Published online Dec 24, 2015. doi: 10.5500/wjt.v5.i4.209
Peer-review started: July 3, 2015
First decision: July 30, 2015
Revised: August 23, 2015
Accepted: September 25, 2015
Article in press: September 28, 2015
Published online: December 24, 2015
Processing time: 176 Days and 21.2 Hours
Several diseases have been successfully modeled since the development of induced pluripotent stem cell (iPSC) technology in 2006. Since then, methods for increased reprogramming efficiency and cell culture maintenance have been optimized and many protocols for differentiating stem cell lines have been successfully developed, allowing the generation of several cellular subtypes in vitro. Gene editing technologies have also greatly advanced lately, enhancing disease-specific phenotypes by creating isogenic cell lines, allowing mutations to be corrected in affected samples or inserted in control lines. Neurological disorders have benefited the most from iPSC-disease modeling for its capability for generating disease-relevant cell types in vitro from the central nervous system, such as neurons and glial cells, otherwise only available from post-mortem samples. Patient-specific iPSC-derived neural cells can recapitulate the phenotypes of these diseases and therefore, considerably enrich our understanding of pathogenesis, disease mechanism and facilitate the development of drug screening platforms for novel therapeutic targets. Here, we review the accomplishments and the current progress in human neurological disorders by using iPSC modeling for Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, spinal muscular atrophy, amyotrophic lateral sclerosis, duchenne muscular dystrophy, schizophrenia and autism spectrum disorders, which include Timothy syndrome, Fragile X syndrome, Angelman syndrome, Prader-Willi syndrome, Phelan-McDermid, Rett syndrome as well as Nonsyndromic Autism.
Core tip: Several diseases have been successfully modeled using induced pluripotent stem cell (iPSC) technology. Neurological disorders are frequent targets of iPSC-disease modeling for its ability to generate in vitro disease-relevant cell types from the central nervous system, such as neurons and glial cells. Patientspecific iPSC-derived neural cells can recapitulate the phenotypes of these diseases, unveiling mechanisms and providing drug screening platforms for novel therapeutic targets. Here, we review the accomplishments and the current progress achieved in human neurological disorders by using iPSC modeling for Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, spinal muscular atrophy, amyotrophic lateral sclerosis, duchenne muscular dystrophy, schizophrenia and autism spectrum disorders.
- Citation: Russo FB, Cugola FR, Fernandes IR, Pignatari GC, Beltrão-Braga PCB. Induced pluripotent stem cells for modeling neurological disorders. World J Transplant 2015; 5(4): 209-221
- URL: https://www.wjgnet.com/2220-3230/full/v5/i4/209.htm
- DOI: https://dx.doi.org/10.5500/wjt.v5.i4.209
Induced pluripotent stem cell (iPSC) technology was first described in 2006 by Takahashi and Yamanaka[1], when murine fibroblast cells were reprogrammed to a pluripotent stage, with the protocol being successfully applied to human fibroblast cells on the following year by the same group[2]. Since then, iPSCs have been greatly used by many laboratories for pathobiology studies, discovery of disease mechanisms and potential drug-screening platforms[3,4].
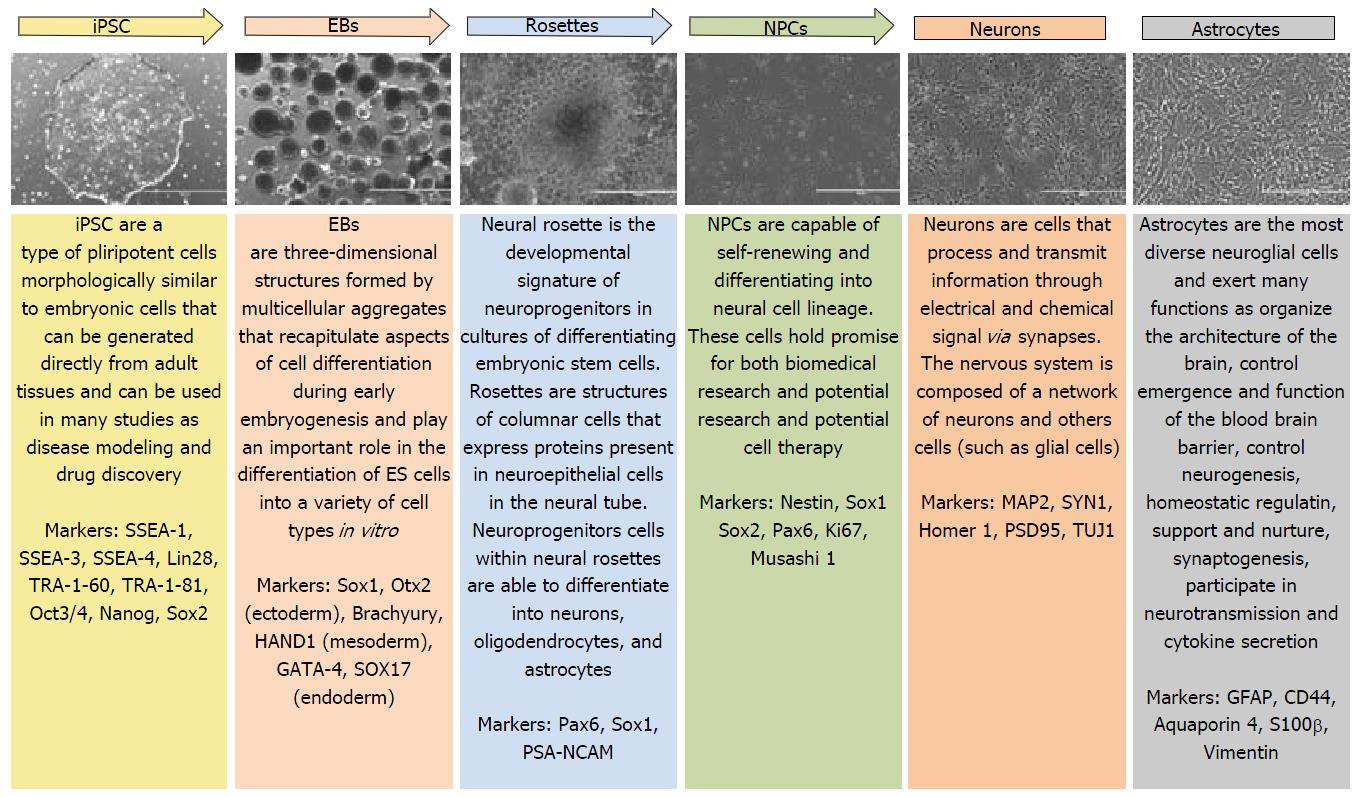
Neurological diseases have benefited enormously from iPSC technology for it allowing in vitro production of human cells that wouldn’t be accessible otherwise, such as the brain, and protocols for generating well-defined neural cell types are already available, being used by several research groups. In our laboratory, the protocol described by Marchetto et al[5] for generating cortical neurons has been successfully reproduced. The steps for neuron generation are represented in Figure 1.
In this review, we introduce an overview of the use of iPSC technology for Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington disease, Spinal muscular atrophy (SMA), amyotrophic lateral sclerosis (ALS), duchenne muscular dystrophy (DMD), autism (syndromic and nonsyndromic) and schizoprhenia as well as its application as a drug screening platform and potential therapeutic application.
AD is the most common progressive neurodegenerative disease affecting the aging population in which patients display gradual memory loss and cognitive impairment. AD can be classified as sporadic late onset (S-AD), which mostly occur after the age of 65 and accounts for 95% of the cases, or more rarely familiar early onset (F-AD), developing in patients in as early as their 30 s. Both occurrences present similar clinical features and pathological phenotypes. For familial cases of AD, mutations in amyloid precursor protein (APP), presenilin 1 and 2 (PS1, PS2) were identified[6].
The amyloid hypothesis of AD pathogenesis stems from the accumulation and aggregation of plaques in the brain comprised of β-amyloid (Aβ) peptides and a hyper phosphorylated form of microtubule associated protein Tau. Point mutations in PS1 or PS2, which form the major component of the γ-secretase complex, affect the γ-secretase-mediated processing of APP, increasing formation of Aβ42 within the neurons, wielding a toxic effect, obstructing neuronal communication and causing oxidative stress[7-9]. Nevertheless, it has been reported contradictory results in animal models for the role of APP in AD[10] and most drugs candidates in clinical trials have failed, implying that to prevent functional and cognitive decline, aiming Aβ alone may not be enough. Utilizing iPSCs in AD modeling allow to further investigate if the cause of neurodegeneration is due to accumulation of Aβ and provide a new method to relate S-AD pathogenesis and newly identified genetic risk variants[11].
Several groups have already successfully generated AD patient specific iPSC-derived neuron lines, providing a novel strategy to investigating the pathogen pathways of the disease[12-14]. Yagi et al[12] first generated neurons from iPSCs from F-AD patients carrying PS1 or PS2 mutations, which revealed elevated levels of Aβ, thus confirming the amyloid cascade hypothesis. Israel et al[14] generated iPSC from two F-AD patients harboring duplications of the APP gene and two S-AD patients and found higher levels of the pathological marker Aβ40, phosphorylated tau (Thr231) and active glycogen synthase kinase-3β, when compared to matched control iPSCs, in both F-AD patients and one S-AD patient. Further treatment of the cells with β-secretase inhibitor improved levels of Thr231 and GSK-23, indicating an APP-tau relationship. Although only one of the S-AD lines recapitulated F-AD phenotype (APP duplication), the autosomal-dominant mechanism forms of F-AD may provide insight into the pathogenesis of S-AD in future studies. Nevertheless, larger numbers of samples will be required in order to fully access their genetic heterogeneity.
Additional studies approaching drug and toxicity screenings in AD, used neuronal cells-iPSC derived, positive for forebrain markers and able to secrete functional proteins involved in Aβ, as well as APP, β-secretase and γ-secretase[15]. After treatment with β- and γ-secretase inhibitors, differences in susceptibility to drugs between the early and late differentiation stages of the cells were reported. Another group used AD iPSC-derived neurons to test for molecules effective against Aβ42 toxicity and revealed that cyclin-dependent kinase 2 inhibitor block Aβ toxicity in the differentiated neural cells[16]. Both studies show the potential that iPSC technology represents in modeling AD and allow to examine patient-specific phenotypes in vitro reflecting the familial and sporadic forms of Alzheimer’s disease, as they are often indistinguishable clinically.
PD is the second most common neurodegenerative disease, behind only to AD, and it’s characterized by progressive loss of dopaminergic neurons (DA) from substantia nigra. Patients display progressive motor dysfunction, such as tremor, rigidity, akinesia and bradykinesia. Most cases of PD are sporadic, but about 20% of patients display familial monogenic forms of the disease[17]. Pathological hallmarks of PD are characterized by presence of Lewy bodies composed of alpha-synuclei (α-syn) protein beyond the nigra and the cortex.
The first dominantly inherited familial PD genetic cause identified was linked to alpha synuclei encoded by the SNCA gene[18], with four mutations currently described[19-22], which causes a misfolding of the protein leading to neuronal dysfunction. Alpha-synuclei is believed to participate in pre-synaptic functions of DA neurons, though the complete actual role of α-SYN is still unknown. DA neurons were generated from iPSCs from a family who carried a triplication of the SNCA locus and expressed double the amount of α-SYN when compared to healthy controls[18]. Further analysis on iPSC-derived DA neurons from the same family, showed increases in mRNA for genes associated with oxidative stress, such as haemoxygenase 2 and monoamine oxidase, and when these neurons were exposed to hydrogen peroxide, increased activation of caspase-3 was detected, suggesting that high levels of α-SYN may present a toxic effect on DA neurons under stress[23].
Another mutation, in A52T SNCA gene, was corrected using zinc finger nuclease (ZFN) technology, both in mutated and control iPSC lines in order to correct the mutation and generate isogenic control lines, respectively. However, the iPSC-derived DA neurons generated were not evaluated, but authors showed the proof of principle that isogenic cell lines are important to evaluate consequences of mutated genes[24].
Two other dominant forms later characterized were linked to mutations in glucocerebrosidase and leucine rich repeat kinase 2 (LRRK2) genes[25-27]. Mutations in LRRK2 gene, usually G2019S, are the most common cause of familial PD, being intensively investigated with use of iPSC technology[28-32]. Increased expression of alpha-synuclein in iPSC-derived DA neurons from LRRK2-mutant lines was found[28], fact observed by other studies[29,32], suggesting a connection between these risk genes, as well as increased expression of oxidative stress genes and increased activation of caspase-3 after treatment with H2O2. Another study used ZFN technology in G2019S-iPSC and health control iPSC lines to correct and add the G2019S mutation, respectively, observed the reversal of the pathogenic phenotype associated with the G2019S mutations[33].
There are three early onset autosomal recessive forms of PD, caused by mutations in Parkin (PARK2), PTEN induced kinase 1 (PINK1) and DJ1 (PARK7)[34-36]. Parkin is believed to mediate mitophagy on a system dependent on PINK1 and account for most cases of early-onset PD[37]. Studies done by different groups in PD iPSC-derived neurons found impaired Parkin recruitment after mitochondria depolarization and observed indications that mutations in PARK2 may predispose neurons to oxidative stress, though details of the exact phenotype remains unclear[38-41].
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disease, affecting approximately 1:10000 persons[42]. Mutations in the huntingtin gene (HTT) lead to poliglutamine repetitions (CAG), causing psychiatric and physiologic alterations[43,44]. Patients with HD display progressive motor and cognitive impairments, change in personality, loss of function along with a decrease in number of neurons, among other symptoms[44,45].
The development of iPSC technology applied to human cells[2] helped elucidate the mechanisms of several devastating neurologic diseases, as HD. Cells from HD patients were first reprogrammed into iPSC in 2010[46], and alterations in electrophysiology, cell metabolism, adherence and toxicity were reported. Expansion of a CAG repeat alters the transport and release of BDFN and increases glutamate receptors, producing toxicity and oxidative stress in neuron and glial cells[44,46,47]. HD iPSC-derived astrocytes displayed 34% more vacuoles when compared to healthy control astrocyte cell lines[42] and on HDN177-82Q mice model, it was observed that mutation in gene HTT causes severe neurological phenotypes and dysfunction in glia cells[48].
Another study created genetically corrected HD iPSCs lines and further differentiated them into neural stem cells (NSC), which displayed normalized pathogenic TGF-β and cadherin signaling pathways. When these genetically corrected NSCs were transplanted into a transgenic HD mice model, it was observed that they were able to populate the striatum after a two week post-transplantation period, uncovering advancements for a potential stem cell replacement therapy[49].
SMA is an autosomal recessive neurodegenerative disease caused by mutations in survival of motor neuron gene (SMN-1), characterized by a selective and progressive loss of lower motor neurons resulting in degeneration of motor neurons in the spinal cord and muscular atrophy on limbs and trunk[50-52].
In order to uncover what is really happening from an inside perspective of the patient’s body, iPSC technology can be used to elucidate this disease mechanism[53]. This was first demonstrated by Ebert et al[50] using fibroblast cells from SMA patients, which were reprogrammed into iPSCs by lentiviral infection carrying Oct4, Sox2, Nanog and Lin28 factors. When these iPSCs were further differentiated into motor neurons, it was observed they displayed smaller soma size and incomplete synapses formation. Valproic acid (1 mmol/L) and Tobramycin (320 umol/L) drugs, both previously described in the treatment of SMA patients[54], were tested and appeared to increase the production of SMN protein in iPSC-derived motor neurons. Valproic acid and anti-sense oligo treatment help improve defects in AChR clustering, increasing levels of SMN transcripts[55].
The neuronal differentiation of SMA iPSCs show reduced capacity to produce motor neurons[51], therefore, applying gene correcting technology may aid in overcoming these methodological shortcomings. The correction of SMN gene, using single-stranded oligonucleotide, was shown to restore the SMN gene profile in neurons derived from SMA-iPSC, converting SMN2 in SMN1[56]. Furthermore, these corrected-gene cells were transplantated in SMA rat models, improving the animals’ disease phenotype and life extension. The possibility of generating genetically corrected, patient-specific SMA-iPSC derived motor neurons and the positive results observed from transplantation in this study, open the path for therapeutic application of autologous cell therapy for SMA patients[57].
ALS is a late adult onset neurodegenerative disease characterized by a progressive degeneration of motor neurons in the cortex, brainstem and bone marrow[58,59]. ALS is a devastating disease; the loss of motor neurons and muscle atrophy confine patients to a wheelchair very rapidly, followed by respiratory failure. The cause of ALS is not yet elucidated, however, mutations in genes SOD1, C9orf72, TDP-43, FUS/TLS, angiogenin, Matrin 3[60-65] and others, have been associated with ALS. Moreover, familial inheritance accounts for about 10% of the cases of patients diagnosed with ALS[65].
Several studies using reprogrammed cells generated from patients of different diseases have been described since 2008[66,67] and they have and still contribute to the understanding, from a physiological point of view to prospective treatments, of these diseases. The first group to generate ALS-derived iPSCs reprogrammed fibroblast cells and further differentiated them into motor neuron cells, opening the path to studies on ALS pathogenesis, yield in a model for testing novel compounds and for autologous cell replacement therapy[67].
iPSC-derived motor neuron cells have been shown to be physiologically active in vitro after reprogramming[68,69] and were immunopositive for ISL+ (motor neuron marker)[68], MNX1 (motor neuron and pancreas homebox protein 1)[69] and also, displaying a phenotype for cholinergic transmitters, positive for ChAT (acetylcholine marker)[68,69].
Neural progenitor cells, which can be generated from iPSC, have become a promising source for cell therapy for ALS. These cells have been transplanted in the lumbar spinal cord in ALS mice models, further differentiated into neurons and astrocytes, and were shown to be able to improve the quality and lifespan of these mice[70,71].
Recently, the world has drawn attention to the ALS “Ice bucket” campaign[72], gaining scientific research strength and raising public awareness about the disease. ALS iPSC research can contribute as a platform to developing new therapeutics, clinical application with cell and gene therapies, enabling new opportunities for future patients’ treatments.
Mutations in the dystrophin gene, located on X chromosome in region p21, lead to dysfunctions in the production of dystrophin, resulting in a misfolded protein. Partial expression or total loss of the dystrophin cause weakness and progressive degeneration of skeletal muscles, reported symptoms of the DMD, whose prevalence is high, affecting approximately 1 in 3300 males[73].
Dystrophin provides support between the actin filaments and cell membrane (sarcolemma) in muscle cells but may also be found in other cellular types, such as in the retina, liver, heart, brain, etc.[74]. Moreover, dystrophin appears to act in the central nervous system. Some studies have reported that DMD patients have difficulties in tests requiring attention and verbal repetition, as well as deficits in speech processing and reading, suggesting DMD may be a cerebellar disorder[75,76]. Approximately one third of DMD patients show cognitive impairment[77,78], in which the mutations in the dystrophin gene seem to alter the efficiency of the brain-cerebellum path, as well as change the neuronal and brain architecture, leading to cognitive deficits in these patients[75-77].
Modeling DMD in vitro will help disclose the neurological mechanism of this disease and even allow to correct the dystrophin deficit in the muscle. To date, cardiomyoblast cells, muscle cells and neurons have been generated from iPSC cells[79-82]. The first group to reprogram cells from DMD patients was Park et al[66] in 2008, followed by other groups modeling DMD in vitro and whose primary objective was to correct the dystrophin in muscle cells. Furthermore, studies applying human artificial chromosome, CRISPR/Cas9 and TALEN technologies[82-84] reported to have restored the expression of dystrophin, observed in vitro and in vivo.
Neuromuscular diseases like DMD have been the focus of iPSC modeling disease studies, which allow the creation of platforms to correct genetic mutations as well as for drug discovery, opening doors to personalized medicine.
Autism spectrum disorder (ASD) is a group of complex neurodevelopmental disorders, affecting 1% of the world’s population, characterized by qualitative communication impairment, atypical social interaction and restricted and repetitive patterns of behavior[85-87]. Autism can be categorized in syndromic and nonsyndromic types. Syndromic autism is definied by an identified neurological disorder, harboring a set of associated phenotypes, where the genetic cause is known and gene mutation is identified. Syndromic forms of ASD are Timothy syndrome (TS), Fragile X syndrome (FXS), Angelman syndrome (AS), Prader-Willi syndrome (PWS), Phelan-McDermid and Rett syndrome (RTT)[5,88-91]. Studies using iPSC technology have already been reported for all of these diseases. Nonsyndromic autism, or simple called ASD, is a group of comorbidities whose genetic cause is not well defined yet, although some genes involved are known, and accounts for the majority of autism cases.
TS is a rare genetic disorder caused by de novo missense mutation in the CACNA1C gene[92,93] and it is associated with developmental delay and autism[92]. This gene encodes the α-subunit of the voltage-gated calcium channel Cav1.2. This channel plays a central role in regulating and signaling network that is essential for neuronal function[94-96].
Cortical neuronal precursor cells and neurons were first differentiated from iPSCs generated from patients with Timothy syndrome by Pasca et al[88]. Intracellular calcium (Ca2+) signals were examined in these cells and a significant increase in TS neurons was observed. Furthermore, TS patient specific-iPSCs were generated to study the effects of the mutation on dendritic arbors. The results found in these cells were then compared to a TS rodent model and revealed an aberrant activity-dependent dendritic retraction in both human derived neurons and animal neurons[97].
Mutations in ion channel genes have been associated with cardiac arrhythmias and TS, but the pathophysiological process is little known. TS iPSC-derived cardiomyocyte cells displayed an erratic and slow contraction behaviour when compared to healthy controls, as well as abnormal calcium handling and irregular and prolonged action potential patterns[98].
FXS is the most common form of syndromic ASD and mental retardation[89]. FXS is caused by loss of expression of the fragile X mental retardation gene 1 (FMR1) located in the X-chromosome, where an expanded CGG repeats in the 5’-untranslated region of the FMR1 gene is present[89,99]. FXS has no cure and patients display developmental impairment, learning and cognitive disabilities, as well as physical and behavioral phenotypes such as stereotypic movements[100,101].
FMR1 gene is associated with synaptogenesis and the FMRP protein can be detected at synapses and dendritic spines[102]. The first FXS iPSC model was derived from fibroblasts and described by Urbach et al[89]. Their findings reported the FMR1 gene remained inactive and highlighted crucial differences between ES and iPS cells. Another study reported variable levels of FMR1 silencing and expression in multiple FXS iPSC lines. Furthermore, these lines showed reduced FRM1 expression during neuronal differentiation[99].
FMRP expression works as an indicator for drug discovery for FXS. In a recent drug screening study, 6 compounds were shown to increase FMR1 gene expression in neural stem cells differentiated from a FXS iPSC line. Despite none of these compounds resulted in clinically relevant levels of FMR1, these findings support the idea this assay can be used as a drug screening platform for FXS[101].
Another study showed that iPSC-derived neurons from FXS patients displayed fewer synaptic protein levels and synapses, reduced neurite length and abnormal functionality, with increased calcium transients[103]. Reduced neurite was also observed in forebrain neurons derived from FXS iPSCs[104].
AS and PWS are neurodevelopmental disorders associated with autism caused by deletions in chromosome 15q11-q13[105]. AS is caused by reduced expression of the ubiquitin-protein ligase E3A gene (UBE3A) of the maternal chromosome[106-108] whereas PWS occurs by the same deletion on the paternally inherited allele[109]. They both share same behavioral and neurological phenotypes. However, cognitive and neurologic impairments are more severe in AS, including seizures, while behavioral problems are more severe in PWS[109].
The first study to model AS and PWS using iPSC-derived from patients was done by Chamberlain et al[105]. Although the authors found no phenotypic differences between AS and control neurons, they observed the UBE3A imprinting occurred during neuronal differentiation in AS cells.
Recently, iPSCs from a PWS patient with an atypical microdeletion on paternal chromosome 15q11-q13 were generated[90], revealing they expressed UBE3A-ATS, typically restricted to neurons as is, consequently, the imprinted expression of UBE3A observed in these iPSCs, as well[90].
Another study generated iPSCs from patients with duplications of chromosome 15q11-q13.1 (Dup15q syndrome) and were further differentiated into functional neurons. Gene expression analysis was performed and compared to AS neurons, revealing they shared common neuronal pathways disrupted in both Angelman and Dup15q syndromes[110].
Phelan-McDermid syndrome (PMDS) is a rare disorder associated with deletions in chromosome 22q13[91,111]. PMDS is a monogenic form of ASD with a frequency of at least 0.5% of ASD cases and is resulted by deletions in SH3 and multiple ankyrin repeat domains 3 (SHANK3)[112]. This gene plays an important role in synaptic function and is involved in the organization of postsynaptic density[113,114]. PMDS patients display some autistic features as severe language delay and intellectual disability[115]. Animal models for ASD carrying SHANK3 mutations display synaptic dysfunction, abnormal social behavior, repetitive and communication behavior patterns and deficient learning and memory[116].
Recently, Shcheglovitov et al[117] generated iPSC-derived neurons from individuals with PMDS carrying large 22q13 deletions that included SHANK3. These neurons displayed fewer synapses and altered electrophysiology. The group reported that excitatory synaptic transmission in PMDS neurons can be corrected by restoring SHANK3 expression or by treating neurons with insulin-like growth factor 1[117].
RTT is a progressive neurodevelopmental disorder caused by mutations in the X-linked gene methyl CpG-binding protein 2 (MeCP2)[5,118]. RTT syndrome affects more females with an incidence of 1 in 10000[118]. Rett patients display a normal development until 18 mo of age, but thereafter, progressive neurological abnormalities begin to emerge[119]. Neurologic pathologies as autistic behavior, stereotypies, loss of speech, microcephaly, seizures and hypotonia have been described in RTT patients[120].
Several studies utilizing RTT-derived iPSC have been published in the past years. The first RTT-derived iPSC lines were generated by the Ellis group[121], however, the first group to make use of iPSC for disease modeling of RTT syndrome was by Marchetto et al[5]. In this work, iPSC-derived neurons from four different RTT patients were generated. Neuronal phenotypes displayed reduced dendritic spine density, smaller soma size, altered electrophysiology, alterations in Ca2+ influx and fewer synapses. Furthermore, insulin-like growth factor 1 (IGF-1) was able to rescue the synaptic defects in these neurons after treatment[5]. Reduced soma and nuclear size phenotypes from RTT iPSC-derived neurons were also observed by another group[122] as well as defects in neuronal maturation[123].
IPSC-derived neurons from heterozygous Mecp2308 mice showed defects in glutamatergic synaptic transmission and generation of action potentials and decreased action potential amplitude. These phenotypes were observed in neurons derived from WT and hemizygous mutant iPSC lines, indicating that these deficits are caused by MeCP2 deficiency[124].
The first isoform-patient specific iPSC model of RTT was reported by Dijuric et al[125]. iPSC-derived neurons from RTTe1 maintain an inactive X-chromosome and express only the mutant allele. Mutant neurons exhibited reduced dendritic complexity, decreased soma size and cell capacitance.
Recently, astrocytes derived from RTT iPSCs were generated by William et al[126]. The group demonstrated that these mutant astrocytes can affect directly the neurons and induce abnormalities. IGF-1 and GPE (an IGF-1 peptide) can partially rescue the morphological defects[126].
RTT syndrome has become a popular target for iPSC studies and this technology has greatly contributed to a better understanding of the disease.
Research on syndromic autism provides us with data that can contribute to the understanding of nonsyndromic autism cases, where the genetic causes are still unknown. Furthermore, animal models provide valuable information on ASD, with recent studies showing similar synaptic phenotypes in nonsyndromic and syndromic mouse models of autisms[127].
The first iPSC model of nonsyndromic autism was recently generated by Griesi-Oliveira et al[128]. In this study, the group investigated the molecular and cellular phenotypes in iPSC-derived neurons from an ASD individual carrying a mutation in the TRPC6 gene, which encodes for protein channel transient receptor potential Canonical 6. TRPC6 protein operates in a calcium channel in the brain, controlling the functioning of neurons, in particular neuronal synapses[128]. In vitro analysis revealed that this mutation leads to a reduction of synapses and morphological changes in mouse neurons. These data showed phenotypes in common with findings from syndromic autism[5,88,89,105,117], where the studies demonstrated neuronal abnormalities such as altered morphology and synaptic deficits. The group was also able to rescue some of the neuronal abnormalities using candidate drugs as, IGF-1 and hyperforin. This study brings valuable information to the understanding of autism disorder, despite this mutation occurs in less than 1% of patients with ASD and the genetics of autism is quite complex and involves several genes[129].
Schizophrenia (SCZD), like nonsyndromic autism, is a complex neurological disorder where the genetic causes are still unclear, affecting a large number of individuals (1.1% of the world’s population)[130,131]. It is considered to stem from a polygenic basis, with an estimated heritability of approximately 80%[130,132,133], and genetic and epigenetic processes underlying the disease, as it was observed in a discordant monozygotic twin study[133]. Moreover, environmental stressors like drug use, being cannabis the most frequently studied, birth complications, maternal immune response, among others, may contibute to SCZD[134-137].
People with SCZD have a lower life expectancy average, mostly to increased health problems and higher suicide rate, and individuals may experience symptoms like hallucinations, delusions, abnormal social behavior (inability to speak, express emotions or find pleasure) and cognitive impairment (deficits in attention, memory and planning)[131,132].
The very first study published with iPSC derived from SCZD patients did not produce neurons[138]. A different group published that same year a study using iPSC technology for SCZD modeling. In this study, iPSC-derived neurons were characterized and revealed defects in neuronal connectivity, reduced outgrowth from soma, reduced PSD95 dendritic protein levels and some altered gene expression. Furthermore, phenotypes in SCZD neurons were ameliorated after treatment with Loxapine, an antipsychotic drug[139].
Another work using SCDZ iPSC-derived neurons carrying 22q11 deletions observed a high L1 copy number in these cells, confirmed by neuronal genome analysis, validating the use of iPSC technology in the study of SCZD condition[140]. Notwithstanding these evidences and taking into consideration SCZD heterogeneity, more studies should be carried out bearing in mind the use of more homogeneous populations, by selecting subjects with rare genetic variants or with similar clinical manifestations[141].
The path for disease treatment and prevention is through the unveiling of pathogenesis and physiological mechanisms that ultimately result in the phenotypic symptoms of diseases. Analysis of live and post-mortem samples, as well as animal models, are great sources for disease study outlines. Despite the importance and relevance of the use of animal models in research, they sometimes are inadequate to fully recapitulate the pathology as it is in humans, and consequently, many drug candidates that once showed to be therapeutically promising in animal models, failed in clinical trials in humans[142].
The development of iPSC technology has come to aid to fill in the gap between pathogenesis and in vivo phenotypes. Since the first human iPSC line was established, this methodology has been used by many laboratories for the study of neurological and psychiatric disorders.
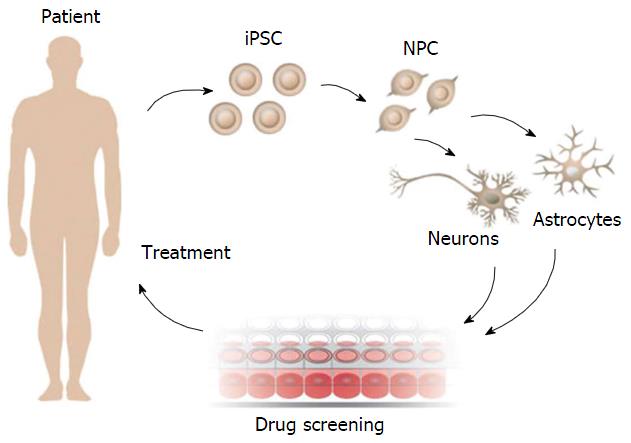
Neuroscience research has taken a significant step with iPSC disease modeling. The possibility of generating patient-specific cell lines and differentiating them into various cellular subtypes in vitro, allow the creation of future personalized therapeutical treatments. This procedure is represented in Figure 2.
Although iPSC technology holds great potential for disease modeling and research, it is still in its initial phase. This promising technology provides a useful platform for a better understanding of neurological diseases mechanisms, drug discovery and future therapeutical applications.
Our acknowledgement is to Lenon Della Rovere for figure design support.
P- Reviewer: Gary LD, Kim YB
S- Editor: Qiu S L- Editor: A E- Editor: Li D
| 1. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18178] [Article Influence: 956.7] [Reference Citation Analysis (0)] |
| 2. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14305] [Article Influence: 841.5] [Reference Citation Analysis (0)] |
| 3. | Grieshammer U, Shepard KA. Proceedings: consideration of genetics in the design of induced pluripotent stem cell-based models of complex disease. Stem Cells Transl Med. 2014;3:1253-1258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Santostefano KE, Hamazaki T, Biel NM, Jin S, Umezawa A, Terada N. A practical guide to induced pluripotent stem cell research using patient samples. Lab Invest. 2015;95:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 5. | Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1082] [Cited by in RCA: 998] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 6. | St George-Hyslop PH. Genetic factors in the genesis of Alzheimer’s disease. Ann N Y Acad Sci. 2000;924:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 7. | Zhang YW, Thompson R, Zhang H, Xu H. APP processing in Alzheimer’s disease. Mol Brain. 2011;4:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 487] [Cited by in RCA: 634] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 8. | Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4534] [Cited by in RCA: 5130] [Article Influence: 155.5] [Reference Citation Analysis (0)] |
| 9. | Wolfe MS. When loss is gain: reduced presenilin proteolytic function leads to increased Abeta42/Abeta40. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:136-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 153] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 10. | Khandekar N, Lie KH, Sachdev PS, Sidhu KS. Amyloid precursor proteins, neural differentiation of pluripotent stem cells and its relevance to Alzheimer’s disease. Stem Cells Dev. 2012;21:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Bettens K, Sleegers K, Van Broeckhoven C. Genetic insights in Alzheimer’s disease. Lancet Neurol. 2013;12:92-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 296] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 12. | Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20:4530-4539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 449] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 13. | Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12:487-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 532] [Cited by in RCA: 575] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 14. | Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 839] [Cited by in RCA: 920] [Article Influence: 70.8] [Reference Citation Analysis (0)] |
| 15. | Yahata N, Asai M, Kitaoka S, Takahashi K, Asaka I, Hioki H, Kaneko T, Maruyama K, Saido TC, Nakahata T. Anti-Aβ drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer’s disease. PLoS One. 2011;6:e25788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 137] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 16. | Xu X, Lei Y, Luo J, Wang J, Zhang S, Yang XJ, Sun M, Nuwaysir E, Fan G, Zhao J. Prevention of β-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of Cyclin-dependent kinases and associated cell cycle events. Stem Cell Res. 2013;10:213-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Lesage S, Brice A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48-R59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 651] [Cited by in RCA: 661] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 18. | Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3195] [Cited by in RCA: 3353] [Article Influence: 152.4] [Reference Citation Analysis (0)] |
| 19. | Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045-2047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5908] [Cited by in RCA: 6034] [Article Influence: 215.5] [Reference Citation Analysis (0)] |
| 20. | Krüger R, Kuhn W, Müller T, Woitalla D, Graeber M, Kösel S, Przuntek H, Epplen JT, Schöls L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2931] [Cited by in RCA: 2957] [Article Influence: 109.5] [Reference Citation Analysis (0)] |
| 21. | Zarranz JJ, Alegre J, Gómez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atarés B. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1988] [Cited by in RCA: 2095] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 22. | Appel-Cresswell S, Vilarino-Guell C, Encarnacion M, Sherman H, Yu I, Shah B, Weir D, Thompson C, Szu-Tu C, Trinh J. Alpha-synuclein p. H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov Disord. 2013;28:811-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 448] [Cited by in RCA: 496] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 23. | Byers B, Cord B, Nguyen HN, Schüle B, Fenno L, Lee PC, Deisseroth K, Langston JW, Pera RR, Palmer TD. SNCA triplication Parkinson’s patient’s iPSC-derived DA neurons accumulate α-synuclein and are susceptible to oxidative stress. PLoS One. 2011;6:e26159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 242] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 24. | Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 631] [Cited by in RCA: 584] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 25. | Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2004;351:1972-1977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 434] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 26. | Neumann J, Bras J, Deas E, O’Sullivan SS, Parkkinen L, Lachmann RH, Li A, Holton J, Guerreiro R, Paudel R. Glucocerebrosidase mutations in clinical and pathologically proven Parkinson’s disease. Brain. 2009;132:1783-1794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 576] [Cited by in RCA: 532] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 27. | Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2078] [Cited by in RCA: 2265] [Article Influence: 113.3] [Reference Citation Analysis (0)] |
| 28. | Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schüle B, Dolmetsch RE, Langston W. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 569] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 29. | Sánchez-Danés A, Richaud-Patin Y, Carballo-Carbajal I, Jiménez-Delgado S, Caig C, Mora S, Di Guglielmo C, Ezquerra M, Patel B, Giralt A. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol Med. 2012;4:380-395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 421] [Cited by in RCA: 438] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 30. | Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med. 2012;4:141ra90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 401] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 31. | Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603-607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 274] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 32. | Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16:394-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 485] [Cited by in RCA: 492] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 33. | Reinhardt P, Schmid B, Burbulla LF, Schöndorf DC, Wagner L, Glatza M, Höing S, Hargus G, Heck SA, Dhingra A. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 400] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 34. | Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2010] [Cited by in RCA: 2037] [Article Influence: 92.6] [Reference Citation Analysis (0)] |
| 35. | Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3646] [Cited by in RCA: 3756] [Article Influence: 139.1] [Reference Citation Analysis (0)] |
| 36. | Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2438] [Cited by in RCA: 2592] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 37. | Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1280] [Cited by in RCA: 1341] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 38. | Seibler P, Graziotto J, Jeong H, Simunovic F, Klein C, Krainc D. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J Neurosci. 2011;31:5970-5976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 305] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 39. | Jiang H, Ren Y, Yuen EY, Zhong P, Ghaedi M, Hu Z, Azabdaftari G, Nakaso K, Yan Z, Feng J. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat Commun. 2012;3:668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 201] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 40. | Imaizumi Y, Okada Y, Akamatsu W, Koike M, Kuzumaki N, Hayakawa H, Nihira T, Kobayashi T, Ohyama M, Sato S. Mitochondrial dysfunction associated with increased oxidative stress and α-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain. 2012;5:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 286] [Cited by in RCA: 311] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 41. | Brundin P, Strecker RE, Lindvall O, Isacson O, Nilsson OG, Barbin G, Prochiantz A, Forni C, Nieoullon A, Widner H. Intracerebral grafting of dopamine neurons. Experimental basis for clinical trials in patients with Parkinson’s disease. Ann N Y Acad Sci. 1987;495:473-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 42. | Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, Ming GL, Song H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 2012;5:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 183] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 43. | A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6006] [Cited by in RCA: 6119] [Article Influence: 191.2] [Reference Citation Analysis (0)] |
| 44. | Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1048] [Cited by in RCA: 1194] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 45. | Sawa A, Tomoda T, Bae BI. Mechanisms of neuronal cell death in Huntington’s disease. Cytogenet Genome Res. 2003;100:287-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 46. | Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2010;RRN1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 193] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 47. | Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171:1001-1012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 315] [Cited by in RCA: 353] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 48. | Bradford J, Shin JY, Roberts M, Wang CE, Sheng G, Li S, Li XJ. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J Biol Chem. 2010;285:10653-10661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 124] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 49. | An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, Melov S, Ellerby LM. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 281] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 50. | Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1169] [Cited by in RCA: 1039] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 51. | Chang T, Zheng W, Tsark W, Bates S, Huang H, Lin RJ, Yee JK. Brief report: phenotypic rescue of induced pluripotent stem cell-derived motoneurons of a spinal muscular atrophy patient. Stem Cells. 2011;29:2090-2093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 52. | Wu CY, Whye D, Glazewski L, Choe L, Kerr D, Lee KH, Mason RW, Wang W. Proteomic assessment of a cell model of spinal muscular atrophy. BMC Neurosci. 2011;12:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 53. | Saporta MA, Grskovic M, Dimos JT. Induced pluripotent stem cells in the study of neurological diseases. Stem Cell Res Ther. 2011;2:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 54. | Brichta L, Hofmann Y, Hahnen E, Siebzehnrubl FA, Raschke H, Blumcke I, Eyupoglu IY, Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet. 2003;12:2481-2489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 277] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 55. | Yoshida M, Kitaoka S, Egawa N, Yamane M, Ikeda R, Tsukita K, Amano N, Watanabe A, Morimoto M, Takahashi J. Modeling the early phenotype at the neuromuscular junction of spinal muscular atrophy using patient-derived iPSCs. Stem Cell Reports. 2015;4:561-568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 56. | Corti S, Nizzardo M, Simone C, Falcone M, Nardini M, Ronchi D, Donadoni C, Salani S, Riboldi G, Magri F. Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med. 2012;4:165ra162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 155] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 57. | Frattini E, Ruggieri M, Salani S, Faravelli I, Zanetta C, Nizzardo M, Simone C, Magri F, Corti S. Pluripotent stem cell-based models of spinal muscular atrophy. Mol Cell Neurosci. 2015;64:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 58. | Papadeas ST, Maragakis NJ. Advances in stem cell research for Amyotrophic Lateral Sclerosis. Curr Opin Biotechnol. 2009;20:545-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 59. | Thomsen GM, Gowing G, Svendsen S, Svendsen CN. The past, present and future of stem cell clinical trials for ALS. Exp Neurol. 2014;262 Pt B:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 60. | Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 789] [Cited by in RCA: 857] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 61. | Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 292] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 62. | Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4:145ra104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 417] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 63. | Padhi AK, Jayaram B, Gomes J. Prediction of functional loss of human angiogenin mutants associated with ALS by molecular dynamics simulations. Sci Rep. 2013;3:1225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 64. | Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664-666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 364] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 65. | Li Y, Balasubramanian U, Cohen D, Zhang PW, Mosmiller E, Sattler R, Maragakis NJ, Rothstein JD. A comprehensive library of familial human amyotrophic lateral sclerosis induced pluripotent stem cells. PLoS One. 2015;10:e0118266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 66. | Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1775] [Cited by in RCA: 1608] [Article Influence: 94.6] [Reference Citation Analysis (0)] |
| 67. | Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1445] [Cited by in RCA: 1404] [Article Influence: 82.6] [Reference Citation Analysis (0)] |
| 68. | Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279-286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 410] [Cited by in RCA: 372] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 69. | Du ZW, Chen H, Liu H, Lu J, Qian K, Huang CL, Zhong X, Fan F, Zhang SC. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun. 2015;6:6626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 284] [Cited by in RCA: 308] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 70. | Popescu IR, Nicaise C, Liu S, Bisch G, Knippenberg S, Daubie V, Bohl D, Pochet R. Neural progenitors derived from human induced pluripotent stem cells survive and differentiate upon transplantation into a rat model of amyotrophic lateral sclerosis. Stem Cells Transl Med. 2013;2:167-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 71. | Kondo T, Funayama M, Tsukita K, Hotta A, Yasuda A, Nori S, Kaneko S, Nakamura M, Takahashi R, Okano H. Focal transplantation of human iPSC-derived glial-rich neural progenitors improves lifespan of ALS mice. Stem Cell Reports. 2014;3:242-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 72. | Vaidya M. Ice bucket challenge cash may help derisk ALS drug research. Nat Med. 2014;20:1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 73. | Wingeier K, Giger E, Strozzi S, Kreis R, Joncourt F, Conrad B, Gallati S, Steinlin M. Neuropsychological impairments and the impact of dystrophin mutations on general cognitive functioning of patients with Duchenne muscular dystrophy. J Clin Neurosci. 2011;18:90-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 74. | Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 665] [Cited by in RCA: 766] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 75. | Hinton VJ, Fee RJ, Goldstein EM, De Vivo DC. Verbal and memory skills in males with Duchenne muscular dystrophy. Dev Med Child Neurol. 2007;49:123-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 76. | Cyrulnik SE, Hinton VJ. Duchenne muscular dystrophy: a cerebellar disorder? Neurosci Biobehav Rev. 2008;32:486-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 78] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 77. | Anderson JL, Head SI, Rae C, Morley JW. Brain function in Duchenne muscular dystrophy. Brain. 2002;125:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 208] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 78. | Daoud F, Angeard N, Demerre B, Martie I, Benyaou R, Leturcq F, Cossée M, Deburgrave N, Saillour Y, Tuffery S. Analysis of Dp71 contribution in the severity of mental retardation through comparison of Duchenne and Becker patients differing by mutation consequences on Dp71 expression. Hum Mol Genet. 2009;18:3779-3794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 123] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 79. | Guan X, Mack DL, Moreno CM, Strande JL, Mathieu J, Shi Y, Markert CD, Wang Z, Liu G, Lawlor MW. Dystrophin-deficient cardiomyocytes derived from human urine: new biologic reagents for drug discovery. Stem Cell Res. 2014;12:467-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 80. | Luo Y, Fan Y, Chen X, Yue L, Yu B, Li Q, Chen Y, Sun X. Modeling induced pluripotent stem cells from fibroblasts of Duchenne muscular dystrophy patients. Int J Neurosci. 2014;124:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 81. | Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S, Bett GC, Rasmusson RL, Denning C, Yang L. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy. Dis Model Mech. 2015;8:457-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 82. | Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 390] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 83. | Kazuki Y, Hiratsuka M, Takiguchi M, Osaki M, Kajitani N, Hoshiya H, Hiramatsu K, Yoshino T, Kazuki K, Ishihara C. Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol Ther. 2010;18:386-393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 84. | Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 360] [Cited by in RCA: 339] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 85. | Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341-355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1404] [Cited by in RCA: 1193] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 86. | Quaak I, Brouns MR, Van de Bor M. The dynamics of autism spectrum disorders: how neurotoxic compounds and neurotransmitters interact. Int J Environ Res Public Health. 2013;10:3384-3408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 87. | Acab A, Muotri AR. The Use of Induced Pluripotent Stem Cell Technology to Advance Autism Research and Treatment. Neurotherapeutics. 2015;12:534-545. [PubMed] |
| 88. | Paşca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Paşca AM, Cord B, Palmer TD, Chikahisa S, Nishino S. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657-1662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 465] [Cited by in RCA: 443] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 89. | Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 294] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 90. | Martins-Taylor K, Hsiao JS, Chen PF, Glatt-Deeley H, De Smith AJ, Blakemore AI, Lalande M, Chamberlain SJ. Imprinted expression of UBE3A in non-neuronal cells from a Prader-Willi syndrome patient with an atypical deletion. Hum Mol Genet. 2014;23:2364-2373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 91. | Sarasua SM, Dwivedi A, Boccuto L, Chen CF, Sharp JL, Rollins JD, Collins JS, Rogers RC, Phelan K, DuPont BR. 22q13.2q13.32 genomic regions associated with severity of speech delay, developmental delay, and physical features in Phelan-McDermid syndrome. Genet Med. 2014;16:318-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 92. | Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19-31. [PubMed] |
| 93. | Boczek NJ, Miller EM, Ye D, Nesterenko VV, Tester DJ, Antzelevitch C, Czosek RJ, Ackerman MJ, Ware SM. Novel Timothy syndrome mutation leading to increase in CACNA1C window current. Heart Rhythm. 2015;12:211-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 94. | Bading H. Nuclear calcium signalling in the regulation of brain function. Nat Rev Neurosci. 2013;14:593-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 284] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 95. | Catterall WA, Leal K, Nanou E. Calcium channels and short-term synaptic plasticity. J Biol Chem. 2013;288:10742-10749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 96. | Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 483] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 97. | Krey JF, Paşca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, Dolmetsch RE. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16:201-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 201] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 98. | Yazawa M, Dolmetsch RE. Modeling Timothy syndrome with iPS cells. J Cardiovasc Transl Res. 2013;6:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 99. | Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 237] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 100. | Rogers SJ, Wehner DE, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr. 2001;22:409-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 335] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 101. | Kumari D, Swaroop M, Southall N, Huang W, Zheng W, Usdin K. High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem Cells Transl Med. 2015;4:800-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 102. | Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 823] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 103. | Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, Mitchell G, Nacey C, Githinji J, McGee J, Garcia-Arocena D. Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet. 2012;21:3795-3805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 104. | Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang SC, Abbeduto L, Bhattacharyya A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 105. | Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci USA. 2010;107:17668-17673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 246] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 106. | Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 287] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 107. | Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 212] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 108. | Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409-412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 336] [Cited by in RCA: 385] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 109. | Cassidy SB, Dykens E, Williams CA. Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet. 2000;97:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 110. | Germain ND, Chen PF, Plocik AM, Glatt-Deeley H, Brown J, Fink JJ, Bolduc KA, Robinson TM, Levine ES, Reiter LT. Gene expression analysis of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11-q13.1. Mol Autism. 2014;5:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 111. | Phelan K, McDermid HE. The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome). Mol Syndromol. 2012;2:186-201. [PubMed] |
| 112. | Kolevzon A, Bush L, Wang AT, Halpern D, Frank Y, Grodberg D, Rapaport R, Tavassoli T, Chaplin W, Soorya L. A pilot controlled trial of insulin-like growth factor-1 in children with Phelan-McDermid syndrome. Mol Autism. 2014;5:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 113. | Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 701] [Cited by in RCA: 811] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 114. | Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1052] [Cited by in RCA: 1139] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 115. | Phelan MC. Deletion 22q13.3 syndrome. Orphanet J Rare Dis. 2008;3:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 116. | Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093-3108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 431] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 117. | Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, Krawisz A, Froehlich W, Bernstein JA, Hallmayer JF. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 353] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 118. | Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3531] [Cited by in RCA: 3551] [Article Influence: 136.6] [Reference Citation Analysis (0)] |
| 119. | Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 869] [Cited by in RCA: 930] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 120. | Percy AK. Rett syndrome: exploring the autism link. Arch Neurol. 2011;68:985-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 121. | Hotta A, Cheung AY, Farra N, Vijayaragavan K, Séguin CA, Draper JS, Pasceri P, Maksakova IA, Mager DL, Rossant J. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods. 2009;6:370-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 226] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 122. | Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103-2115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 123. | Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci USA. 2011;108:14169-14174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 124. | Farra N, Zhang WB, Pasceri P, Eubanks JH, Salter MW, Ellis J. Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations. Mol Psychiatry. 2012;17:1261-1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 125. | Djuric U, Cheung AY, Zhang W, Mok RS, Lai W, Piekna A, Hendry JA, Ross PJ, Pasceri P, Kim DS. MECP2e1 isoform mutation affects the form and function of neurons derived from Rett syndrome patient iPS cells. Neurobiol Dis. 2015;76:37-45. [PubMed] |
| 126. | Williams EC, Zhong X, Mohamed A, Li R, Liu Y, Dong Q, Ananiev GE, Mok JC, Lin BR, Lu J. Mutant astrocytes differentiated from Rett syndrome patients-specific iPSCs have adverse effects on wild-type neurons. Hum Mol Genet. 2014;23:2968-2980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 152] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 127. | Baudouin SJ. [Mouse models of autism: a common basis for syndromic and non syndromic autisms. Med Sci (Paris). 2013;29:121-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 128. | Griesi-Oliveira K, Acab A, Gupta AR, Sunaga DY, Chailangkarn T, Nicol X, Nunez Y, Walker MF, Murdoch JD, Sanders SJ. Modeling non-syndromic autism and the impact of TRPC6 disruption in human neurons. Mol Psychiatry. 2014;Nov 11; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 129. | De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209-215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1776] [Cited by in RCA: 1976] [Article Influence: 179.6] [Reference Citation Analysis (0)] |
| 130. | Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187-1192. [PubMed] |
| 131. | Brennand KJ, Gage FH. Concise review: the promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem Cells. 2011;29:1915-1922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 132. | Carpenter WT, Buchanan RW. Schizophrenia. N Engl J Med. 1994;330:681-690. [PubMed] |
| 133. | Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12-17. [PubMed] |
| 134. | Brown AS. The environment and susceptibility to schizophrenia. Prog Neurobiol. 2011;93:23-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 469] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 135. | Torrey EF, Bartko JJ, Yolken RH. Toxoplasma gondii and other risk factors for schizophrenia: an update. Schizophr Bull. 2012;38:642-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 262] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 136. | Scherr M, Hamann M, Schwerthöffer D, Froböse T, Vukovich R, Pitschel-Walz G, Bäuml J. Environmental risk factors and their impact on the age of onset of schizophrenia: Comparing familial to non-familial schizophrenia. Nord J Psychiatry. 2012;66:107-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 137. | Connor CM, Dincer A, Straubhaar J, Galler JR, Houston IB, Akbarian S. Maternal immune activation alters behavior in adult offspring, with subtle changes in the cortical transcriptome and epigenome. Schizophr Res. 2012;140:175-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 138. | Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 139. | Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 1042] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 140. | Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:306-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 141. | Brennand KJ, Landek-Salgado MA, Sawa A. Modeling heterogeneous patients with a clinical diagnosis of schizophrenia with induced pluripotent stem cells. Biol Psychiatry. 2014;75:936-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 142. | Hermann A, Storch A. Induced neural stem cells (iNSCs) in neurodegenerative diseases. J Neural Transm. 2013;120 Suppl 1:S19-S25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |