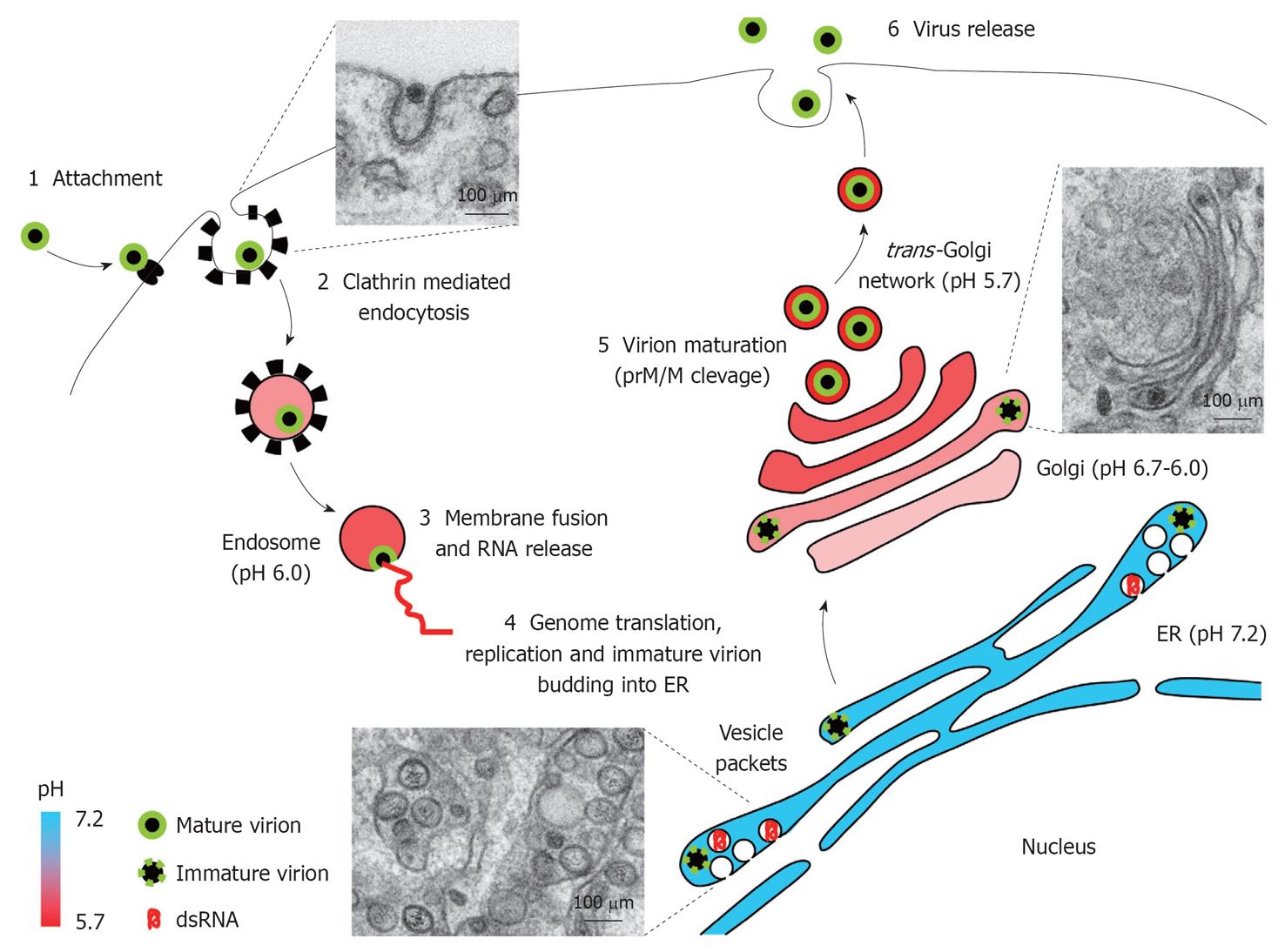
Figure 3 West Nile virus replication cycle.
Schematic view of West Nile virus replication cycle in an infected cell. Electron micrographs of West Nile virus-infected Vero cells illustrate distinct snapshots from infectious cycle: entry of virions, vesicle packets formation, and virion trafficking through Golgi complex. See text for details. ER: Endoplasmic reticulum; prM/M: Premembrane/membrane protein.
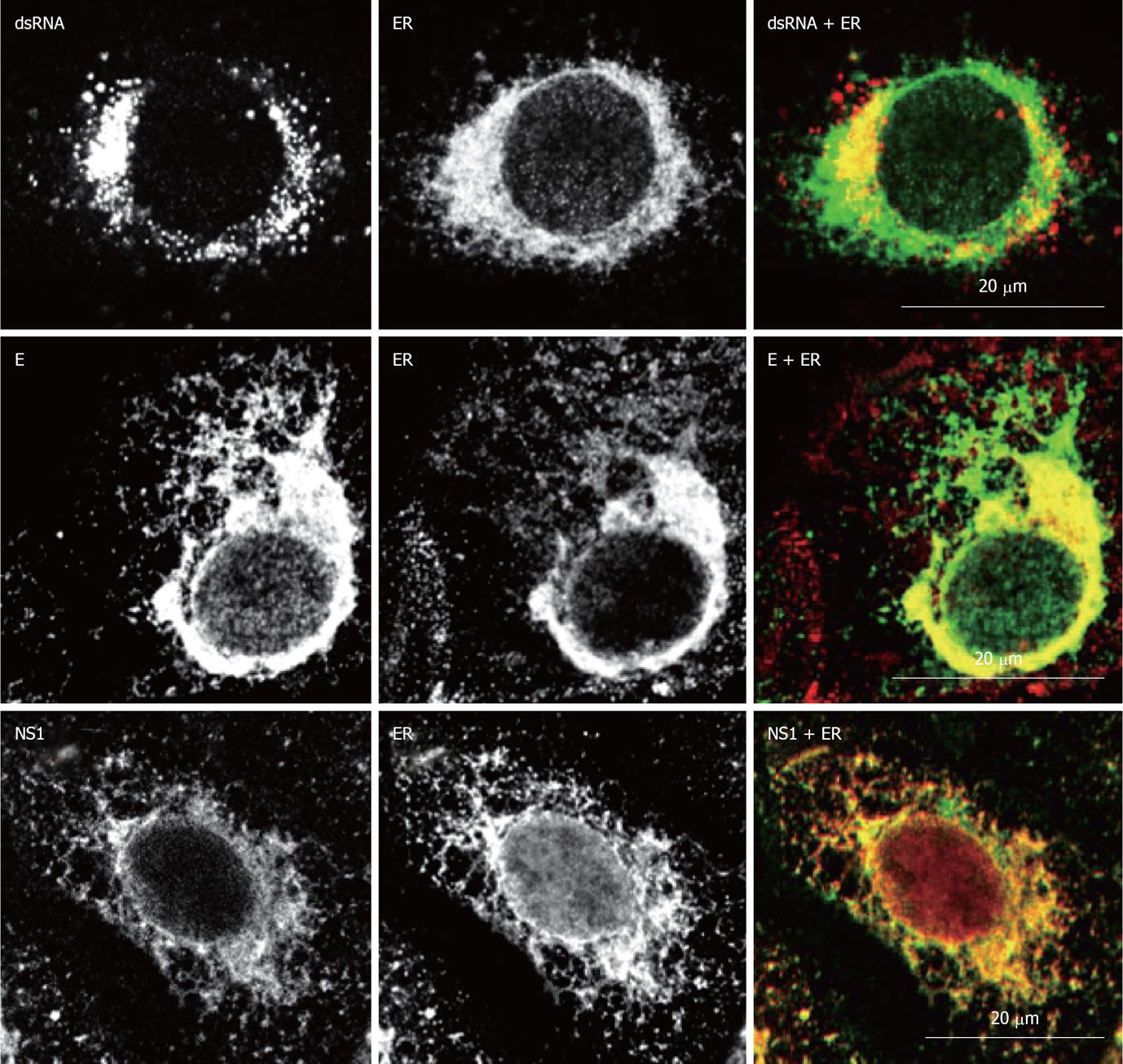
Figure 4 Localization of West Nile virus dsRNA, structural and non-structural proteins at the endoplasmic reticulum of infected cells.
Double immunofluorescence labeling of Vero cells 24h post-infection with West Nile virus (MOI of 1 PFU/cell) and stained for dsRNA, using monoclonal antibody J2 (English and Scientific Consulting Bt., Hungary), E glycoprotein or NS1, using monoclonal antibody 3.67G and 3.1112G (Millipore, Temecula, CA), respectively, in combination with rabbit polyclonal antibody against calnexin (Abcam, Cambridge UK) as an specific marker for endoplasmic reticulum. Procedures for immunostaining have been previously described[21].
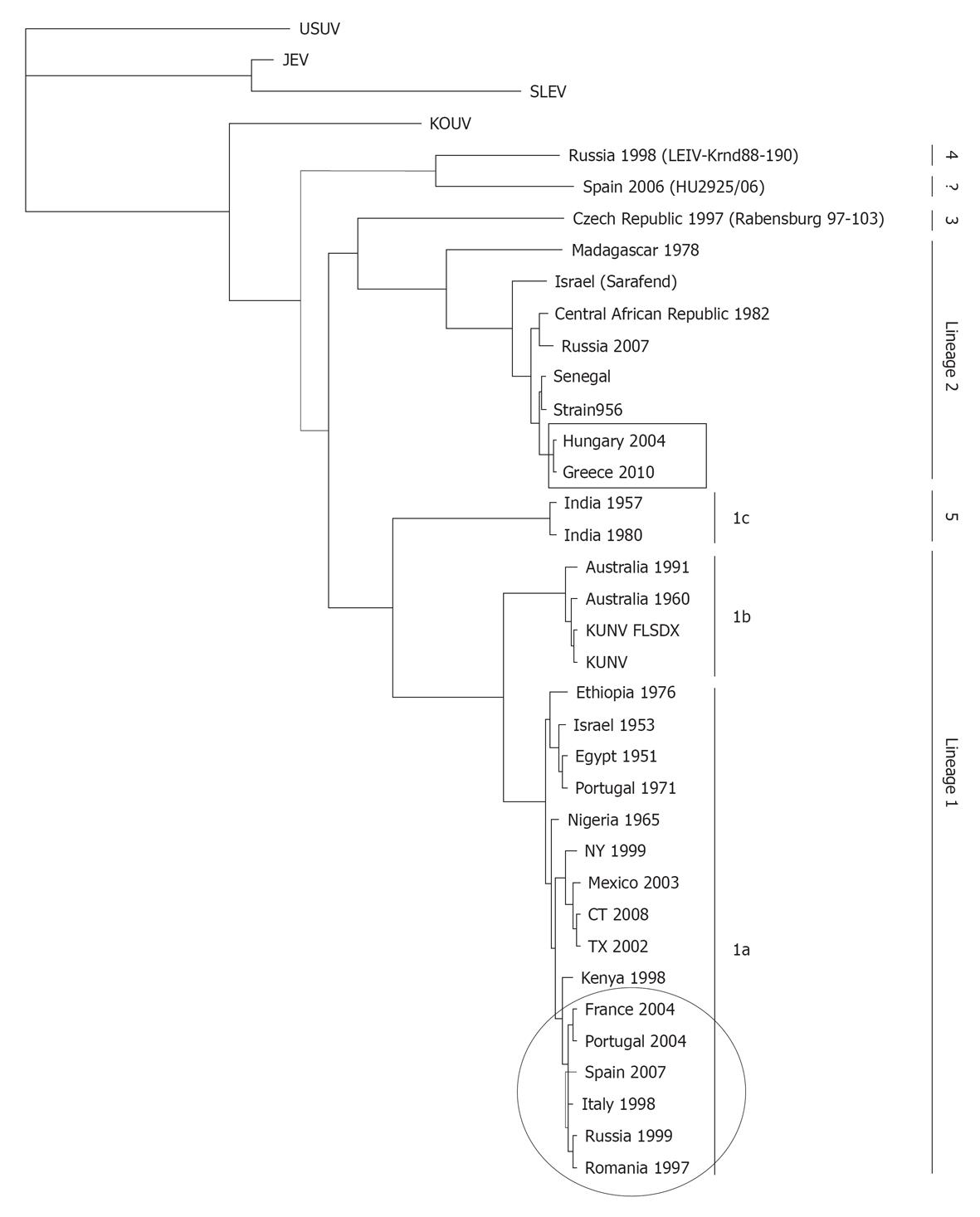
Figure 5 Representative phylogram showing the relationships between West Nile virus strains.
The country of origin and year of isolation is displayed when available. The scale indicates 0.1 substitutions/site. The tree was based on complete NS5 nucleotide sequence (except for HU2925/06), built from a multiple alignment using ClustalW and Phylogeny.fr[231] and visualized with TreeView. Bootstrap values correspond to 500 replications. Squared and circled isolates correspond to lineage 1 and 2 recently circulating in Europe. GeneBank Accesions: Usutu virus (USUV) (AY453412), Japanese encephalitis virus (JEV) (NC_001437), St. Louis encephalitis virus (SLEV) (AY167443), KOUV (EU082200), Russia 1998 (AY277251), Spain 2006 (GU047875), Czech Republic 1997 (AY765264), Madagascar 1978 (ABB01532), Sarafend (AY688948), Central African Republic 1982 (DQ318020), Russia 2007 (FJ425721), Senegal (DQ318019), Strain 956 (NC_001563), Hungary 2004 (DQ116961.1), Greece 2010 (HQ537483), India 1957 (GQ851605), India 1980 (ABC40712), Australia 1991 (GQ851603), Australia 1960 (GQ851602), KUNV_FLSDX (AY274504.1), KUNV (D00246.1), Ethiopia 1976 (AY603654), Israel 1953 (HM051416), Egypt 1951 (EU081844), Portugal 1971 (AM404308), Nigeria 1965 (GQ851607), NY 1999 (AF196835), Mexico 2003 (AY660002), CT 2008 (AEF33448), TX 2002 (DQ176637), Kenya 1998 (AAP20887), France 2004 (DQ786573), Portugal 2004 (AJ965626), Spain 2007 (FJ766332), Italy 1998 (AF404757), Russia 1999 (AF317203), Romania 1997 (AF260969), Italy 2008 (HM641229), and Italy 2009 (HM641225).
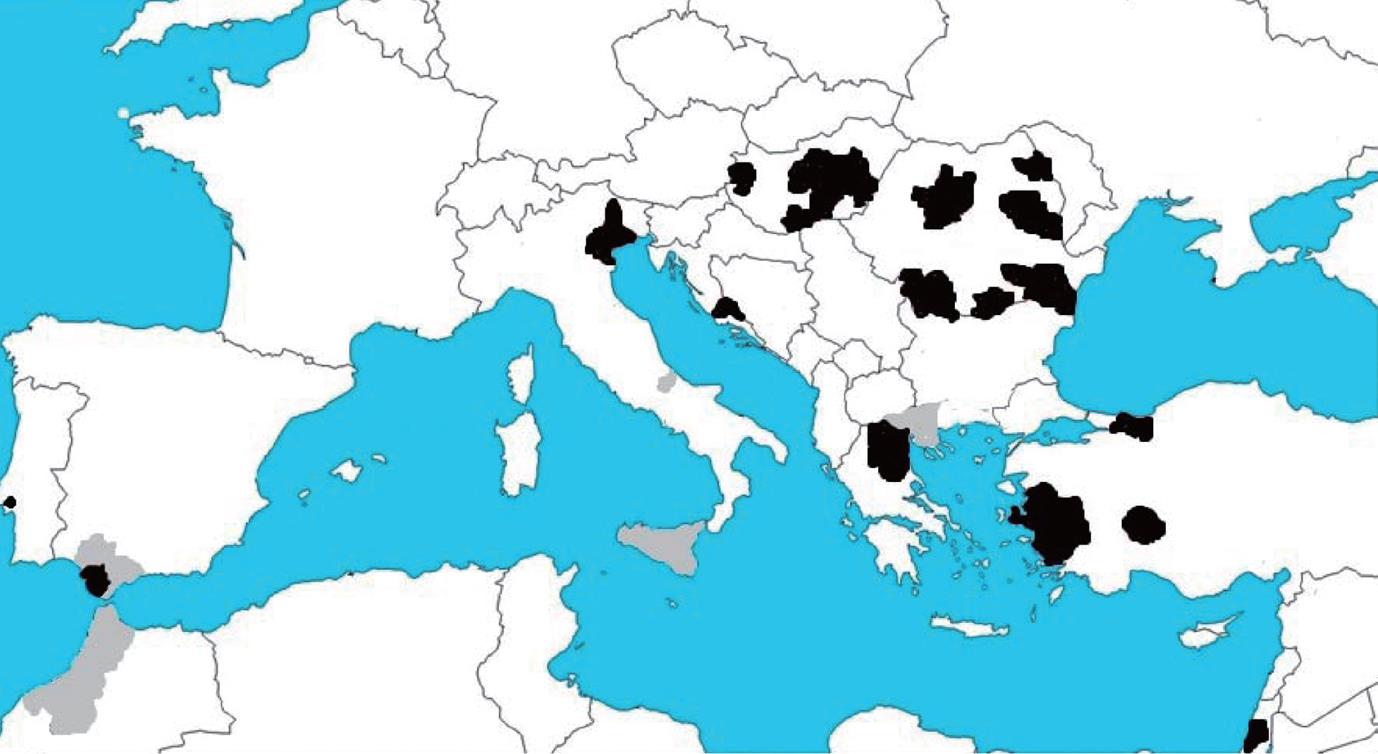
Figure 6 West Nile activity in Europe and the Mediterranean basin in 2010.
Areas with West Nile virus confirmed cases in horses (gray) and humans (black). Data collected from http://ecdc.europa.eu.
- Citation: Martín-Acebes MA, Saiz JC. West Nile virus: A re-emerging pathogen revisited. World J Virol 2012; 1(2): 51-70
- URL: https://www.wjgnet.com/2220-3249/full/v1/i2/51.htm
- DOI: https://dx.doi.org/10.5501/wjv.v1.i2.51