Published online Sep 19, 2025. doi: 10.5498/wjp.v15.i9.108222
Revised: May 13, 2025
Accepted: July 23, 2025
Published online: September 19, 2025
Processing time: 140 Days and 10.3 Hours
A recent meta-analysis has suggested a 5-HTR1A promoter variant may predict antidepressant response. The present review comments on the claims made in view of sensitivity issues and issues pertaining to genetic exposure. We also alert to errors in the original data that had been carried over. Specifically, primers meant to amplify the HTR1A gene aligned to the BDNF gene sequence. Alleles had been confounded owing to DNA strand ambiguities and demographic information proved inaccurate. In the light of these findings, adherence to PRISMA guidelines and use of the Newcastle-Ottawa Scale did not safeguard against bias. More after action reviews are encouraged to identify factors likely to interfere with estimates of genetic risk in large data sets. These may result from pooling of ethnic groups, the use of binary data or other formats that are not human-readable, the introduction of surrogate identifiers and a failure to reverse-engineer previously published experimental protocols. Unless the above challenges are met, sequence variants are unlikely to inform personalized medicine strategies in psychiatry.
Core Tip: A recent meta-analysis has suggested a 5-HTR1A promoter variant may predict antidepressant response. However, re-analysis of the original data unveiled numerous shortcomings pertaining to genetic exposure and interpretation of genetic findings. We alert to ambiguities that are likely to have introduced bias in the calculation of overall effects by muddling of alleles or entire genes, and by demographic inaccuracies. In part, these may be explained by specific features of genetic information that are not encoded in the DNA sequence, such as transcriptional orientation.
- Citation: Sand PG, Poeppl TB, Roessler V. Unmet needs in precision psychiatry. World J Psychiatry 2025; 15(9): 108222
- URL: https://www.wjgnet.com/2220-3206/full/v15/i9/108222.htm
- DOI: https://dx.doi.org/10.5498/wjp.v15.i9.108222
We read with interest the recent article by Wu et al[1] on the role of the 5-HTR1A gene in predicting the response to antidepressants. From a voluminous literature, and amidst a snowstorm of conflicting data, the authors have sought to distill the overall effects and thereby enhance the robustness of pharmacogenetic predictions. Upon second glance, however, the conclusions warrant further scrutiny. Reference is made to PRISMA and the Newcastle-Ottawa Scale (NOS), but the actual genotype counts obtained from pooling primary research are not listed. The data shown indicate discrepancies for: (1) The number of cases and controls in 5/11 studies[2-6]; (2) Male and female gender in 4/11 studies[2-4,6]; (3) Diagnostic criteria in 4/11 studies[2,4,7,8]; and (4) Demographic information in 3/11 studies[6,9,10].
From a clinical point of view, an approach that indiscriminately pools all currently available antidepressants and multiple definitions of response to these agents, is already at odds with precision medicine. Spurious variability is further enhanced by multicentre recruitment[5], a 6-fold difference in duration of treatment[3,11], ethnic admixture[7] and naturalistic settings[11]. The inclusion/exclusion of lithium and other active co-medications in a subset of populations is also liable to have impacted on effect size. Numerous eligible investigations were excluded[12-23] reducing statistical power and lowering sensitivity of the meta-analysis (Nexaminedvs Ntotal = 1436/3359).
From a basic science point of view, roughly 15% of human single nucleotide polymorphisms, including C-1019G, are population-specific, and require separate analyses for different ethnic groups. The study by Wu et al[1] did not conduct a separate analysis despite a bimodal allelic distribution in the total sample (781 Asian participants of whom 25% carry the C allele vs 635 Caucasian participants of whom 50% carry the C allele). Moreover, at least one study never even addressed the 5-HTR1A promoter variant of interest[2]. Instead of rs6295 on chromosome 5, the authors appear to have assessed an alternative polymorphism, rs6265 in the BDNF gene on chromosome 11. Wu et al[11] are invited to consult the methods section for details and may consider reranking this publication, rated 9/9 on the NOS.
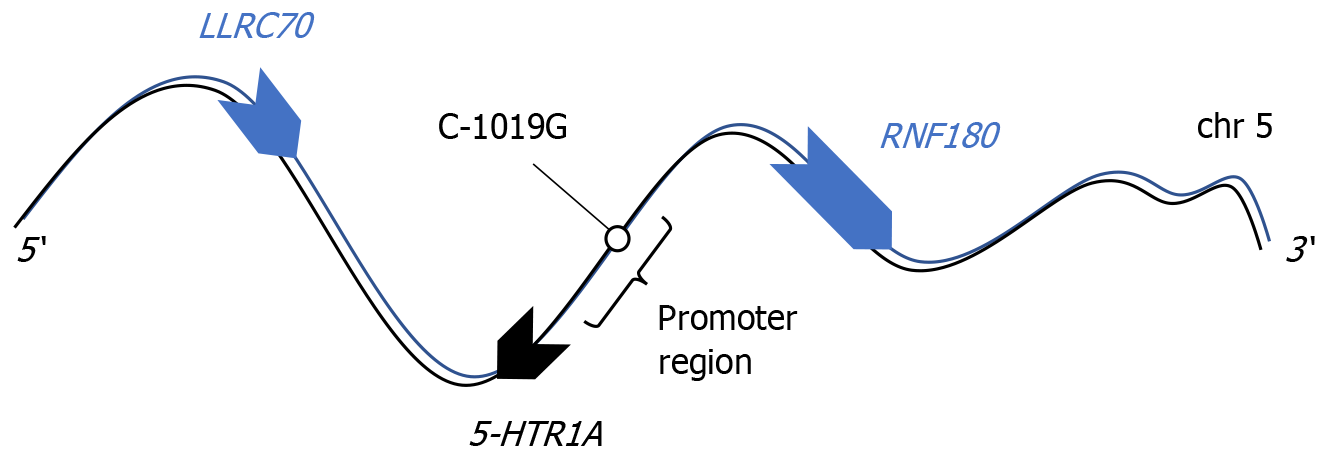
Which brings us to measures of bias. Unlike ratings of standard clinical trials, the assessment of genetic association analyses calls for a thorough understanding of potential pitfalls specific to DNA code, to the selection, arrangement and operation of alleles, the modes of inheritance etc. To meet some of these additional requirements, an adaptation of the NOS to genetic studies has been proposed[24]. Still, many key issues remain unresolved, e.g., a minimum requirement is to ensure the allele frequencies obtained match those of reference populations. When we apply this criterion to studies by Chang et al[3] or by Kato et al[11], that both served for the above meta-analysis, we note major inconsistencies. In sharp contrast to the remaining studies[10], and to other Asian reference populations in public databases, the reported C allele frequency is in the range of 0.75, rather than 0.25. Allele frequencies that fail to align with expected values strongly suggest the presence of bias. It is worth noting that multiple alerts have been published following incorrect readout of 5-HTR1A alleles[25-27]. These artefacts persist in 3/10 of current 5-HTR1A genotyping protocols[3,6,11] and have been missed (Figure 1).
In summary, the present meta-analysis illustrates the need to ascertain exposure, to narrow the phenotype, and to take into account additional confounders. We propose that the cumulative effects of C-1019G on response to antidepressants be reassessed. Subgroup analyses by ethnicity are advocated, as are standardized antidepressant response criteria across studies and formal power calculations to assess sample sufficiency.
| 1. | Wu HN, Zhu SY, Zhang LN, Shen BH, Xu LL. Association between 5-HTR1A gene C-1019G polymorphism and antidepressant response in patients with major depressive disorder: A meta-analysis. World J Psychiatry. 2024;14:1573-1582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 2. | Ramesh V, Venkatesan V, Ramasamy B. Role of serotonin transporter and receptor gene polymorphisms in treatment response to selective serotonin reuptake inhibitors in major depressive disorder. Hum Psychopharmacol. 2022;37:e2830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Chang HS, Lee HY, Cha JH, Won ES, Ham BJ, Kim B, Lee MS. Interaction of 5-HTT and HTR1A gene polymorphisms in treatment responses to mirtazapine in patients with major depressive disorder. J Clin Psychopharmacol. 2014;34:446-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Serretti A, Fabbri C, Pellegrini S, Porcelli S, Politi P, Bellino S, Menchetti M, Mariotti V, Demi C, Martinelli V, Cappucciati M, Bozzatello P, Brignolo E, Brambilla P, Pae CU, Balestrieri M, De Ronchi D. No effect of serotoninergic gene variants on response to interpersonal counseling and antidepressants in major depression. Psychiatry Investig. 2013;10:180-189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 5. | Noro M, Antonijevic I, Forray C, Kasper S, Kocabas NA, Lecrubier Y, Linotte S, Mendlewicz J, Montgomery S, Snyder L, Souery D, Verbanck P, Zohar J, Massat I. 5HT1A and 5HT2A receptor genes in treatment response phenotypes in major depressive disorder. Int Clin Psychopharmacol. 2010;25:228-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Serretti A, Artioli P, Lorenzi C, Pirovano A, Tubazio V, Zanardi R. The C(-1019)G polymorphism of the 5-HT1A gene promoter and antidepressant response in mood disorders: preliminary findings. Int J Neuropsychopharmacol. 2004;7:453-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 88] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 7. | Scutt G, Overall A, Scott R, Patel B, Hachoumi L, Yeoman M, Wright J. Does the 5-HT(1A) rs6295 polymorphism influence the safety and efficacy of citalopram therapy in the oldest old? Ther Adv Drug Saf. 2018;9:355-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Basu A, Chadda RK, Sood M, Kaur H, Kukreti R. Association of serotonin transporter (SLC6A4) and receptor (5HTR1A, 5HTR2A) polymorphisms with response to treatment with escitalopram in patients with major depressive disorder: A preliminary study. Indian J Med Res. 2015;142:40-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Illi A, Setälä-Soikkeli E, Viikki M, Poutanen O, Huhtala H, Mononen N, Lehtimäki T, Leinonen E, Kampman O. 5-HTR1A, 5-HTR2A, 5-HTR6, TPH1 and TPH2 polymorphisms and major depression. Neuroreport. 2009;20:1125-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Lin E, Chen PS, Chang HH, Gean PW, Tsai HC, Yang YK, Lu RB. Interaction of serotonin-related genes affects short-term antidepressant response in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:1167-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Kato M, Fukuda T, Wakeno M, Okugawa G, Takekita Y, Watanabe S, Yamashita M, Hosoi Y, Azuma J, Kinoshita T, Serretti A. Effect of 5-HT1A gene polymorphisms on antidepressant response in major depressive disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Lemonde S, Du L, Bakish D, Hrdina P, Albert PR. Association of the C(-1019)G 5-HT1A functional promoter polymorphism with antidepressant response. Int J Neuropsychopharmacol. 2004;7:501-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Arias B, Catalán R, Gastó C, Gutiérrez B, Fañanás L. Evidence for a combined genetic effect of the 5-HT(1A) receptor and serotonin transporter genes in the clinical outcome of major depressive patients treated with citalopram. J Psychopharmacol. 2005;19:166-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 72] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Parsey RV, Olvet DM, Oquendo MA, Huang YY, Ogden RT, Mann JJ. Higher 5-HT1A receptor binding potential during a major depressive episode predicts poor treatment response: preliminary data from a naturalistic study. Neuropsychopharmacology. 2006;31:1745-1749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 15. | Yu YW, Tsai SJ, Liou YJ, Hong CJ, Chen TJ. Association study of two serotonin 1A receptor gene polymorphisms and fluoxetine treatment response in Chinese major depressive disorders. Eur Neuropsychopharmacol. 2006;16:498-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Levin GM, Bowles TM, Ehret MJ, Langaee T, Tan JY, Johnson JA, Millard WJ. Assessment of human serotonin 1A receptor polymorphisms and SSRI responsiveness. Mol Diagn Ther. 2007;11:155-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 17. | Baune BT, Hohoff C, Roehrs T, Deckert J, Arolt V, Domschke K. Serotonin receptor 1A-1019C/G variant: impact on antidepressant pharmacoresponse in melancholic depression? Neurosci Lett. 2008;436:111-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Villafuerte SM, Vallabhaneni K, Sliwerska E, McMahon FJ, Young EA, Burmeister M. SSRI response in depression may be influenced by SNPs in HTR1B and HTR1A. Psychiatr Genet. 2009;19:281-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Zhu YZ, Zhang Y, Ma H, Xie SF, Jiang WY, Sun GW, Liu Y. [Relationship between HTR1A Gene-1019C/G Polymorphism and Clinical Response of Fluoxetine in the Treatment of Major Depressive Disorder]. Zhongguo Yike Daxue Xuebao. 2010;6:467-473. |
| 20. | Badamasi IM, Lye MS, Ibrahim N, Abdul Razaq NA, Ling KH, Stanslas J. Serotonergic receptor gene polymorphism and response to selective serotonin reuptake inhibitors in ethnic Malay patients with first episode of major depressive disorder. Pharmacogenomics J. 2021;21:498-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Dong WT, Ji YH. [Relationship between 5-HTR1A, 5-HTR2A gene polymorphism and antidepressant efficacy of SSRIs]. Zhongguo Yishi Zazhi. 2021;23:1857-1859. [DOI] [Full Text] |
| 22. | Si RGL, Liu XC, Yang F, Yang H. [Study on the Association of 5-HTR1A/2A Receptor Gene Polymorphism with Antidepressant Efficacy of SSRIs]. Zhongguo Yaofang. 2018;17:2394-2398. [DOI] [Full Text] |
| 23. | Xu Z, Chen Z, Shen T, Chen L, Tan T, Gao C, Chen B, Yuan Y, Zhang Z. The impact of HTR1A and HTR1B methylation combined with stress/genotype on early antidepressant efficacy. Psychiatry Clin Neurosci. 2022;76:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 24. | Norris JM, Simpson BS, Ball R, Freeman A, Kirkham A, Parry MA, Moore CM, Whitaker HC, Emberton M. A Modified Newcastle-Ottawa Scale for Assessment of Study Quality in Genetic Urological Research. Eur Urol. 2021;79:325-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 25. | Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry. 2010;15:473-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 329] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 26. | Sand PG. The serotonin 1A receptor gene in mood disorders: a tale of missed opportunities. Eur Arch Psychiatry Clin Neurosci. 2013;263:445-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 27. | Sand PG. A functional 5-HT1A variant and comorbid anxiety. Epilepsy Res. 2012;100:199-200; author reply 201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |