Published online Oct 27, 2022. doi: 10.5496/wjmg.v10.i2.7
Peer-review started: March 5, 2022
First decision: June 16, 2022
Revised: June 28, 2022
Accepted: October 12, 2022
Article in press: October 12, 2022
Published online: October 27, 2022
Processing time: 235 Days and 21.9 Hours
Epigenetic disruptions have been implicated in some cases of infertility and can serve as therapeutic targets. However, the involvement of epigenetics in infertility has not received adequate attention.
This study aimed to determine the epigenetic basis of infertility in order to enhance public knowledge.
Relevant articles on the subject were collected from PubMed, RCA, Google Scholar, SpringerLink, and Scopus. The articles were pooled together and duplicates were removed using Endnote software.
Available information shows that epigenetic mechanisms, mainly DNA methylation, histone modification, and microRNA interference are necessary for normal gametogenesis and embryogenesis. As a result, epigenetic disruptions in genes that control gametogenesis and embryogenesis, such as DDX3X, ADH4, AZF, PLAG1, D1RAS3, CYGB, MEST, JMJD1A, KCNQ1, IGF2, H19, and MTHFR may result in infertility. Aberrant DNA methylation during genomic imprinting and parental epigenetic mark erasures, in particular, may affect the DNA epigenomes of sperm and oocytes, resulting in reproductive abnormalities. Histone epigenetic dysregulation during oocyte development and histone-protamine replacement in the sperm may also cause reproductive abnormalities. Furthermore, overexpression or repression of certain microRNAs embedded in the ovary, testis, embryo, as well as granulosa cells and oocytes may impair reproduction. Male infertility is characterized by spermatogenesis failure, which includes oligozoospermia, asthenozoospermia, and teratozoospermia, while female infertility is characterized by polycystic ovary syndrome. Some epigenetic modifications can be reversed by deactivating the regulatory enzymes, implying that epigenetic reprogramming could help treat infertility in some cases. For some disorders, epigenetic drugs are available, but none have been formulated for infertility.
Some cases of infertility have an epigenetic etiology and can be treated by reversing the same epigenetic mechanism that caused it. As a result, medical practitioners are urged to come up with epigenetic treatments for infertility that have an epigenetic cause.
Core Tip: This article reviews the role of epigenetics in the etiology of infertility, which can be used as a therapeutic target. Some cases of infertility are due to epigenetic disruptions, and this is probably the cause of unknown etiology in some cases of infertility. However, there is little awareness on this subject, hindering its application in mainstream medicine.
- Citation: Yahaya TO, Bashar DM, Oladele EO, Umar J, Anyebe D, Izuafa A. Epigenetics in the etiology and management of infertility. World J Med Genet 2022; 10(2): 7-21
- URL: https://www.wjgnet.com/2220-3184/full/v10/i2/7.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v10.i2.7
Infertility is defined as a couple’s inability to conceive after a year of consistent copulation without the use of contraception[1]. Infertility is becoming more prevalent worldwide and is now a serious public health concern[2,3]. At the very least, roughly 15% of couples are infertile[4], with males accounting for 40%, females also account for 40%; and both jointly contributing to the remaining 20%[5]. The most common feature of male infertility is spermatogenesis failure, which is responsible for half of all human infertility[6]. Spermatogenesis failure is characterized by an abnormal sperm count (oligozoospermia), weak sperm motility (asthenozoospermia), and abnormal sperm morphology (teratozoospermia)[6,7]. The most common features of female infertility are amenorrhea and irregular menstruation[8].
Infertility is often devastating and affects all aspects of life, including physical, mental, and social health[9,10]. Infertility causes enormous psychological problems, poor sexual satisfaction, and a low quality of life[10]. Women are often more affected by the effects of infertility than men, as they are deprived of financial support and basic needs by their husbands, families, and communities[11]. In cultures that prioritize child-bearing, childless couples are stigmatized and mocked[1]. In some cases, childlessness causes infidelity, polygamy, and divorce or separation. Infertility treatment can also be expensive, especially in developing countries like Nigeria where people with this problem often have to pay for their own medical care[3].
The pathophysiology of infertility is complex. It may be caused by specific or multiple physical and physiological factors, including hormonal and homeostatic disruptions, environmental and genetic alterations[3]. Recently, epigenetic alterations have been implicated in some cases of infertility[3]. "Epigenetics" refers to biological processes that regulate gene expression without altering the genetic material[12]. The most common epigenetic mechanisms are DNA methylation, histone modification, and microRNA (miRNA) interference[12]. Biological processes, including gametogenesis and embryogenesis, require epigenetic modifications[13]. However, epigenetic modifications, apart from normal cellular functions or responses to external factors, can cause heritable epigenetic mutations and thus, diseases, including infertility[7,12]. By inhibiting the enzymes that modulate epigenetic mechanisms, epigenetic changes and normal functions of the affected genes can be restored[12]. This suggests that epigenetic reprogramming can be used to treat infertility with an epigenetic origin in some cases. This study, therefore, provides an update on the role of epigenetics in the etiology and management of infertility.
Reputable academic repositories, namely PubMed, Google Scholar, RCA, SpringerLink, and Scopus, were searched separately for peer-reviewed articles on the subject. The keywords used for the search were: "epigenetics," "infertility," "male infertility," "female infertility," "DNA methylation," "histone modifications," "microRNAs," "epigenetic tests for infertility," and "epigenetic drugs for infertility." Other keywords used include "epigenetic mechanisms," "role of DNA methylation in infertility," "role of histone modification in infertility," and "role of microRNAs in infertility." The articles retrieved were sorted using EndNote software, and double citations were removed.
Included articles were those that were available in the English language, those that focused on the epigenetic basis of infertility and management, and those that were published between the years 2000 and 2021, this was to obtain up-to-date information.
Excluded articles were those that were not available in the English language, articles written before the year 2000, and articles for which only abstracts were available.
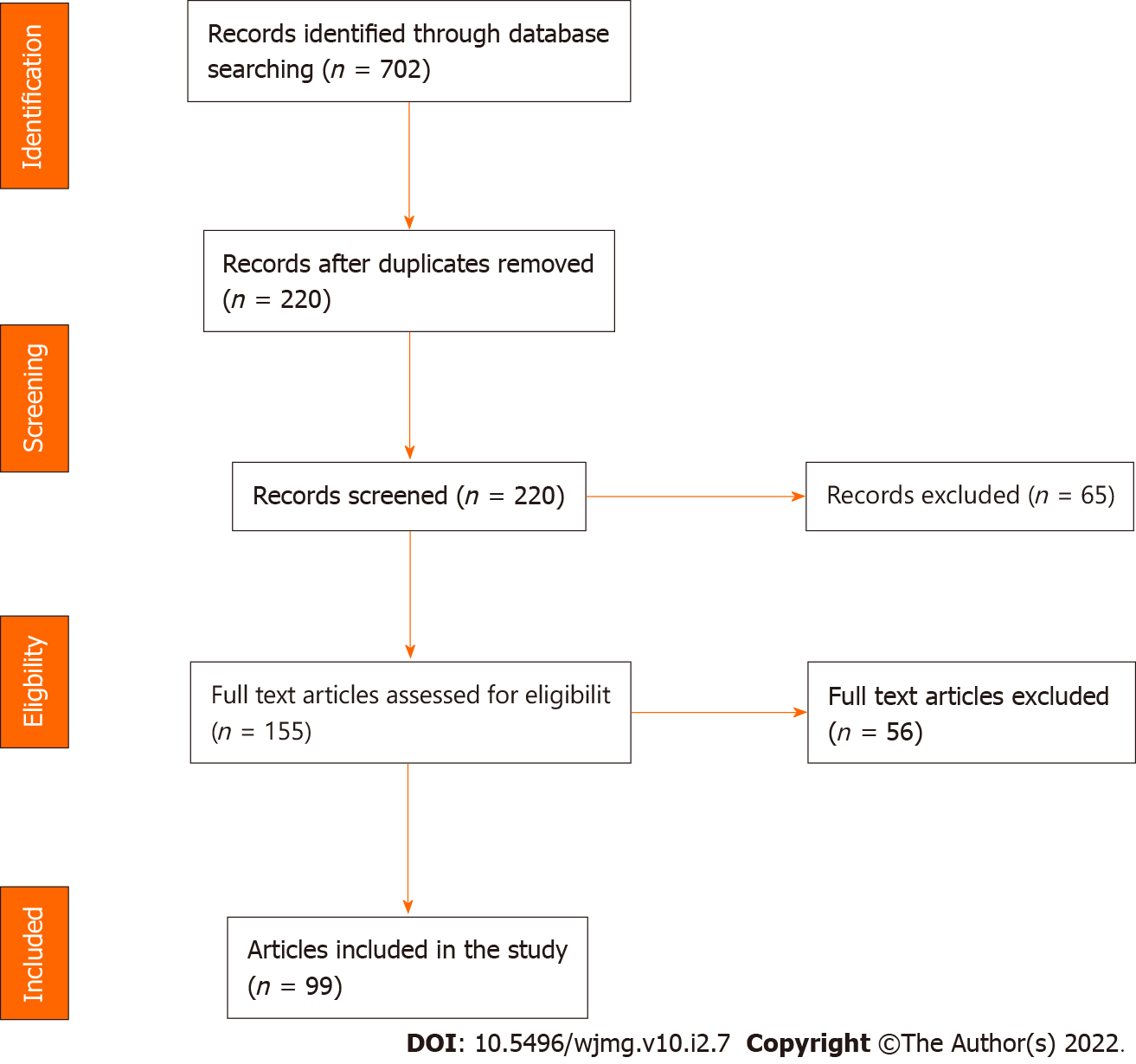
In all, 702 articles were retrieved from the databases searched (Figure 1), but 220 articles were retained after removing duplicates. The retained articles were subjected to the eligibility test, and 155 passed. Of the 155 eligible articles, 99 fitted the study objectives and thus made the final selection.
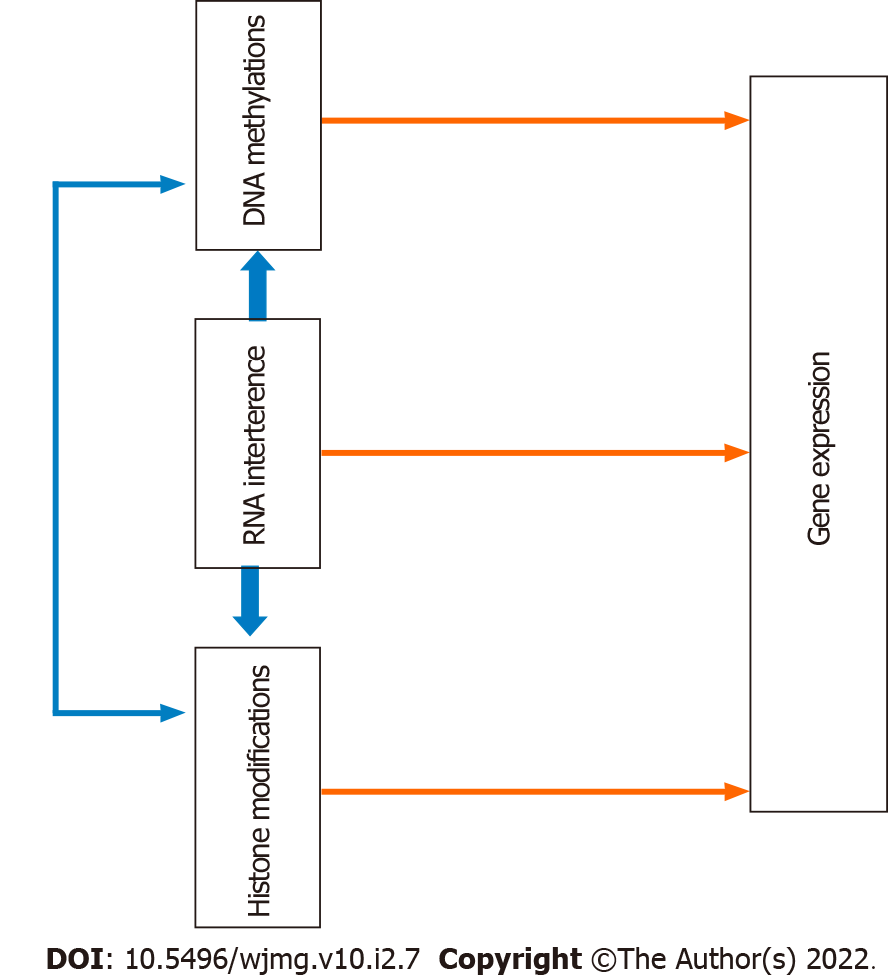
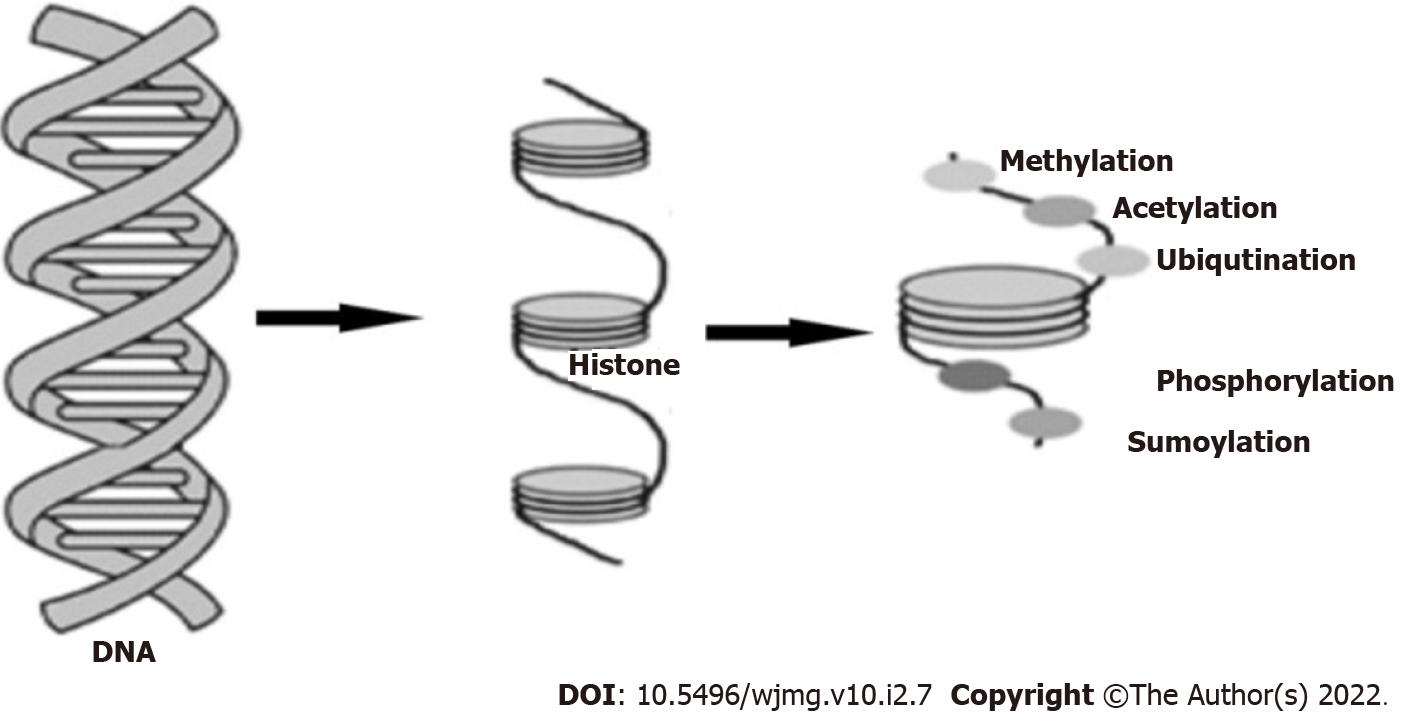
The word "epigenetics" was previously employed to describe the relationship between the genome and the environment that takes part in the development of mammals and some other organisms[14]. However, it is currently defined as heritable alterations in DNA accessibility and chromatin structure, affecting gene expression without changing the DNA sequence[14,15]. Epigenetics plays an important role in normal development, cell differentiation, and disease pathologies[14,15]. There are several epigenetic mechanisms. However, the most common epigenetic mechanisms are DNA methylation, histone modifications, and microRNA (miRNA) interference[12,16]. These mechanisms may alter gene expressions individually or interact to control gene expressions[12]. Figure 2 depicts interactions among epigenetic mechanisms, and Table 1 summarizes the mechanistic links between epigenetic disruptions and infertility.
| Epigenetic mechanisms | Links (Pathophysiology) | Ref. |
| DNA methylation | Hypermethylation or hypomethylation disrupts genomic imprinting and parental epigenetic mark erasure, resulting in abnormal expression of some genes and imprinted genes involved in gametogenesis and embryogenesis | [3,4,12,13,17,20,25,29-42,44-49] |
| Histone post-translational modification | Abnormal histone modification alters the expression of certain genes important in gametogenesis and embryogenesis. Also, it disrupts sperm DNA protamination, causing sperm abnormalities | [7,43,56-63] |
| miRNA | Up-regulation or down-regulation alters the expression of certain genes important in gametogenesis and embryogenesis | [15,68-75] |
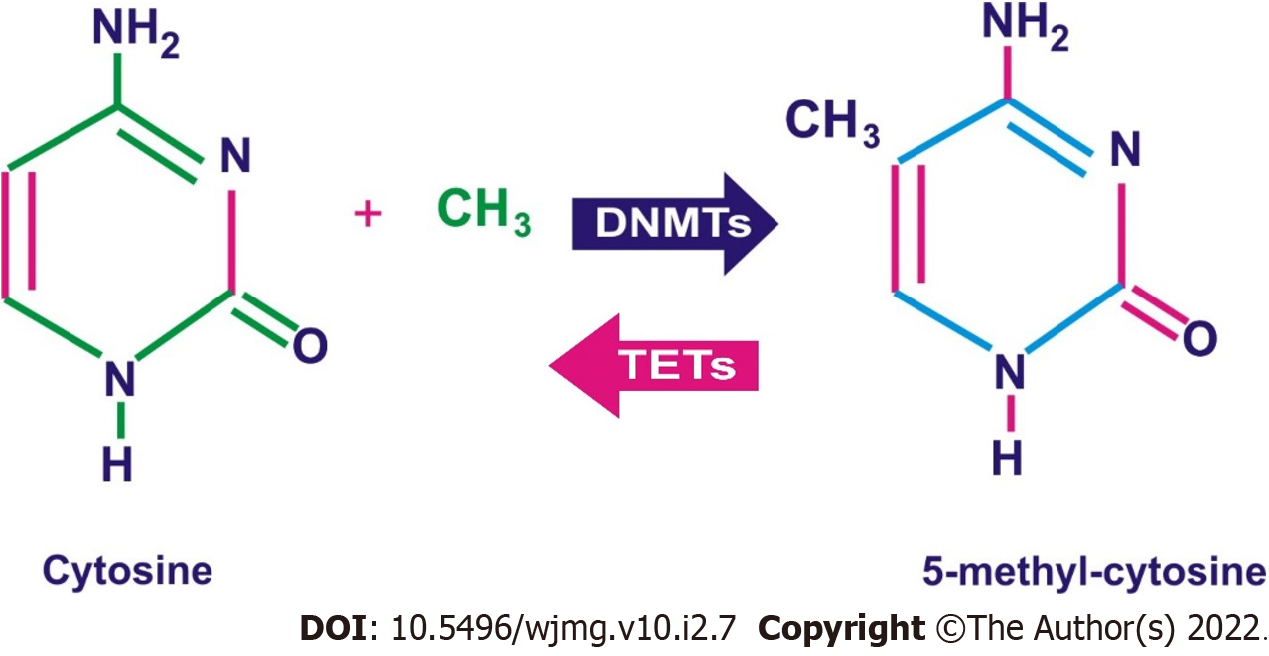
DNA methylation is the most researched epigenetic mechanism and often results in gene silencing[17,18]. DNA methylation involves the binding of a methyl group to the DNA, resulting in a change of the expression and functions of the embedded genes (Figure 3).
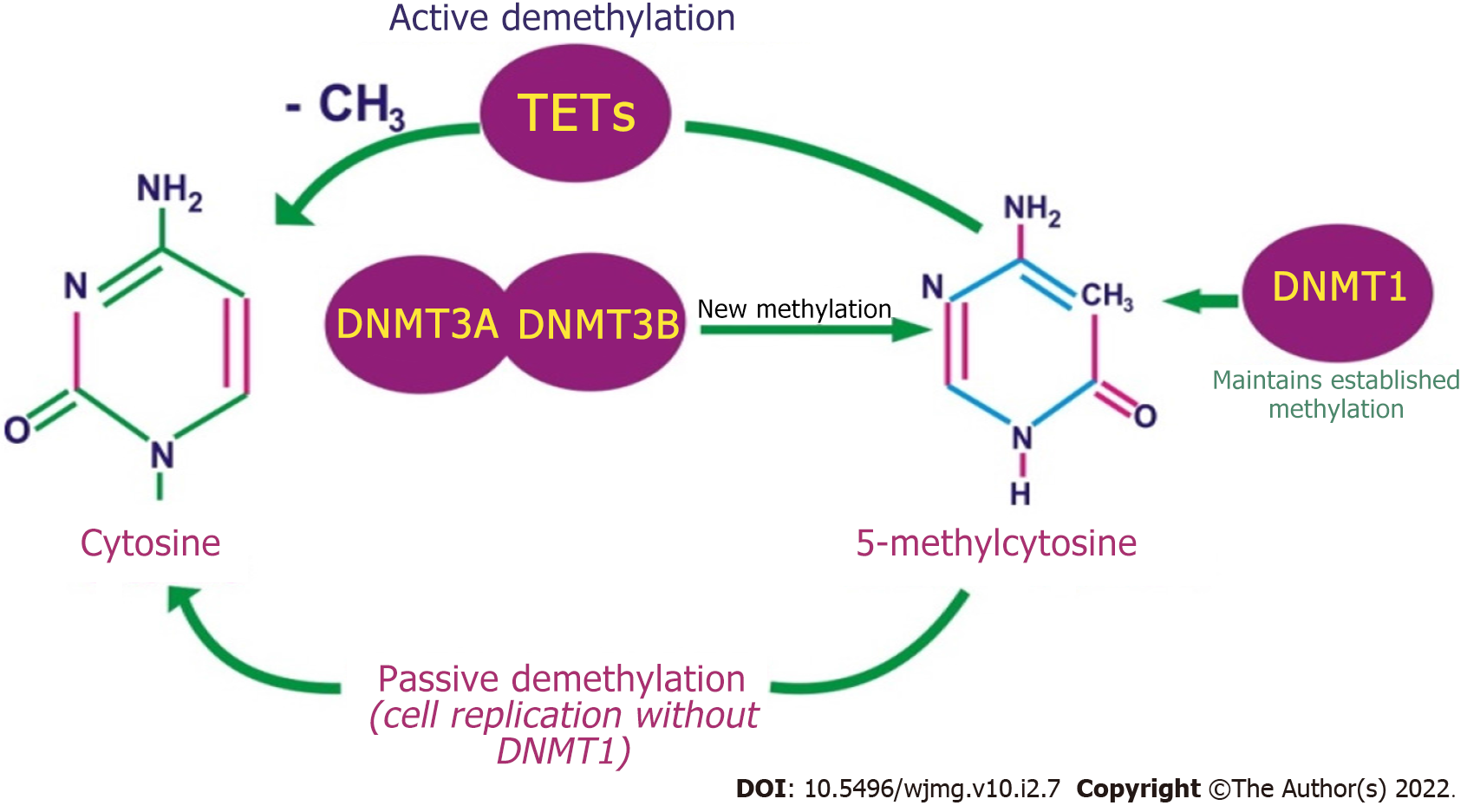
In somatic cells, the binding occurs mainly close to the CpG sites, while in gamete cells occurs near the non-CpG sites[15,19]. CpG sites are DNA sections where a cytosine nucleotide is adjacent to a guanine nucleotide. During DNA methylation, S-adenosyl-L-methionine releases a methyl group and binds to the 5-carbon of the cytosine ring, resulting in 5-methylcytosine (5-mC)[15,20]. The methyl group is then thrusted into the DNA and alters gene transcription. DNA methylation is mediated by a family of enzymes known as the DNA methyltransferases (DNMTs), and members of these enzymes include: DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L[15,21]. DNMT1 regulates established methylated DNA, while DNMT3a and DNMT3b regulate new DNA methylation processes (Figure 4). However, in diseased cells, DNMT1, DNMT3a, and DNMT3b combine to cause DNA over-methylation. Furthermore, during epigenetic reprogramming, DNMT1 prevents the methylation of new DNA, while a group of enzymes called the ten-eleven translocation (TET) modulates the de-methylation of already methylated DNA. DNMT2 inhibits the mutation of small RNA molecules[22]. DNMT3L is similar to DNMT3A and 3B, but does not catalyze epigenetic changes[23]. Instead, DNMT3L enhances the functions of DNMT3A and B[23]. DNMT3L also identifies un-methylated histone H3-lysine 4 (H3K4) nucleosomes and stimulates cells to produce more DNMT3A and DNMT3B to methylate them[24].
DNA methylation is important in reproduction, particularly during genomic imprinting[12,17]. Genomic imprinting is an epigenetic phenomenon in which only one parental allele is expressed while the other allele is imprinted or silenced[25]. Thus, genomic imprinting maintains the parent-of-origin expression of genes. However, some genes may not be fully imprinted; instead of one allele being completely expressed and the other repressed, the two alleles show varied expressions[26]. As of 2019, 228 imprinted genes have been reported in the human genome[27]. Normal imprinting of some genes is necessary for healthy development as it protects the genome’s integrity[25,28]. Abnormal imprinting, often caused by alterations in DNA methylation, is associated with many diseases, including impaired spermatogenesis and infertility[3,29]. In a study that analyzed the DNA methylation patterns of seven differently methylated regions (DMRs), in the sperm of 97 infertile men, 14 showed abnormal paternal DNA methylation at H19 and GTL2, and 20 had abnormal maternal DNA methylation at PEG1, LIT1, ZAC, PEG3, and SNRPN[30]. These DMRs contain imprinted genes that regulate spermatogenesis, and at least half of the genes show maternal and paternal imprint abnormalities in infertile men[30]. In another study, methylation and imprinting errors were observed in the IGF2/H19 imprinting control region 1 (ICR1) and MEST DMRs in the spermatozoa of 148 idiopathic infertile men compared with 33 normozoospermic controls[31]. The idiopathic infertile men (sperm motility below 40% and normal sperm morphology below 5%) displayed hypermethylation of the MEST DMRs and hypomethylation of the IGF2/H19 ICR1, while the control showed the opposite. Thus, in the study, infertility was clearly linked with IGF2/H19 ICR1 hypomethylation and MEST hypermethylation[32,33]. In another study, seven out of 15 (46.7%) individuals with low sperm count (below 10 × 106/mL) showed defective methylation of H19 and/or MEST imprinted genes[34]. Of the seven patients that expressed imprinting errors, two had both H19 hypomethylation and MEST hypermethylation, while five had only one of the impaired imprinted genes[34]. This again proved that imprinted genes in H19 and MEST play an important role in spermatogenesis, fetal growth and development, and placental function[35,36]. Similarly, in a study that compared the DNA methylation at DMRs of maternally imprinted genes extracted from stillborn pups and control embryos, hypermethylations were observed at Zac1 imprinting genes in the stillborn pups[37]. Zac1 regulates an imprinted gene network that is important in the regulation of embryonic growth[38]. Aberrant DNA methylation has also been implicated in some genomic imprinting disorders, which, in severe cases, can cause recurrent molar pregnancy, mis-carriage, or infertility[39]. These disorders include Prader-Willi syndrome and Angelman syndrome, which are caused by loss of function of imprinted genes on chromosome 15 in females and males, respectively[40]. Beckwith-Wiedemann syndrome and Russell-Silver syndrome are two others. Both are caused by the loss of function of imprinted genes on chromosomes 7 or 11[41,42].
Aside from genomic imprinting, DNA methylation is also involved in parental epigenetic mark erasures in which DNA methylation undertakes two rounds of epigenetic reprogramming during gametogenesis and embryogenesis[13]. One reprogramming occurs immediately after fertilization, in which sperm and oocyte DNA are stripped of the parental methylation marks (DNA demethylation)[4]. Some DNA demethylation occurs specifically in paternally inherited imprinted genes[43]. The erasure of DNA methylation continues until new imprints are formed[43]. The stripping allows the totipotent zygote to start new gene transcription and new cell methylation[4]. Because of this, most epigenetic modifications that occur in sperm and egg cells when the two merge to form a fertilized egg are removed[3]. Thus, epigenetic reprogramming enables the fetus’s cells to start afresh and determine their own epigenome[3]. However, some of the epigenetic modifications in parents’ sperm and egg cells may escape the reprogramming and be transmitted to the next generation[3]. Another genome-wide stripping of DNA methylation and subsequent new DNA methylation occurs in the primordial germ cells (gamete precursors), which subsequently differentiate into the gametes (sperm and eggs)[17,44]. Overall, this showed the importance of DNA methylation in gametogenesis and embryogenesis and, hence, fertility. In fact, DNMT1, DNMT3a, and DNMT3b have been shown to be highly expressed in the early embryonic stage[20]. Furthermore, it has been shown that more than 150 genes are associated with mammalian spermatogenesis, and if the normal expression of any of these genes is altered, the reproductive success of males could be compromised[25]. Thus, aberrant DNA methylation may cause dysfunctional gametogenesis and embryogenesis, resulting in infertility[4,25]. In a study, 696 differentially methylated CpGs, comprising 184 (26%) hypomethylations and 512 (74%) hypermethylations associated with 501 genes, were identified between the spermatozoa of 19 fertile men and 42 infertile men[45]. The CpGs are home to 13 processes related to spermatogenesis. Moreover, 17 differentially methylated genes related to spermatogenesis were observed between the fertile and infertile groups[45]. In another study that compared 46 sperm samples obtained from 17 normospermic fertile men and 29 normospermic infertile men, 2752 CpGs showing aberrant DNA methylation patterns were observed in the sperm of infertile men[46]. Importantly, these differentially methylated CpGs were significantly associated with CpG sites that are involved in spermatogenesis[46]. Additionally, 48 imprinted genes were abnormally methylated in the altered CpGs of the infertile patients. In a related study that compared the sperm of 12 fertile and 45 infertile men, reactive oxygen species were found to cause DNA fragmentation and abnormal methylation in the infertile group’s sperm[47]. Similar to infertile men, abnormal DNA methylation has also been reported in the germ cells or reproductive tract of infertile women. For instance, in a genome-wide methylation study of the endometrium of women expressing endometriosis, compared with a matched control, 59 genes were hypermethylated and 61 genes were hypomethylated[48]. It was observed in the study that aberrant methylation and expression of these genes contributed to abnormal endometrial cell proliferation and function in women[48]. In another genome-wide study involving 85 women expressing polycystic ovary syndrome, the CpG sites of luteinizing hormone/choriogonadotropin receptor promoter regions were hypomethylated compared with the control[49]. The hypomethylation of the luteinizing hormone/choriogonadotropin receptor caused its overexpression in women with polycystic ovary syndrome compared with that in control women.
Histones are the ‘cylindrical’ protein building-blocks of chromatin around which DNA winds and shortens the DNA[50]. Thus, post-translational modifications of histones restructure the chromatin (condensed or non-condensed), which determines the transcriptional status of the associated DNA and genes[15,51]. Non-condensed or loose chromatin (euchromatin) is active and transcribes DNA, while condensed chromatin (heterochromatin) is inactive and thus lacks the ability to transcribe genes[15,52]. The genes in the condensed chromatin are tightly bonded to the DNA and are thus silenced due to the inability of the transcription factors to gain access to the promoters of the genes[15,52]. There are five main classes of histones, which are: H1/H5, H2A, H2B, H3, and H4[53,54]. The core histones are histones H2A, H2B, H3, and H4, while the linker histone is histone H1/H5[53]. Histones can be modified through several mechanisms, such as methylation, acetylation, phosphorylation, sumoylation, and ubiquitylation (Figure 5). However, methylation and acetylation are the most common mechanisms[15,55]. Acetylation binds an acetyl group to the amino acid lysine in the histone, while methylation binds a methyl group to the amino acids of histone proteins, primarily lysine and arginine residues[15,55]. As lysine and arginine are the most abundant amino acids in histones, they are frequently acetylated and methylated[50]. Acetylation ideally takes place in non-condensed chromatin, while deacetylation usually takes place in condensed chromatin[15]. Histone methylation can take place in both forms of chromatin. Histone acetyltransferases and histone methyltransferases catalyze histone acetylation and methylation, respectively[12], whereas histone deacetylases (HDACs) and histone demethylases catalyze deacetylation and demethylation[12]. Aside from the structural state of chromatin mentioned earlier, the effects of histone post-translational modifications on gene expression also depend on the mechanisms and degree of methylation or acetylation, which could be mono-, di-, or tri-methylated[12]. Sometimes, both DNA methylation and histone post-translational modification combine to cause epigenetic changes[12]. DNA methylation plays a part in ensuring high levels of chromatin structure[45].
Histone modifications play an important role in gametogenesis and embryogenesis, as well as in fertility[56]. To successfully transfer a sperm’s genetic and epigenetic materials to an egg, the chromatin must be very condensed for proper motility and protection of the paternal DNA and epigenome from external stimuli[56,57]. This is guaranteed by the replacement of histones with protamines (a unique, sperm-specific protein) by sperm DNA protamination[56,57]. Despite this, some regions, particularly the sperm head, retain histones, which are prone to modifications[56,57]. Protamination and some other biological events during spermatogenesis are controlled by epigenetic mechanisms in which abnormal histone modifications may cause the sperm to lose its oocyte fertilizing capacity[7]. Defects in either the replacement or the modification of histones might cause male infertility, characterized by azoospermia, oligozoospermia, or teratozoospermia[58]. In females, during oocyte development, histone methylation and acetylation increase significantly, resulting in the global restructuring of chromatin and the silencing of many embedded genes[43]. This global change is mediated by the increased production of methyltransferases and acetyltransferases[43]. Thus, histone epigenetic dysregulation can disrupt oogenesis, leading to aneuploidy in fertilized oocytes, culminating in embryonic death[7]. In a study that compared the sperm transcriptomes of 3 oligozoospermic infertile men with 8 fertile men, the former showed a 17-fold down-regulation in genes involved in histone modifications[59]. In the study, 157 transcripts were either overexpressed or repressed in the sperm of oligozoospermic infertile men as compared to normozoospermic fertile individuals[59]. Importantly, the histone dysregulation in infertile men caused up to a 43-fold reduction in the expression of some genes involved in spermatogenesis and sperm motility, such as DDX3X and JMJD1A[59]. Furthermore, a 17-fold increase was observed in the expression of some genes that prevent oxidative stress and abortive spermatogenesis[59]. These genes include: ADH4, HSD17B7, CYGB, and NXNL1[59]. It is noteworthy that at the start of the mentioned study, the patients were screened and confirmed negative for known causes of infertility, including chromosome anomalies and Y chromosome AZF deletions[59]. This suggests that the observed epigenetic changes were responsible for the reproductive abnormalities in the infertile men. In a transgenic mouse study, overexpression of KDM1A (a histone demethylase) during spermatogenesis reduces histone H3 Lysine 4 dimethylation (H3K4me2) in sperm at more than 2300 genes[60]. Some of these genes regulate development, and the reduction of H3K4 dimethylation in the mice sperm severely impaired the fertility, development, and survivability of the offspring[60]. The defects were observed across multiple generations in the absence of KDM1A germline expression and were linked to altered RNA profiles in sperm and offspring[60]. In a study that determined the cause of idiopathic early miscarriage in 3 pregnant women, 81 genes were overexpressed in the chorionic villous of the affected compared with controls[61]. These genes take part in several important physiological processes, such as cell proliferation, nuclear division, chromatic assembly, DNA packing, and modification[61]. Furthermore, 231 genes that are functionally involved in histone modifications and cell cycle control were down-regulated in the chorionic villous of the affected women compared with controls[61]. In a study of histone locations and modifications, in the semen of seven infertile patients, unlike fertile men, five infertile men had non-programmatic (randomly distributed) histone retention genome-wide[62]. Although the methylation patterns of H3K4me and H3K27me in infertile men were similar to those in the control group, the amounts of histones retained by developmental transcription factors and certain imprinted genes were decreased[62]. In a study that monitored the effects of chlordecone exposure on the epigenome of the ovaries of mice, compared with the control, reduced H3K4me3 and H4ac in fully grown oocytes were observed. This reduction caused repression of genes associated with estrogen signaling and oocyte maturation in adult ovaries[63]. Furthermore, gene expression analysis revealed that RCBTB2 and RBPMS genes were not expressed in the embryonic gonads[63]. Reproductive abnormalities observed in the exposed mice included compromised meiotic double-strand break repair in female embryos, puberty delay, decreased primordials, and increased atretic follicles[63]. The study showed that exposure to a low dose of chlordecone during pregnancy impaired female reproductive functions, which are mediated by abnormal histone modifications[63].
MicroRNAs (miRNAs) are small, single-stranded non-coding RNA molecules of between 19 and 25 nucleotides[15,64]. MicroRNAs interact with transcriptional and epigenetic regulators in cells to maintain lineage-specific gene expression[15,65]. Specifically, miRNAs control the expression of genes during transcription by disrupting the translation of target messenger RNA. However, in diseased cells, miRNA expression is changed, leading to altered expression, mostly overexpression of the target genes[15,66]. Approximately 1% of the human genome is made up of genes that contain miRNAs[67], which shows how important they are.
MicroRNAs play an active role in many cellular functions, including cell cycle control, cell differentiation, intra and intercellular communication (cell-to-cell communication), and apoptosis[15,68]. In mammalian reproduction, miRNAs are embedded in the tissues of the ovary, testis, and embryo, as well as granulosa cells and oocytes[68]. MicroRNAs are actively involved in mammalian sex differentiation, gametogenesis, fertilization, zygotic genome activation and early development, implantation, germ layer specification, and pregnancy[69,70]. These mentioned reproductive functions and others show that impairing miRNAs may result in reproductive anomalies such as infertility and pregnancy failure[15]. It has been demonstrated that the loss of one or both components of the miRNA processing machinery (Dicer and Drosha) severely impairs gametogenesis, resulting in male and female infertility[70]. In an experiment, deletion of Dicer1 at the early stage of male gamete cell development in six transgenic mice caused infertility compared with matched controls[71]. The infertility was caused by several cumulative defects at the meiotic and post-meiotic stages, culminating in the absence of functional spermatozoa[71]. Increased apoptosis in spermatocytes, fewer spermatids, and spermatozoa with abnormal morphology were also observed in the tested rats, unlike the controls[71]. Furthermore, the expression of transposable elements of the SINE family was overexpressed in the Dicer1-deficient spermatocytes[71]. In another study that examined the expression of 736 miRNAs in the spermatozoa of 10 fertile men, 221 miRNAs were frequently present in all the participants[72]. Additionally, 452 miRNAs were present in some participants, and 63 were absent in all the participants[72]. Further analysis showed that these miRNAs take part in processes related to cell differentiation, development, morphogenesis, and embryogenesis[72]. This shows that human sperm contains many miRNAs, which functionally promote embryogenesis and spermatogenesis[72]. In a study of human spermatozoa from 27 patients with various spermatogenic abnormalities, 50 miRNAs were up-regulated and 27 miRNAs were down-regulated in asthenozoospermic males compared with controls (Table 2). In the oligoasthenozoospermic participants, 42 miRNAs were up-regulated and 44 miRNAs were down-regulated when compared with normozoospermic males[73]. The most overexpressed miRNAs in asthenozoospermic men were miR-34b, miR-122, and miR-1973, whereas in oligoasthenozoospermic men were miR-34b, miR-34b*, miR-15b, miR-34c-5p, miR-122, miR-449a, miR-1973, miR-16, and miR-19a[73]. These miRNAs play an essential role in male germ cell development and spermatogenesis, and, hence, their imbalances may cause male infertility[73]. The regulatory role of miRNAs in oogenesis has also been demonstrated in several studies. In a female mouse study, the removal of the miR-17-92 cluster in the ovaries caused overexpression of several genes involved in apoptotic pathways compared with controls[74]. These genes include pro-apoptotic BH3-only genes (Noxa, Bmf, Bid, Bik, Bad, and Bim) and the pro-apoptotic effector protein genes (Bax and Bak)[74]. Other genes are initiator caspases (Caspase 8 and Caspase 9), executioner caspase (Caspase 3), and some follicular atresia-related genes (Cyp1a1 and Egr-1)[74]. This suggests that apoptosis is the major mechanism involved in the reproductive anomalies observed in the miR-17-92 deficient mice[74]. The reproductive anomalies caused by these epigenetic alterations include increased oocyte degradation and follicular atresia, decreased ovulation, perturbed oogenesis, and ultimately culminate in subfertility and reduced fecundity[74]. Overall, the study showed that the miR-17-92 cluster is an important regulator of oogenesis[74]. Similarly, in a study that compared the endometrium of patients with repeated implantation failure with controls, 13 differentially expressed miRNAs that regulate 3800 genes were identified in the affected patients[75]. Ten of the miRNAs were overexpressed (including miR 145, 23b, and 99a), and three were repressed (Table 2). These miRNAs target genes are involved in important implantation processes, such as adherens junctions, cell adhesion molecules, Wnt-signaling, p53 signaling, and cell cycle pathways[75].
| miRNAs | Status | Effect | Ref. |
| miR-34b | Up-regulated | Asthenozoospermia | [73] |
| miR-122 | Up-regulated | Asthenozoospermia | [73] |
| miR-1973 | Up-regulated | Asthenozoospermia | [73] |
| miR-15b | Up-regulated | Asthenozoospermia | [73] |
| miR-34c-5p | Up-regulated | Oligoasthenozoospermia | [73] |
| miR-449a | Up-regulated | Oligoasthenozoospermia | [73] |
| miR-16 | Up-regulated | Oligoasthenozoospermia | [73] |
| miR-19a | Up-regulated | Oligoasthenozoospermia | [73] |
| miR-17-92 | Deficient | Abnormal oogenesis | [74] |
| miR-145 | Up-regulated | Implantation failure | [75] |
| miR-23b | Up-regulated | Implantation failure | [75] |
| miR-99a | Up-regulated | Implantation failure | [75] |
| hsa-miR-32 | Down-regulated | Implantation failure | [75] |
| hsa-miR-628-5p | Down-regulated | Implantation failure | [75] |
| hsa-miR-874 | Down-regulated | Implantation failure | [75] |
Currently, there is no standard epigenetic-based test for infertility. This could be due to the relative newness of the field, thus, the field is not yet fully understood. However, as of the time of writing this review, only one commercially available epigenetic-based infertility test called "Seed" has been announced. The epigenetic test is a male infertility test that was developed in 2016 by reputable reproductive scientists and computational biologists at Episona Incorporation, California, United States[76]. Seed identifies alterations in the sperm’s DNA that provide an insight into why some pregnancies fail[76]. Seed focuses mainly on DNA methylation and examines at least 480000 regions of sperm DNA for unusual methylation at certain gene sites important to fertility[76]. Each abnormal region detected is scored as a risk for either male factor infertility or poor embryo development[76]. The results of the test determine the type of reproductive assistance the person needs, which could be either intrauterine insemination or in vitro fertilization (IVF)[76].
The manufacturers of Seed believe the test is more effective than the available infertility tests, including traditional semen analysis. According to them, while semen analysis gives useful information on sperm counts, motility, and morphology, Seed goes further to identify problems related to sperm function and embryo development[76]. Seed combines modern discoveries in science and technology to provide patients with previously unknown information about their fertility[76]. Seed increases the chances of pregnancy; it is more cost-effective and can be used to personalize fertility treatment for the affected persons[76]. The precision of Seed has been validated in two clinical studies; one was a retrospective study involving 127 IVF patients and 36 fertile controls[76], the second was a prospective study involving over 200 patients from several clinics and 96 fertile controls[76].
As epigenetic changes are dynamic and reversible, they can thus be used as therapeutic targets in diseases that have an epigenetic etiology[77-81]. This can be achieved by blocking or deleting the enzymes that modulate the epigenetic alterations in the affected individuals, thereby preventing or reversing the associated disease[82-84]. Complementary single-stranded oligonucleotides (otherwise called anti-miRNAs) can also be used to silence overexpressed genes or boost repressed genes[85-87].
Currently, there is no particular epigenetic drug for treating infertility. However, epigenetic drugs have been developed for some diseases, such as cancer and diabetes mellitus (Table 3). Notably, a HDAC3 inhibitor known as RGFP966 has been shown to reverse Type 1 diabetes and its complications in transgenic mice fed for three months[88]. The DNA methylation inhibitor known as 5-Azacytidine destroys cancer cells[89]. As epigenetic mechanisms are the same in all biological processes, including disease pathologies, it can be hypothesized that some available epigenetic drugs may also be helpful in the treatment of infertility. Alternatively, infertility epigenetic drugs can be formulated from the bioactive components of existing epigenetic drugs or from entirely different bioactive substances. Thus, infertility caused by DNA hypomethylation in both males and females can potentially be reversed or reduced by methyl-donating compounds and epigenetic drugs such as folate, methionine, choline, betaine, and vitamin B-12[15]. Hypomethylation in infertile persons can also be corrected by epigenetic drugs that block DNA-demethylating enzymes (TETs)[15]. These drugs include a cytosine-based lead compound known as Bobcat339 (though not approved yet), which has been shown to inhibit TET1 and TET2[90]. A small molecule known as C35 is another inhibitor that has been demonstrated to target the TET catalytic domain and decrease the 5hmC concentration in the genome[91]. Similarly, infertility caused by DNA hypermethylation can potentially be treated by DNA methylation inhibitors, which include zebularine, disulfiram, decitabine, azacitidine, and chaetocin[92]. The mentioned epigenetic drugs work by inhibiting the catalyzing enzymes of DNA methylation and inducing activation of genes silenced by methylation[93,94]. Moreover, infertility caused by histone post-translational modification can potentially be treated by histone modification inhibitors such as RGFP966, vorinostat, romidepsin, garcinol, and belinostat[95,96]. Infertility caused by abnormal expression of miRNAs can be corrected by anti-miRNA oligonucleotides such as locked nucleic acid, antagomirs, morpholinos, byetta, victoza, trulicity, janu-via, onglyza, and tradjenta[15,97]. RG108 and MG98, for example, bind to the 3′ untranslated region of DNMT1, preventing gene transcription[98,99].
| Epigenetic drug | Target | Effect | Ref. |
| Choline | HDAC3 | Increases DNA methylation | [15] |
| Betaine | HDAC3 | Increases DNA methylation | [15] |
| Bobcat339 | TETs and TET2 | Increases DNA methylation | [15] |
| C35 | TET | Increases DNA methylation | [91] |
| Zebularine | DNMTs | Reduces hypermethylation | [92] |
| Disulfiran | DNMTs | Reduces hypermethylation | [92] |
| Decitabine | DNMTs | Reduces hypermethylation | [92] |
| Azacitidine | DNMTs | Reduces hypermethylation | [92] |
| Chaeton | DNMTs | Reduces hypermethylation | [92] |
| RGFP966 | HDAC3 | Inhibits histone modification | [95,96] |
| RG108 | Anti-miRNA | Reduces gene expression | [98,99] |
Abnormal epigenetic modifications in genes that control gametogenesis and embryogenesis such as DDX3X, ADH4, AZF, PLAG1, D1RAS3, CYGB, MEST, JMJD1A, KCNQ1, IGF2, H19, and MTHFR can cause infertility. This suggests that some cases of infertility have epigenetic etiologies. The most common epigenetic mechanisms regarding infertility are DNA methylation, histone post-translational modification, and microRNA interference. Dysregulation of these mechanisms in reproductive tissues and cells can disrupt genomic imprinting as well as oocyte and sperm epigenomes. Fortunately, epigenetic changes are reversible by blocking the mediating enzymes such as HDAC3, TET, TET2, and DNMTs. This indicates that infertility induced by epigenetic alterations can be treated by reversing the same mechanisms that caused them. There are some certified epigenetic drugs currently in use, including Choline, Betaine, Zebularine, Disulfiran, Decitabine, Azacitidine, Chaeton, RGFP966, and RG108, but none have been formulated specifically for infertility. It is theorized that some of the available epigenetic drugs could be helpful in infertility as epigenetic mechanisms are the same in all disease pathologies. Epigenetic drugs for infertility can also be formulated from the bioactive compounds of existing epigenetic drugs or from entirely different bioactive substances.
Medical practitioners are advised to formulate treatment procedures and epigenetic drugs for infertility having an epigenetic etiology.
Epigenetic disruption is involved in some cases of infertility.
Epigenetic disruptions in genes that control gametogenesis and embryogenesis, such as DDX3X, ADH4, AZF, PLAG1, D1RAS3, CYGB, MEST, JMJD1A, KCNQ1, IGF2, H19, and MTHFR may result in infertility.
Relevant information was collected from notable academic repositories and the articles collected were sorted using Endnote software.
The study was aimed at articulating and disseminating the epigenetic basis of infertility to raise public awareness.
The study was motivated by the desire to reduce the incidence and burden of infertility.
Abnormal epigenetic modifications have been implicated in some cases of infertility and can be used as therapeutic targets. However, the role of epigenetics in infertility has not been given adequate attention.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Genetics and heredity
Country/Territory of origin: Nigeria
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Lei XH, China; Sukocheva OA, Australia S-Editor: Liu JH L-Editor: Webster JR P-Editor: Liu JH
| 1. | Yahaya TO, Liman UU, Abdullahi H, Koko YS, Ribah SS, Adamu Z, Abubakar S. Genes Predisposing to Syndromic and Nonsyndromic Infertility: a narrative review. Egypt J Med Hum Genet. 2020;21:46. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 2. | Yahaya TO, Oladele EO, Anyebe D, Obi C, Bunza MDA, Sulaiman R, Liman UU. Chromosomal Abnormalities Predisposing to Infertility, Testing and Management: A Narrative Review. Bull Natl Res Cent. 2021;45:65. [DOI] [Full Text] |
| 3. | Das L, Parbin S, Pradhan N, Kausar C, Patra SK. Epigenetics of reproductive infertility. Front Biosci (Schol Ed). 2017;9:509-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Carroll M, Nevin C. Sperm DNA Methylation, Infertility and Transgenerational Epigenetics. J Genet Genomic Sci. 2015;1:004. [DOI] [Full Text] |
| 5. | Pouresmaeili F. Epigenetics and fertility. Urol Nephrol Open Access J. 2019;7:45-48. [DOI] [Full Text] |
| 6. | Kitamura A, Miyauchi N, Hamada H, Hiura H, Chiba H, Okae H, Sato A, John RM, Arima T. Epigenetic alterations in sperm associated with male infertility. Congenit Anom (Kyoto). 2015;55:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Lestari SW, Rizki MD. Epigenetic: A new approach to etiology of infertility. Med J Indones. 2017;25:255-262. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 8. | Cedars M, Jaffe RB. Infertility and Women. J Clin Endocrinol Metab. 2005;90:E2. [DOI] [Full Text] |
| 9. | McSwiggin HM, O'Doherty AM. Epigenetic reprogramming during spermatogenesis and male factor infertility. Reproduction. 2018;156:R9-R21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 10. | Bakhtiyar K, Beiranvand R, Ardalan A, Changaee F, Almasian M, Badrizadeh A, Bastami F, Ebrahimzadeh F. An investigation of the effects of infertility on Women's quality of life: a case-control study. BMC Womens Health. 2019;19:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 11. | Dyer SJ, Patel M. The economic impact of infertility on women in developing countries a systematic review. Facts Views Vis Obgyn. 2012;4:102-109. [PubMed] |
| 12. | Yahaya T, Ufuoma BS. Role of epigenetics in aetiology and therapies for Type 1 Diabetes Mellitus: A narrative review. J Health Soc Sci. 4:199-212. [DOI] [Full Text] |
| 13. | Dada R, Kumar M, Jesudasan R, Fernández JL, Gosálvez J, Agarwal A. Epigenetics and its role in male infertility. J Assist Reprod Genet. 2012;29:213-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 14. | Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145-2156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 798] [Cited by in RCA: 662] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 15. | Yahaya T, Oladele E, Shemishere U, Abdulrau’f M. Role of Epigenetics in the Pathogenesis and Management of Type 2 Diabetes Mellitus. UTJMS. 6:20-28. [DOI] [Full Text] |
| 16. | Nowacka-Zawisza M, Wiśnik E. DNA methylation and histone modifications as epigenetic regulation in prostate cancer (Review). Oncol Rep. 2017;38:2587-2596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 17. | Ingouff M, Selles B, Michaud C, Vu TM, Berger F, Schorn AJ, Autran D, Van Durme M, Nowack MK, Martienssen RA, Grimanelli, D. (2017). Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev. 31 (1):72-83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 18. | Oppermann U. Why is epigenetics important in understanding the pathogenesis of inflammatory musculoskeletal diseases? Arthritis Res Ther. 2013;15:209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3346] [Cited by in RCA: 3374] [Article Influence: 210.9] [Reference Citation Analysis (0)] |
| 20. | Cui X, Jing X, Wu X, Yan M, Li Q, Shen Y, Wang Z. DNA methylation in spermatogenesis and male infertility. Exp Ther Med. 2016;12:1973-1979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Gujar H, Weisenberger DJ, Liang G. The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome. Genes (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 22. | Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 23. | Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 515] [Article Influence: 36.8] [Reference Citation Analysis (1)] |
| 24. | Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 305] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 25. | Cisneros FJ. DNA Methylation and Male Infertility. Front Biosci. 2002;7:d752-764. [RCA] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Morcos L, Ge B, Koka V, Lam KC, Pokholok DK, Gunderson KL, Montpetit A, Verlaan DJ, Pastinen T. Genome-wide assessment of imprinted expression in human cells. Genome Biol. 2011;12:R25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 27. | Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC; Erice Imprinting Group. Genomic Imprinting and Physiological Processes in Mammals. Cell. 2019;176:952-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 349] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 28. | Macdonald WA. Epigenetic mechanisms of genomic imprinting: common themes in the regulation of imprinted regions in mammals, plants, and insects. Genet Res Int. 2012;2012:585024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Rotondo JC, Selvatici R, Di Domenico M, Marci R, Vesce F, Tognon M, Martini F. Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males. Epigenetics. 2013;8:990-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542-2551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 293] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 31. | Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 91] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 32. | Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94:585-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 33. | Tang D, Huang Y, Liu W, Zhang X. Up-Regulation of microRNA-210 is Associated with Spermatogenesis by Targeting IGF2 in Male Infertility. Med Sci Monit. 2016;22:2905-2910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 34. | Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 257] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 35. | Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363:1700-1702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 228] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 36. | Liu JH, Zhu JQ, Liang XW, Yin S, Ola SI, Hou Y, Chen DY, Schatten H, Sun QY. Diploid parthenogenetic embryos adopt a maternal-type methylation pattern on both sets of maternal chromosomes. Genomics. 2008;91:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 37. | Mahadevan S, Sathappan V, Utama B, Lorenzo I, Kaskar K, Van den Veyver IB. Maternally expressed NLRP2 links the subcortical maternal complex (SCMC) to fertility, embryogenesis and epigenetic reprogramming. Sci Rep. 2017;7:44667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 38. | Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, Pavlidis P, Journot L. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 350] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 39. | Tomizawa S, Sasaki H. Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. J Hum Genet. 2012;57:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 40. | MedlinePlus 2020. Prader-Willi syndrome. (Accessed May 2, 2021). Available from: https://medlineplus.gov/genetics/condition/prader-willi-syndrome/#causes. |
| 41. | MedlinePlus 2020. Beckwith-Wiedemann syndrome. (Accessed May 2, 2021). Available from: https://medlineplus.gov/genetics/condition/beckwith-wiedemann-syndrome/#causes. |
| 42. | MedlinePlus 2020. Russell-Silver syndrome. (Accessed May 2, 2021). Available from: https://medlineplus.gov/genetics/condition/russell-silver-syndrome/#causes. |
| 43. | Wongtawan T. The importance of epigenetics in embryonic development and reproductive biotechnology. J Appl Anim Sci. 2012;1-18. |
| 44. | Montorsi F, Gandaglia G, Fossati N, Briganti A. Re: The Magnetic Resonance Imaging in Active Surveillance (MRIAS) Trial: Use of Baseline Multiparametric Magnetic Resonance Imaging and Saturation Biopsy to Reduce the Frequency of Surveillance Prostate BiopsiesA. Amin, M. J. Scheltema, R. Shnier, A. Blazevski, D. Moses, T. Cusick, A. Siriwardena, B. Yuen, P. J. van Leeuwen, A. M. Haynes, J. Matthews, P. Brenner, G. O'Neill, C. Yuen, W. Delprado, P. Stricker and J. Thompson J Urol 2020; 203: 910-917. J Urol. 2020;204:843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 45. | Camprubí C, Salas-Huetos A, Aiese-Cigliano R, Godo A, Pons MC, Castellano G, Grossmann M, Sanseverino W, Martin-Subero JI, Garrido N, Blanco J. Spermatozoa from infertile patients exhibit differences of DNA methylation associated with spermatogenesis-related processes: an array-based analysis. Reprod Biomed Online. 2016;33:709-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 46. | Urdinguio RG, Bayón GF, Dmitrijeva M, Toraño EG, Bravo C, Fraga MF, Bassas L, Larriba S, Fernández AF. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum Reprod 2015; 30: 1014-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 134] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 47. | Tunc O, Tremellen K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet. 2009;26:537-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 180] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 48. | Naqvi H, Ilagan Y, Krikun G, Taylor HS. Altered genome-wide methylation in endometriosis. Reprod Sci. 2014;21:1237-1243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 49. | Wang P, Zhao H, Li T, Zhang W, Wu K, Li M, Bian Y, Liu H, Ning Y, Li G, Chen ZJ. Hypomethylation of the LH/choriogonadotropin receptor promoter region is a potential mechanism underlying susceptibility to polycystic ovary syndrome. Endocrinology. 2014;155:1445-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 50. | Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 554] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 51. | Cruickshank MN, Besant P, Ulgiati D. The impact of histone post-translational modifications on developmental gene regulation. Amino Acids. 2010;39:1087-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 52. | Murakami Y. Heterochromatin and Euchromatin. In: Dubitzky W, Wolkenhauer O, Cho KH, Yokota H. (eds) Encyclopedia of Systems Biology. Springer, New York, NY. 2013. [DOI] [Full Text] |
| 53. | Bhasin M, Reinherz EL, Reche PA. Recognition and classification of histones using support vector machine. J Comput Biol. 2006;13:102-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 54. | Brockers K, Schneider R. Histone H1, the forgotten histone. Epigenomics. 2019;11:363-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 55. | Santoni JR, Santoni Williams CJ. Letter to the editor on a paper by Kimura A, Yoshiro H, Yuasa T. Chronic inflammatory demyelinating polyneuropathy in a patient with hyperIgEemia. J Neurol Sci. 2009;285:270; author reply 271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 56. | Jenkins TG, Carrell DT. The sperm epigenome and potential implications for the developing embryo. Reproduction. 2012;143:727-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 57. | Štiavnická M, García-Álvarez O, Ulčová-Gallová Z, Sutovsky P, Abril-Parreño L, Dolejšová M, Řimnáčová H, Moravec J, Hošek P, Lošan P, Gold L, Fenclová T, Králíčková M, Nevoral J. H3K4me2 accompanies chromatin immaturity in human spermatozoa: an epigenetic marker for sperm quality assessment. Syst Biol Reprod Med. 2020;66:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 58. | Wang T, Gao H, Li W, Liu C. Essential Role of Histone Replacement and Modifications in Male Fertility. Front Genet. 2019;10:962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 59. | Montjean D, De La Grange P, Gentien D, Rapinat A, Belloc S, Cohen-Bacrie P, Menezo Y, Benkhalifa M. Sperm transcriptome profiling in oligozoospermia. J Assist Reprod Genet. 2012;29:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 60. | Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler J, Peters AH, Kimmins S. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350:aab2006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 354] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 61. | Zhu Y, Li B, Wu T, Ye L, Zeng Y, Zhang Y. Cell cycle and histone modification genes were decreased in placenta tissue from unexplained early miscarriage. Gene. 2017;636:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 62. | Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 2011;26:2558-2569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 63. | Legoff L, Dali O, D'Cruz SC, Suglia A, Gely-Pernot A, Hémery C, Kernanec PY, Demmouche A, Kervarrec C, Tevosian S, Multigner L, Smagulova F. Ovarian dysfunction following prenatal exposure to an insecticide, chlordecone, associates with altered epigenetic features. Epigenetics Chromatin. 2019;12:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 64. | Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38:257-268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 194] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 65. | Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 559] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 66. | Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol. 2010;2:161-179. [PubMed] |
| 67. | John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2680] [Cited by in RCA: 2887] [Article Influence: 137.5] [Reference Citation Analysis (0)] |
| 68. | Salilew-Wondim D, Gebremedhn S, Hoelker M, Tholen E, Hailay T, Tesfaye D. The Role of MicroRNAs in Mammalian Fertility: From Gametogenesis to Embryo Implantation. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 69. | Chen X, Li X, Guo J, Zhang P, Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis. J Anim Sci Biotechnol. 2017;8:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 90] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 70. | Reza AMMT, Choi YJ, Han SG, Song H, Park C, Hong K, Kim JH. Roles of microRNAs in mammalian reproduction: from the commitment of germ cells to peri-implantation embryos. Biol Rev Camb Philos Soc. 2019;94:415-438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 71. | Romero Y, Meikar O, Papaioannou MD, Conne B, Grey C, Weier M, Pralong F, De Massy B, Kaessmann H, Vassalli JD, Kotaja N, Nef S. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS One. 2011;6:e25241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 72. | Salas-Huetos A, Blanco J, Vidal F, Mercader JM, Garrido N, Anton E. New insights into the expression profile and function of micro-ribonucleic acid in human spermatozoa. Fertil Steril. 2014;102:213-222.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (1)] |
| 73. | Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, Backes C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril. 2013;99:1249-1255.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 74. | Wang J, Xu B, Tian GG, Sun T, Wu J. Ablation of the MiR-17-92 MicroRNA Cluster in Germ Cells Causes Subfertility in Female Mice. Cell Physiol Biochem. 2018;45:491-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 75. | Revel A, Achache H, Stevens J, Smith Y, Reich R. MicroRNAs are associated with human embryo implantation defects. Hum Reprod. 2011;26:2830-2840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 171] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 76. | Episona Incorporation. 2017. Episona Enters Consumer Market with Epigenetics Test for Male Infertility. (Accessed Jan 22, 2021). Available from: https://www.prnewswire.com/news-releases/episona-enters-consumer-market-with-epigenetics-test-for-male-infertility-300538689.html. |
| 77. | Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 1630] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 78. | Weinhold B. Epigenetics: the science of change. Environ Health Perspect. 2006;114:A160-A167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 322] [Cited by in RCA: 313] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 79. | Aggarwal R, Jha M, Shrivastava A, Jha AK. Natural Compounds: Role in Reversal of Epigenetic Changes. Biochemistry (Mosc). 2015;80:972-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 80. | Schuebel K, Gitik M, Domschke K, Goldman D. Making Sense of Epigenetics. Int J Neuropsychopharmacol. 2016;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 81. | Center for Disease control and Prevention (CDC). Genomics & Precision Health: What is Epigenetics? 2020. (Accessed May 01, 2021). Available from: https://www.cdc.gov/genomics/disease/epigenetics.htm. |
| 82. | Bramswig NC, Kaestner KH. Epigenetics and diabetes treatment: an unrealized promise? Trends Endocrinol Metab. 2012;23:286-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 83. | Wright J. Epigenetics: reversible tags. Nature. 2013;498:S10-S11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 84. | Pop S, Enciu AM, Tarcomnicu I, Gille E, Tanase C. Phytochemicals in cancer prevention: modulating epigenetic alterations of DNA methylation. Phytochem Rev. 2019;18:1005-1024. [DOI] [Full Text] |
| 85. | Mao Y, Mohan R, Zhang S, Tang X. MicroRNAs as pharmacological targets in diabetes. Pharmacol Res. 2013;75:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 86. | Henaoui I, Stoll L, Tugay K, Regazzi R. Therapeutic potential of miRNAs in diabetes mellitus. Expert Rev Endocrinol Metab. 2015;10:285-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 87. | Lima JF, Cerqueira L, Figueiredo C, Oliveira C, Azevedo NF. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018;15:338-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 88. | Xu Z, Tong Q, Zhang Z, Wang S, Zheng Y, Liu Q, Qian LB, Chen SY, Sun J, Cai L. Inhibition of HDAC3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP5-ERK1/2 pathway. Clin Sci (Lond). 2017;131:1841-1857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 89. | Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 358] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 90. | Chua GNL, Wassarman KL, Sun H, Alp JA, Jarczyk EI, Kuzio NJ, Bennett MJ, Malachowsky BG, Kruse M, Kennedy AJ. Cytosine-Based TET Enzyme Inhibitors. ACS Med Chem Lett. 2019;10:180-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 91. | Singh AK, Zhao B, Liu X, Wang X, Li H, Qin H, Wu X, Ma Y, Horne D, Yu X. Selective targeting of TET catalytic domain promotes somatic cell reprogramming. Proc Natl Acad Sci U S A. 2020;117:3621-3626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 92. | Patnaik S, Anupriya. Drugs Targeting Epigenetic Modifications and Plausible Therapeutic Strategies Against Colorectal Cancer. Front Pharmacol. 2019;10:588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 93. | Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 373] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 94. | Momparler RL. Pharmacology of 5-Aza-2'-deoxycytidine (decitabine). Semin Hematol. 2005;42:S9-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 162] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 95. | Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, Kundu TK. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004;279:33716-33726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 408] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 96. | Raha P. Outcome of Combining Epigenetic Drugs with Other Treatments in the Clinic. In Book: Medical Epigenetics. Chapter 40, Pp. 799-824. Elsevier Inc., 2016. [DOI] [Full Text] |
| 97. | Jo S, Chen J, Xu G, Grayson TB, Thielen LA, Shalev A. miR-204 Controls Glucagon-Like Peptide 1 Receptor Expression and Agonist Function. Diabetes. 2018;67:256-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 98. | Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305-6311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 388] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 99. | Amato RJ. Inhibition of DNA methylation by antisense oligonucleotide MG98 as cancer therapy. Clin Genitourin Cancer. 2007;5:422-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |