Published online Feb 18, 2024. doi: 10.5312/wjo.v15.i2.192
Peer-review started: October 26, 2023
First decision: December 12, 2023
Revised: December 29, 2023
Accepted: January 16, 2024
Article in press: January 16, 2024
Published online: February 18, 2024
Processing time: 104 Days and 1.4 Hours
The effectiveness of platelet-rich plasma (PRP) for the treatment of Achilles tendon disorders still needs to be evaluated through a series of prospective studies, but genomic analysis can reveal the existence of complementary PRP treatment options. Based on the 96 platelet activation-related genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, we performed Gene Ontology functional enrichment analysis and KEGG enrichment analysis, path
Core Tip: The cAMP signalling pathway may be the key to enhancing the effectiveness of platelet-rich plasma (PRP) in the treatment of Achilles tendon disease. ADCY1-9 may be the key to activating the cAMP signalling network and is expected to be a complementary target for enhancing the effectiveness of PRP in the treatment of Achilles tendon disease.
- Citation: Sun JY, Li C, Du FY. Adenylate cyclase activates the cAMP signalling pathway to enhance platelet-rich plasma-treated Achilles tendon disease, a theoretical bioinformatics-based study. World J Orthop 2024; 15(2): 192-200
- URL: https://www.wjgnet.com/2218-5836/full/v15/i2/192.htm
- DOI: https://dx.doi.org/10.5312/wjo.v15.i2.192
We read with great interest the study by Huang et al[1], which reported that platelet-rich plasma (PRP) injections alone had no significant efficacy on patients who suffered Achilles tendon rupture or Achilles tendinopathy and that PRP alone improved ankle mobility after 12 months of treatment. Moreover, Dan’s team suggested that the method of PRP preparation and the active ingredient, frequency of injection, injection dose, and injection site may be factors influencing the uncertainty of clinical outcomes.
We agree with the authors that the assessment of the effectiveness of PRP in the treatment of Achilles tendinopathy needs to be advanced by a series of prospective studies, which require blinding, randomization, and control of variables. In theory, the pathophysiologic process of Achilles tendinopathy is associated with localized inflammation of the Achilles tendon, with further degenerative lesions leading to complete Achilles tendon rupture[2,3]. PRP, obtained by centrifugation of whole blood, allows the inclusion of supraphysiologic concentrations of growth factors. The release of these growth factors is effectively activated, leading to increased regeneration and healing at the site of injection[4,5]. Moreover, PRP also treats tendon tissue by inducing type I collagen and glycosaminoglycan deposition[5]. The central key to these beneficial biological processes in the treatment of Achilles tendon disease is platelet activation, and the inability to effectively activate platelets may be one of the reasons why PRP is so ineffective in treating this disease. As the field of genomics has developed and transformed, it has allowed us to more accurately understand the keys to complex biological processes, facilitating the development of more precise and effective therapeutic regimens. With respect to the relevant genes in the “map04611: Platelet activation” (https://www.genome.jp/entry/pathway+map04611) pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, we used bioinformatics analysis to provide a theoretical basis for the potential activation of platelets in PRP.
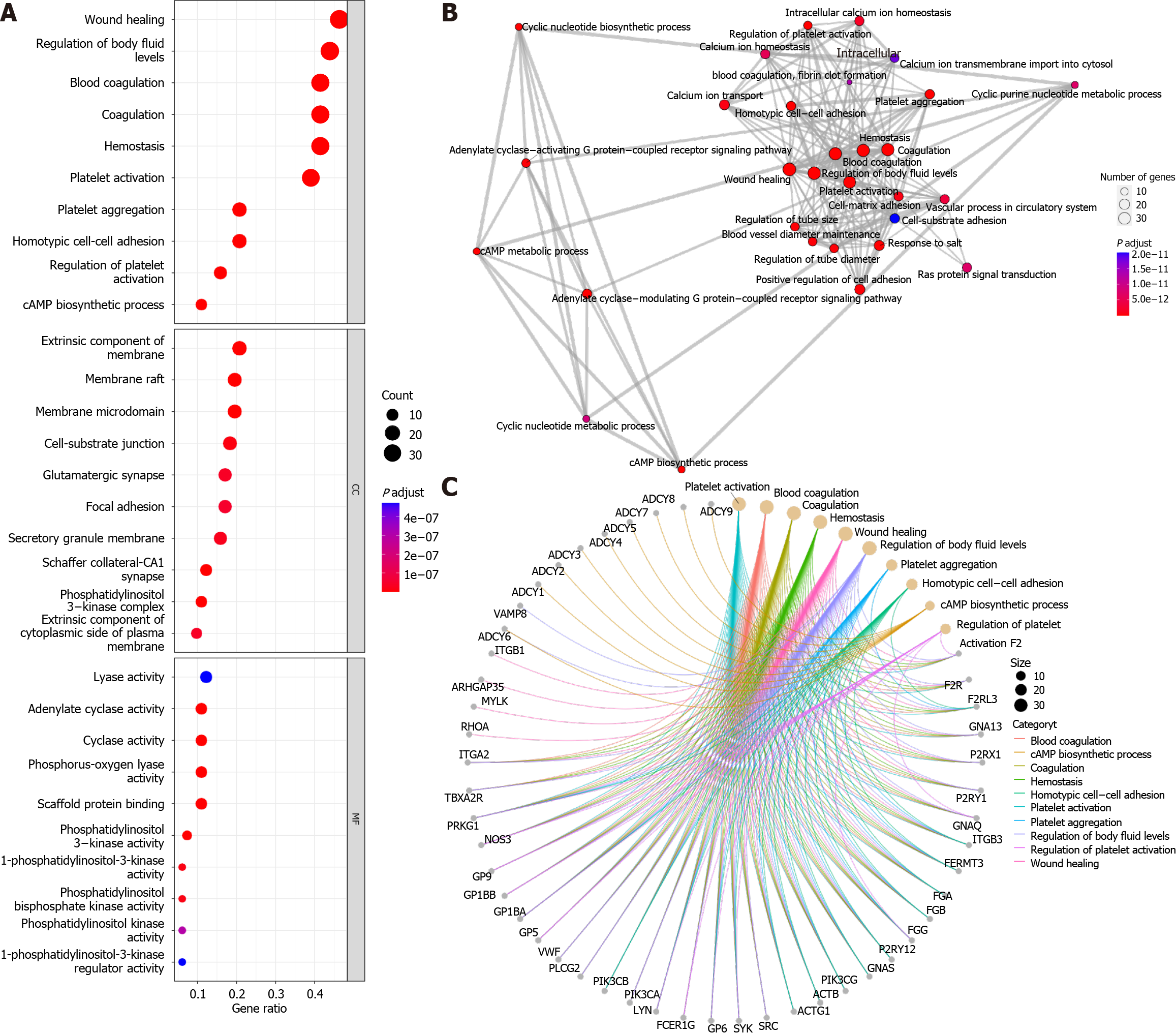
Gene ontology (GO) annotation of 96 genes in the “platelet activation” signalling pathways using the “enrichGO” function of the “clusterProfiler” R package[6] revealed that, under the classification of biological processes, in addition to platelet activation-related pathways (Figure 1A), the “wound healing”, “homotypic cell-cell adhesion” and “cAMP biosynthetic process” pathways were significantly enriched. In terms of cellular components, cell membrane-related pathways were more enriched, which may be a prerequisite for the activation of “cell-substrate junction” pathways. In terms of molecular function, the phosphatidylinositol-related pathway was activated, which may be a preparation for “scaffold protein binding”. Furthermore, using the “pairwise_termsim” function of the “enrichplot” R package to calculate the correlation between the enriched GO pathways and the enrichment map for the enrichment results of the gene set enrichment analysis by the “emapplot” function revealed that “platelet activation” was correlated with “wound healing”, “vascular process in circulatory system”, “cell-matrix-substrate adhesion”, and other biological functions closely related to Achilles tendon recovery (Figure 1B). In addition, cAMP biosynthesis and metabolism appear to constitute another functional subgroup resulting from platelet activation. Plotting the Gene-Concept Network using the “cnetplot” function revealed that none of the pathways, such as “wound healing” and “cAMP biosynthetic process”, had common genes but were associated with “regulation of body fluid levels” (Figure 1C). However, in the KEGG database, all 96 genes were identified as related to “platelet activation”. This result suggested that the specific role of cAMP-related functions in platelet activation is unclear. The cAMP signalling pathway, one of the well-known is the protein kinase A (PKA) system, is a type of cyclic nucleotide system that regulates key physiological processes such as nodal metabolism, secretion, calcium homeostasis, muscle contraction, cell fate and gene transcription[7]. These processes are closely related to the local inflammatory response and immune response of the Achilles tendon, the metabolism and differentiation of fibroblasts, and the senescence and apoptosis of fibroblasts[2,8,9]. Overall, these physiological conditions are known to characterize Achilles tendon inflammation, Achilles tendon recovery, and degenerative Achilles tendon disease.
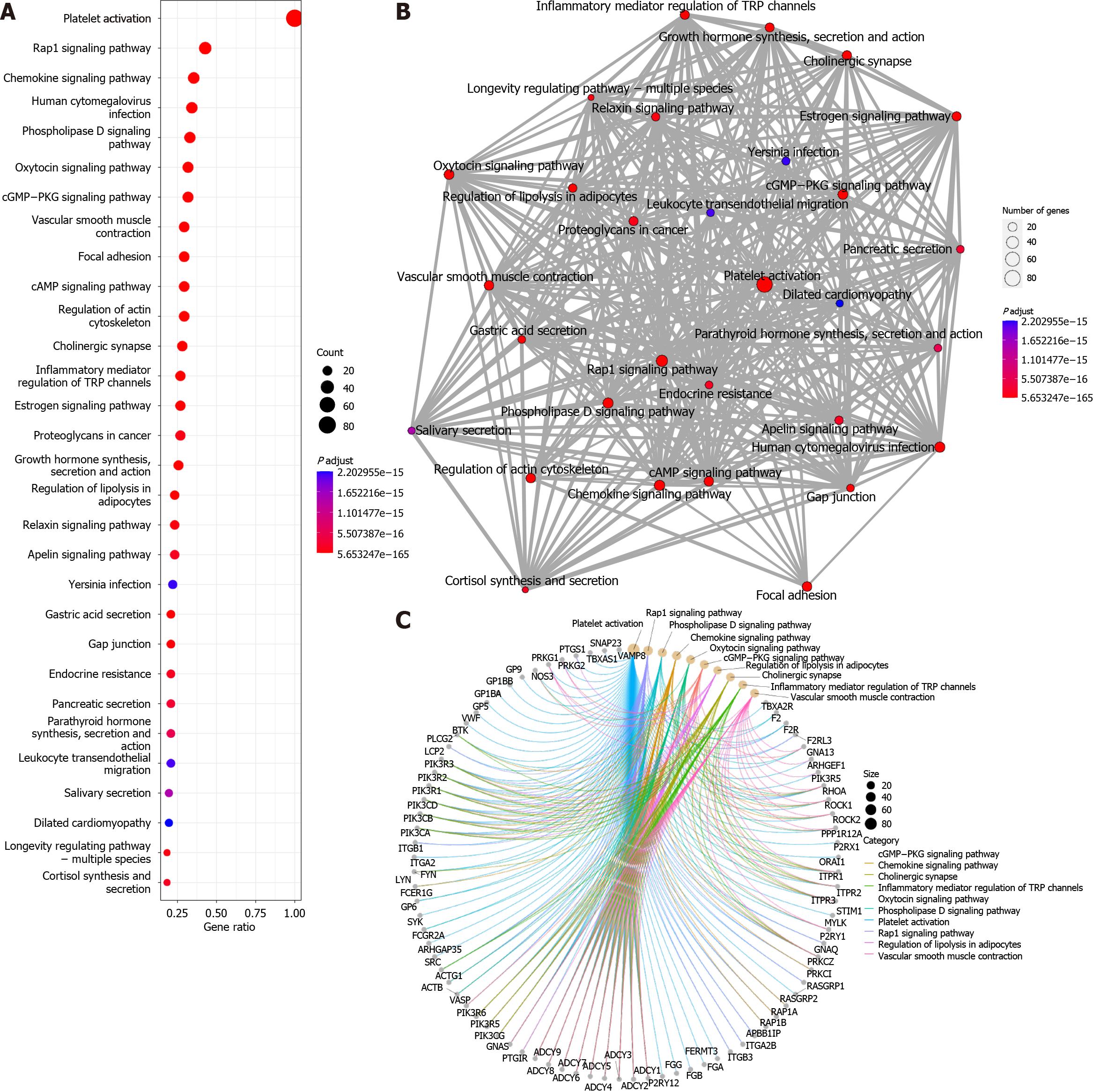
The “enrichKEGG” function was used to analyse the KEGG pathways associated with these 96 platelet activation-related genes (Figure 2A), and the Rap1, cGMP-PKG, and cAMP signalling pathways were found to be enriched. This further verified the results of the GO analysis, suggesting that cAMP-related functions might be involved in the alteration of low platelet activation. The enrichment map generated by the “emapplot” function shown in Figure 2B reveals that platelet activation and the signalling pathways of Rap1, cGMP-PKG, and cAMP are strongly associated and do not exist in relatively large subgroups of the pathways. The Rap1 signalling pathway is involved mainly in the regulation of cell adhesion and cell junctions, as well as the processes of cell migration, polarization, proliferation, and survival[10,11]. In Achilles tendon disease, regulation of the Rap1 signalling pathway may involve aspects of fibroblast metabolism, differentiation, and repair, thereby affecting Achilles tendon development and repair. The second messenger cAMP stimulates Epac1, a Rap guanylate exchange factor. cAMP activates Epac1 migration to the plasma membrane, activates Epac-Rap1 signalling, and enhances integrin-mediated cell adhesion[12,13]. The regulation of the cGMP-PKG signalling pathway may involve inflammatory responses, immune responses, and apoptosis, thus affecting the pathogenesis and pathological processes of diseases such as Achilles tendonitis, Achilles tendon rupture, and Achilles tendon degeneration. The cross-interaction between ATP and GTP also leads to the interaction of cGMP with cAMP, which are also second messengers[14]. Gene-concept network analysis revealed that ADCY1-9 were common genes for platelet activation, Rap1, cGMP-PKG, cAMP, and cytokine activation (Figure 2C), which was also consistent with the results of the GO analysis (Figure 1C). These results suggest that ADCY1-9 may be key to the cAMP pathway and platelet activation pathway and are closely related to a large functional subgroup of the GO pathway beyond “platelet activation”. Interaction network analysis of these 96 platelet-related genes using the STRING database (https://string-db.org/) revealed that ADCY1-9 may be the upstream genes of AKT1 (Figure 3), which may be the key to ameliorating the inability of platelets to be activated efficiently.
The adenylate cyclase (ADCY) family of genes encodes a membrane-associated enzyme that catalyses the formation of cAMP[15], which is in complete agreement with the results of GO and KEGG enrichment analyses. We can further speculate that if activated, ADCY1-9 stimulate the cAMP signalling pathway, which in turn leads to the upregulation of the Rap1 and cGMP-PKG signalling pathways, ultimately completing the global mobilization of the “platelet activation” pathway. ADCY1-9 can be used as an adjunctive target for RPR therapy to enhance its effectiveness in treating Achilles tendon disorders, such as Achilles tendon rupture, Achilles tendonitis, and degenerative Achilles tendon diseases.
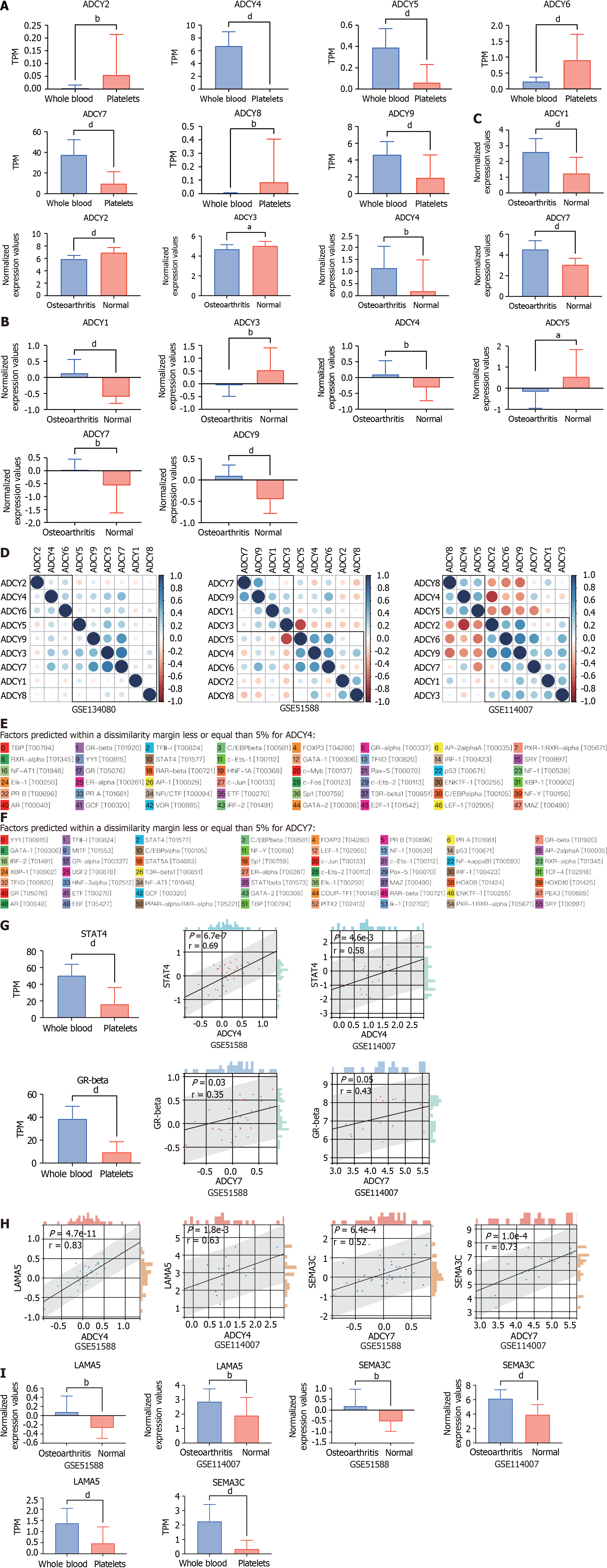
Further, we searched the Gene Expression Omnibus (GEO) database for data on healthy human platelets sequenced in the GSE178158 cohort[16], and 65 samples from the GSE178158 cohort were included, with 100 healthy human whole blood samples from the GSE134080 cohort used as controls[17]. Comparison of the differences in the expression levels of ADCY1-9 in platelets and whole blood revealed (Figure 4A) that ADCY2, 6, and 8 were highly expressed in platelets, while ADCY4, 5, 7, and 9 were lowly expressed in platelets, and the rest did not differ. Cartilage tissue sequencing data from the GEO database for osteoarthritic diseases were also obtained, which included 40 osteoarthritic disease samples and 10 healthy cartilage tissue samples from the GSE51588 cohort[18], and 20 osteoarthritic disease samples and 18 healthy cartilage tissue samples from the GSE114007 cohort were also included[19]. Comparative analysis revealed (Figure 4B and 4C) that ADCY1, 4, and 7 were all highly expressed in osteoarthritic disease samples, while ADCY3 was all highly expressed in healthy cartilage tissue samples. Correlation analysis of ADCY 1-9 expression levels and clustering by the ward.D2 method (Figure 4D) showed that the expression level of adenylate cyclase was synergistic in both blood samples and cartilage tissue samples, which was mainly represented by ADCY 4 and ADCY 7. The low expression of ADCY 4 and ADCY 7 in platelets is consistent with healthy cartilage tissue, and they may be the Achilles heel of osteoarthritic diseases. The promoter sequences of ADCY4 (chr14: 24, 318, 359-24, 335, 071) and ADCY7 (chr16: 50, 266, 551-50, 318, 135) were searched in the UCSC database (https://genome.ucsc.edu/), and the 2000 nt region upstream of the transcriptional start site was set as the selected promoter region. The obtained sequences were entered into the PROMO database (https://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)[20,21], with species selection of Homo sapiens and tolerance set at 5% to retrieve site-bound transcription factors. As shown in Figure 4E and F, 48 transcription factors were retrieved for ADCY4, whereas 56 were retrieved for ADCY7. Differential expression analysis in platelets vs whole blood and correlation analysis in the osteoarthritic disease cohort revealed (Figure 4G) that STAT4 is either the upstream transcription factor of ADCY4, while the upstream transcription factor of ADCY7 may be GR-beta. By The Human Protein Atlas (https://www.proteinatlas.org/) searching which proteins could be secreted and analyzing the correlation of all secreted proteins (Figure 4H)[22], we concluded that ADCY 4 may cause increased secretion of LAMA 5, while ADCY 7 caused SEMA3C. Surprisingly, as shown in Figure 4I, both LAMA 5 and SEMA3C were highly expressed in the osteoarthritic disease samples. These results prove that the STAT 4-ADCY 4-LAMA 5 axis and the GRbeta-ADCY 7-SEMA3C axis may regulate the occurrence in osteoarthritic diseases.
Based on the above bioinformatics analysis, we speculate that the role of adenylate cyclase in osteoarthritic disease is very important, and the poor treatment effect of Achilles tendon disease may be that the cAMP signaling pathway is not effectively activated, and the key point may lie in ADCY 4 and ADCY 7. Both STAT 4-ADCY 4-LAMA 5 axis and GRbeta-ADCY 7-SEMA3C axis are suspected to mediate the occurrence of osteoarthritic disease, and the relevant treatment options can be explored as a theoretical basis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Orthopedics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nezhady MAM, Iran S-Editor: Liu JH L-Editor: A P-Editor: Zhao S
| 1. | Huang D, Vithran DTA, Gong HL, Zeng M, Tang ZW, Rao ZZ, Wen J, Xiao S. Effectiveness of platelet-rich plasma in the treatment of Achilles tendon disease. World J Orthop. 2023;14:485-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Reference Citation Analysis (0)] |
| 2. | Millar NL, Silbernagel KG, Thorborg K, Kirwan PD, Galatz LM, Abrams GD, Murrell GAC, McInnes IB, Rodeo SA. Tendinopathy. Nat Rev Dis Primers 2021; 7: 1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 433] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 3. | Dakin SG, Newton J, Martinez FO, Hedley R, Gwilym S, Jones N, Reid HAB, Wood S, Wells G, Appleton L, Wheway K, Watkins B, Carr AJ. Chronic inflammation is a feature of Achilles tendinopathy and rupture. Br J Sports Med. 2018;52:359-367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 4. | Everts P, Onishi K, Jayaram P, Lana JF, Mautner K. Platelet-Rich Plasma: New Performance Understandings and Therapeutic Considerations in 2020. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 450] [Article Influence: 90.0] [Reference Citation Analysis (0)] |
| 5. | van Buul GM, Koevoet WL, Kops N, Bos PK, Verhaar JA, Weinans H, Bernsen MR, van Osch GJ. Platelet-rich plasma releasate inhibits inflammatory processes in osteoarthritic chondrocytes. Am J Sports Med. 2011;39:2362-2370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 284] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 6. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 4783] [Article Influence: 1195.8] [Reference Citation Analysis (0)] |
| 7. | Ould Amer Y, Hebert-Chatelain E. Mitochondrial cAMP-PKA signaling: What do we really know? Biochim Biophys Acta Bioenerg. 2018;1859:868-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 8. | Rho JH, Ko IG, Jin JJ, Hwang L, Kim SH, Chung JY, Hwang TJ, Han JH. Polydeoxyribonucleotide Ameliorates Inflammation and Apoptosis in Achilles Tendon-Injury Rats. Int Neurourol J. 2020;24:79-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Paula DR, França MS, Leão LKR, Maciel AA, Moura TAA, Moraes SAS, Bahia CP, Borges RS, Batista EJO, Passos ACF, Oliveira KRHM, Herculano AM. Total rupture of Achilles tendon induces inflammatory response and glial activation on the spinal cord of mice. Braz J Med Biol Res. 2023;56:e12391. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Shah S, Brock EJ, Ji K, Mattingly RR. Ras and Rap1: A tale of two GTPases. Semin Cancer Biol. 2019;54:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 11. | Stefanini L, Bergmeier W. RAP1-GTPase signaling and platelet function. J Mol Med (Berl). 2016;94:13-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 12. | Korayem AH, Mujica PE, Aramoto H, Durán RG, Nepali PR, Kim DD, Harris AL, Sánchez FA, Durán WN. Endothelial cAMP deactivates ischemia-reperfusion-induced microvascular hyperpermeability via Rap1-mediated mechanisms. Am J Physiol Heart Circ Physiol. 2017;313:H179-H189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Pannekoek WJ, Vliem MJ, Bos JL. Multiple Rap1 effectors control Epac1-mediated tightening of endothelial junctions. Small GTPases. 2020;11:346-353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Smolenski A. Novel roles of cAMP/cGMP-dependent signaling in platelets. J Thromb Haemost. 2012;10:167-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 214] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 15. | Sanchez-Collado J, Lopez JJ, Jardin I, Salido GM, Rosado JA. Cross-Talk Between the Adenylyl Cyclase/cAMP Pathway and Ca(2+) Homeostasis. Rev Physiol Biochem Pharmacol. 2021;179:73-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Konstorum A, Mohanty S, Zhao Y, Melillo A, Vander Wyk B, Nelson A, Tsang S, Blevins TP, Belshe RB, Chawla DG, Rondina MT, Gill TM, Montgomery RR, Allore HG, Kleinstein SH, Shaw AC. Platelet response to influenza vaccination reflects effects of aging. Aging Cell. 2023;22:e13749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 17. | Aguirre-Gamboa R, de Klein N, di Tommaso J, Claringbould A, van der Wijst MG, de Vries D, Brugge H, Oelen R, Võsa U, Zorro MM, Chu X, Bakker OB, Borek Z, Ricaño-Ponce I, Deelen P, Xu CJ, Swertz M, Jonkers I, Withoff S, Joosten I, Sanna S, Kumar V, Koenen HJPM, Joosten LAB, Netea MG, Wijmenga C; BIOS Consortium, Franke L, Li Y. Deconvolution of bulk blood eQTL effects into immune cell subpopulations. BMC Bioinformatics. 2020;21:243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 18. | Chou CH, Wu CC, Song IW, Chuang HP, Lu LS, Chang JH, Kuo SY, Lee CH, Wu JY, Chen YT, Kraus VB, Lee MT. Genome-wide expression profiles of subchondral bone in osteoarthritis. Arthritis Res Ther. 2013;15:R190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 19. | Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, Sasho T, Koziol JA, Su AI, Lotz MK. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage. 2018;26:1531-1538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 189] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 20. | Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 949] [Cited by in RCA: 1109] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 21. | Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651-3653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 743] [Cited by in RCA: 874] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 22. | Uhlén M, Karlsson MJ, Hober A, Svensson AS, Scheffel J, Kotol D, Zhong W, Tebani A, Strandberg L, Edfors F, Sjöstedt E, Mulder J, Mardinoglu A, Berling A, Ekblad S, Dannemeyer M, Kanje S, Rockberg J, Lundqvist M, Malm M, Volk AL, Nilsson P, Månberg A, Dodig-Crnkovic T, Pin E, Zwahlen M, Oksvold P, von Feilitzen K, Häussler RS, Hong MG, Lindskog C, Ponten F, Katona B, Vuu J, Lindström E, Nielsen J, Robinson J, Ayoglu B, Mahdessian D, Sullivan D, Thul P, Danielsson F, Stadler C, Lundberg E, Bergström G, Gummesson A, Voldborg BG, Tegel H, Hober S, Forsström B, Schwenk JM, Fagerberg L, Sivertsson Å. The human secretome. Sci Signal. 2019;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 298] [Article Influence: 49.7] [Reference Citation Analysis (0)] |