Published online Jun 24, 2025. doi: 10.5306/wjco.v16.i6.105691
Revised: April 6, 2025
Accepted: May 13, 2025
Published online: June 24, 2025
Processing time: 136 Days and 23.4 Hours
Hypoxia-inducible factor 1α (HIF-1α) plays a crucial role in the prognosis of breast cancer, but the current evidence remains inconclusive.
To provide comprehensive evidence about the correlation of altered HIF-1α expression with overall survival (OS) and disease-free survival (DFS) in breast cancer patients.
A systematic search was conducted in PubMed, Embase, and Web of Science databases to collect relevant articles that were published before April 8, 2024. A meta-analysis was used to assess the impact of altered HIF-1α expression on the OS and DFS of breast cancer patients. Subgroup and sensitivity analyses were also performed in this meta-analysis.
This meta-analysis included 40 studies. The average percentage of breast cancer patients with high HIF-1α expression was 39.6%. The overall meta-analysis results demonstrated that high HIF-1α expression is strongly linked to poor outcomes in patients of breast cancer. Compared with low HIF-1α expression, the overall hazard ratio for OS in patients with high HIF-1α expression was 1.47 [95% con
Compared to patients with low HIF-1α expression, those with high expression level had shorter OS and DFS. However, the prognostic significance of high HIF-1α expression varies across molecularly stratified breast cancer cohorts needs to be further elucidated.
Core Tip: Hypoxia is a crucial characteristic of tumors that can influence the phenotype of cancer stem cells and contribute to cancer cell invasion and metastasis, thereby significantly impacting the prognosis of cancer patients. This meta-analysis offers a more exhaustive analysis of the relationship between high hypoxia-inducible factor 1α expression and the prognosis of breast cancer patients, providing clinicians with systematic evidence about the role of high hypoxia-inducible factor 1α expression in predicting breast cancer prognosis.
- Citation: Zheng XD, Li HY, Gao SY, Wang Q, Liu JB. High hypoxia inducible factor-1α expression is associated with reduced survival in patients with breast cancer: A meta-analysis. World J Clin Oncol 2025; 16(6): 105691
- URL: https://www.wjgnet.com/2218-4333/full/v16/i6/105691.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i6.105691
According to 2022 Global Cancer Statistics, it is estimated that breast cancer accounted for 2.3 million incident cases, establishing it as the foremost oncological disease affecting women worldwide[1]. In China, approximately 357000 new breast cancer cases were reported among women in 2022, ranking second among all new cancer cases[1]. In the United States, it is estimated that there were 313000 new breast cancer cases in 2024, and breast cancer ranked first among all new cancer cases in American women[2]. Despite considerable progress in early diagnosis, comprehensive treatment stra
Hypoxia is a critical characteristic of tumors, and it influences the phenotype of tumor stem cells and participates in the invasion and metastasis of cancer cells[4]. Under hypoxic conditions, the expression of hypoxia-inducible factor 1α (HIF-1α) is increased in cancer cells. Subsequently, HIF-1α maintains the stemness of cancer cells through multiple me
Although many clinical prognostic studies have provided evidence supporting the connection between high expression of HIF-1α and poor prognosis in individuals with breast cancer, definitive evidence about the role of HIF-1α expression in predicting the prognosis of breast cancer and guiding treatment decisions is still lacking. Although previous meta-analyses systematically evaluated and quantitatively analyzed published prognostic studies, they failed to examine all relevant studies through a more comprehensive search[12-16]. Moreover, in recent years, the publication of recent studies requires a re-evaluation through a systematic review to analyze the impact of HIF-1α expression on breast cancer prognosis[17-22]. Therefore, this study aimed to execute a systematic review together with meta-analysis, striving to evaluate more accurately how the expression of HIF-1α influences the prognostic significance regarding OS and DFS among individuals with breast cancer. This approach is intended to provide high-level clinical evidence for more precise prediction of prognosis and treatment-related decision-making for patients with breast cancer.
The quantitative evidence synthesis was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines, which provided the methodological framework to ensure reporting standards.
Scholarly resource identification spanned three principal knowledge bases: PubMed, Embase, and Web of Science. A variety of combinations of the following keywords were used to search titles, abstracts, or full texts: “Breast cancer or Human Mammary Carcinoma”, “Cell Hypoxia or hypoxic or Hypoxia-Inducible Factor 1α”, and “Survival or outcome or prognosis or Recurrence”. The search covered publications up to April 8, 2024. Additionally, manual searches and the reference backtracking method were performed to identify further relevant literature.
The retrieved research studies satisfied the inclusion criteria detailed below: (1) A pathological diagnosis of human breast cancer; (2) The detection of HIF-1α expression in breast cancer tissues through immunohistochemistry or immunofluorescence; (3) Study endpoints of DFS/RFS and OS/breast cancer-specific survival (BCSS); (4) The provision of prognostic effect indicators hazard ratio (HR)/logHR and 95% confidence interval (CI)/standard error (SE) or raw data for HIF-1α expression and OS and DFS in breast cancer; and (5) Articles written in either Chinese or English.
Two researchers (Xue-Di Zheng and Huan-Yu Li) initially reviewed the titles and abstracts of the candidate studies independently, eliminating those that did not meet the inclusion criteria. Subsequently, the two researchers meticulously read the full texts of the remaining candidates to determine their eligibility for inclusion based on the established criteria. In cases of uncertainty about a study’s inclusion or a disagreement between the researchers, the study designer (Jiang-Bo Liu) was consulted for review and discussion until a consensus was reached. Additionally, if multiple studies were conducted on the same population after a comprehensive evaluation, the study selected for the meta-analysis was the one with either the largest sample size or the most recent publication.
Two researchers (Xue-Di Zheng and Huan-Yu Li) separately extracted the following data and information from every study that satisfied the inclusion criteria in accordance with a pre-prepared form: First author, publication year, study location, recruitment period, analytical method, study design, sample size, average age, tissue type, molecular subtype, antibodies used for HIF-1α detection, HIF-1α expression cutoff value, study endpoint, HR and 95%CI, and follow-up time.
The endpoints of this systematic review and meta-analysis focused on investigating the connections linking HIF-1α to the prognosis of breast cancer patients, were DFS/RFS and OS/BCSS. These endpoints were defined as follows: DFS/RFS indicated the period from the commencement of treatment (including surgery) to the recurrence or metastasis of the disease, and OS/BCSS represented the duration from the time of diagnosis and surgical intervention to the patient’s death due to any cause/breast cancer metastasis.
Review Manager 5.4 was utilized to conduct the meta-analysis. Data on the prognostic analysis of HIF-1α expression were extracted. If the included studies reported HRs and 95%CIs, the generic inverse variance method was used in Review Manager to calculate the logHR and SE for each study for meta-analysis. If studies did not directly report HRs and CIs, statistical data were obtained using methods such as survival curve data extraction and estimation of study endpoint events to calculate the logHR and SE for each study[23]. This meta-analysis primarily combined the multivariable Cox regression effect sizes (HR and CI) from each study; however, when studies reported only univariate HR and CI, they were included in the pooled analysis as univariate effect sizes. Finally, a pooled HR greater than 1 indicated a higher risk of recurrence, metastasis, or death and poorer prognosis for patients of breast cancer with high HIF-1α expression. If the 95%CI did not overlap with 1, it was considered significant according to statistical analysis (P value < 0.05).
The I2 statistic was utilized to evaluate heterogeneity among the studies selected for the meta-analysis, which quantifies variability on a scale from 0% to 100%. Studies with an I2 value below 50% or a P value exceeding 0.05 were considered to show no or low heterogeneity, justifying the use of a fixed-effects model for meta-analysis. In cases where these thresholds were not met, a random-effects model was employed. To further investigate potential sources of heterogeneity and examine the influence of specific variables, subgroup analyses were systematically performed.
Furthermore, only studies with large sample sizes (≥ 200) were included to investigate whether population sampling affected the sensitivity of this meta-analysis. Funnel plots and Egger’s test were used to investigate publication bias in the included studies. If funnel plot asymmetry was observed or the P value from Egger’s test was less than 0.05, publication bias was present[24]. To address possible publication bias, a Duval and Tweedie[25] trim-and-fill analysis was applied for both detection and adjustment in the meta-analysis. The analytical framework implemented two-tailed verification protocols, establishing 0.05 as the critical significance benchmark.
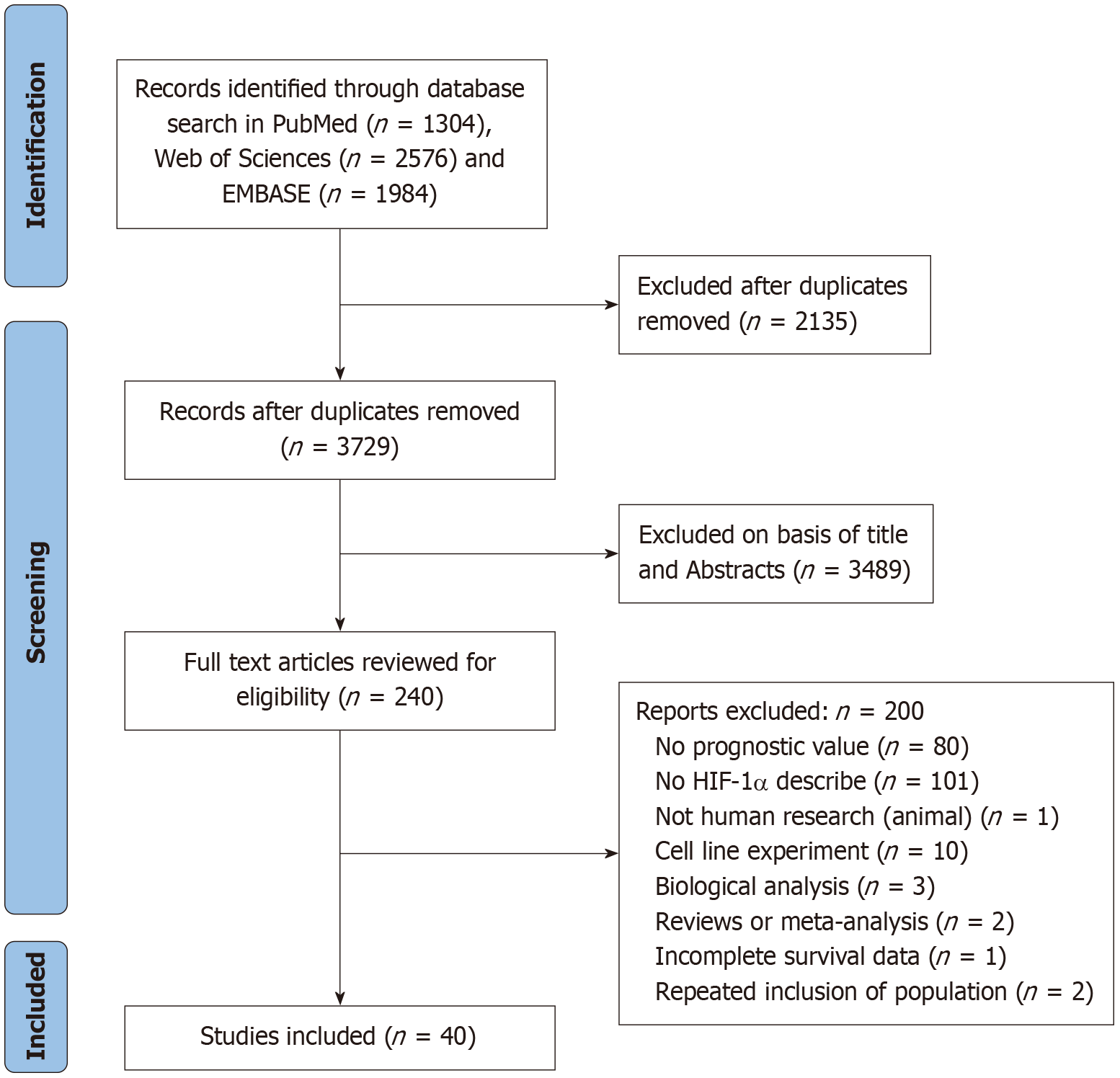
The initial literature search yielded 5864 articles. Through a rigorous screening process based on the established inclusion criteria, 40 eligible studies involving 7738 breast cancer patients were ultimately included in this systematic review and meta-analysis. Based on the Oxford Evidence levels, this provides level 2b evidence regarding prognosis. The process and results of the literature screening are presented in Figure 1.
Detailed characteristics of the included studies are summarized Table 1. Among the 40 studies, 23[12-14,16,20,21,26-41] included European populations, 16[15,18,19,22,42-53] included Asian populations, and 1[17] included an African popu
| Ref. | Recruitment period | Study design | Sample size | Age median mean year (range) | Histological grading, and TNM staging | Percentage of grading and staging | Molecular typing | N of HIFα positive (%), cut-off, and antibody | Survival endpoint (months) | Hazard ratio, 95%CI and P value | HR estimation | Follow-up months (range) |
| El-Guindy et al[17], 2023, Egypt | June 2019-June 2022 | Prospective | 60 | 47.5 | G2: 24, G3: 36; NA | G1/G2: 40%; I/II: NA | TNBC | 28 (46.6); ≥ 10%; EP1215Y | OS | (M) 7.56 (1.21-47.24), 0.03 | Reported | 24 |
| Jögi et al[26], 2019, Sweden | 1977-2007 | NA | 634 of 688 (BC2) | NA | NA; I: 273, II: 1 59, III: 83, Unknow: 119 | G1/G2: NA; I/II: 82.6% | NA | 111 (17.3); ≥ 1%; BD610959 | OS | (U) 1.6 (1.2-2.2), 0.001 | Reported | NA |
| (M) 1.6 (1.0-2.5), 0.03 | ||||||||||||
| Laurinaviciu et al[31], 2015, Lithuania | 1977-2007 | NA | 107 | NA | G1: 25, G2: 51, G3: 31 | G1/G2: 71%; I/II: NA | Luminal A: 60, luminal B: 29, HER2 +: 28 | NA; score 14; EP1215Y | OS | (M) 0.23 (0.08-0.62), 0.002 | Reported | 84 |
| Li et al[48], 2016, China | 2005-2009 | NA | 156 | 50 | G1: 32, G2: 78, G3: 46; I: 23, II: 60, III: 48, IV: 25 | G1/G2: 70%; I/II: 53% | NA | 83 (53.3); score 6; ab82832 | OS | (U) 2.07 (1.58-2.71), < 0.001 | (U) Survival curve (M) reported | 60 |
| (M) 2.37 (1.09-5.15), 0.029 | ||||||||||||
| Zhuang et al[22], 2024, China | May 2014-August 2016 | NA | 197 | NA | G1-G2: 130, G3: 57; I-II: 147, III- IV: 50 | G1/G2: 69%; I/II: 75% | Luminal A: 37, luminal B: 133, HER2 +: 13, TNBC: 14 | 121 (61.4); score 3; ab51608 | OS | (U) 5.36 (2.02-14.22), 0.001 | Reported | NA |
| (M) 4.94 (1.86-13.14), 0.001 | ||||||||||||
| Rajković-Molek et al[38], 2014, Croatia | 2000-2004 | Retrospective | 208 | 65 | G1: 46, G2: 113, G3: 49; I: 67, II: 68, III: 60, IV: 3, Unknown: 10 | G1/G2: 76%; I/II: 68% | Luminal A: 111, luminal B: 46, HER2 +: 19, TNBC: 32 | 82 (39.4); ≥ 10 %; NB100-131 | OS | (U) 1.63 (1.03-2.60), 0.0369 | Reported | NA |
| Malfettone et al[39], 2012, Italy | NA | NA | 187 | 50 | G1: 31, G2: 87, G3: 69; NA | G1/G2: 63%; I/II: NA | NA | 58 (31.0); ≥ 1%; H206 | OS | (U) 1.52 (1.10-2.11), 0.011 | Estimate | NA |
| Ni et al[42], 2013, China | January 2005-October 2006 | NA | 85 of 95 | NA | G1: 16, G2: 35, G3: 24; I-II: 38, III: 37 | G1/G2: 68%; I/II: 51% | NA | 52 (61.2); score ≥ 1; NA | OS | (U) 1.56 (1.01-2.42), 0.045; (M) 2.25 [1.38-6.45], 0.037 | U: Survival curve M: Reported | 60 |
| Peurala et al[54], 2012, Finland | 2002-2005 | NA | 102 | 59 | G1: 30, G2: 35, G3: 37; NA | G1/G2: 63%; I/II: NA | NA | 27 (26.4); score ≥ 2; NA | BCSS | (U) 2.4 (0.75-7.5), 0.126 | Reported | NA |
| Kornegoor et al[13], 2012, Netherlands | 1986-2010 | NA | MBC; 126 | 66 | G1-G2: 81, G3: 44; NA | G1/G2: 50%; I/II: NA | NA | 51 (40.5); ≥ 5%; NA | OS | (M) 2.50 (1.1-5.6), 0.029 | Reported | NA |
| Deb et al[12], 2014, Australia | 1990-2007 | Retrospective | MBC; 286 | 70 | NA | NA; NA | NA | 68 (23.8); score 6; NA | OS | (U) 3.8 (1.5-9.8), 0.006 | Reported | 96 |
| Dales et al[34], 2005, France | 1986-1995 | Retrospective | 745 | 56.1 | NA | NA; NA | NA | 543 (72.9); > 10 %; H206 | OS; DFS | OS: (U) 1.21 (1.03-1.43), 0.019 | U: Survival curve | 162 |
| (M) 1.20 (1.02-1.41), 0.03 | M: Estimate | |||||||||||
| DFS: (M) 1.12 (0.96-1.32), 0.158 | ||||||||||||
| Kronblad et al[37], 2006, Sweden | 1984-1991 | Prospective | 377 | NA | G1: 41, G2: 142, G3: 183, Unknow: 12; NA | NA; NA | NA | 91 (24.1); > 2%; NB100-123H2 | OS; RFS | OS: (U) 1.21 (0.95-1.53), 0.11; RFS: (U) 1.27 (1.00-1.61), 0.048 | Survival curve | 166.8 |
| LN-: 1.17 (0.66-2.09), 0.59 | ||||||||||||
| LN+: 1.69 [1.11-2.57], 0.014 | ||||||||||||
| Bos et al[32], 2003, Netherlands | 1985-1993 | NA | 150 | 60 | G1: 35, G2: 49, G3: 66; NA | G1/G2: 56%; I/II: NA | NA | 51 (34.0); > 5%; H1α67 | OS; DFS | LN+: (M) OS: 6.37 (1.32-30.67), 0.021; (U) 1.21 (1.05-1.39), 0.008; DFS: (M) 4.19 (1.56-12.08), 0.008; (U) 1.38 (1.11-1.72), 0.004 | M: Reported | 106 |
| LN-: (M) OS: 2.16 (0.95-4.89), 0.067; DFS: 1.67 (0.88-3.17), 0.115 | U: Survival curve | |||||||||||
| Gruber et al[29], 2004, Switzerland | August 1988-June 1998 | NA | 77 | 57 | Moderate/G2: 43, poor/G3: 32; NA | G1/G2: 57%; I/II: NA | NA | 43 (55.8); score 1; H1α67 | OS; DFS | (U) OS: 1.34 (0.85-2.11), 0.21 | M: Reported | 36 |
| DFS: 1.61 (1.02-2.55), 0.04 | U: Estimate | |||||||||||
| (M) OS: 2.66 (0.83–8.51), 0.09; DFS: 1.68 (0.62–4.47), 0.30 | ||||||||||||
| Giatromanolaki et al[27], 2022, Greece | 2003-2009 | Prospective | 175 | 61 | NA | NA; NA | Luminal A: 88, luminal B: 47, HER2 +: 24, TNBC: 16 | 39 (22.3); ≥ 50%; ESEE122 | OS, DFS | (U) OS: 2.26 (1.06-4.82), 0.035 | Survival curve | 130 |
| DFS: 1.96 (1.01-3.80), 0.004 | ||||||||||||
| Ong et al[19], 2022, Singapore | 2003-2013 | Retrospective | 307 | 55 | G1/2: 48, G3: 288; NA | G1/G2: 14%; I/II: NA | TNBC | 141 (45.9); score ≥ 1; NA | OS, DFS | (U) OS: 1.06 (0.65-1.75), 0.807 | Estimate | NA |
| DFS: 1.40 (0.90-2.17), 0.137 | ||||||||||||
| Schindl et al[33], 2002, Austria | NA | Prospective | 206 | 52.3 | NA | NA; NA | NA | 48 (23.3); score 3; H1α67 | OS, DFS | OS: (U) 1.89 (1.01-3.53), 0.045; (M) 1.41 (1.12-1.77), 0.003 | M: Reported | 87 |
| DFS: (U) 2.83 (1.54-5.19), 0.0008; (M) 1.40 (1.14-1.72), 0.001 | U: Survival curve | |||||||||||
| Yamamoto et al[43], 2008, Japan | 1993-2001 | NA | 171 | NA | G1: 56, G2: 83, G3: 32 | G1/G2: 81%; I/II: 90% | NA | 63 (36.8); > 5%; H1α67 | OS, DFS | OS: (U) 1.39 (1.17-1.65), 0.0002; (M) 2.15 (1.15-5.76), 0.016 | M: Reported | 60 |
| I: 45, II: 109, III: 17 | DFS: (U) 1.7 (1.67-1.70), < 0.0001 | U: Survival curve | ||||||||||
| 1.59 (1.05-2.43), 0.017 | ||||||||||||
| Lu et al[18], 2022, China | 2017-2019 | NA | 189 | 52.0 | NA; I-II: 91, III- IV: 98 | G1/G2: NA; I/II: 48% | Luminal A: 61, luminal B: 43, HER2 +: 56, TNBC: 29 | 122 (64.6); score ≥ 2; ab51608 | OS; DFS | (M) OS: 1.024 (0.85-1.23), 0.788; DFS: 1.07 (0.88-1.29), 0.506 | Reported | 60 |
| Choi et al[52], 2013, South Korea | January 2000-December 2001 | NA | 276 | 49.0 | G1: 33, G2: 147, G3: 96 | G1/G2: 65%; I/II: NA | Luminal A: 121, luminal B: 33, HER2 +: 25, TNBC: 97 | 13 (4.7); > 10%; EP1215Y | OS; DFS | OS: (U) 1.95 (1.49-2.54), < 0.001; (M) 4.54 (1.46-14.11), 0.009; DFS: (M) 5.21 (1.84-14.78), 0.002 | M: Reported | 67 |
| NA | U: Estimate | |||||||||||
| Dong et al[53], 2013, China | April 1999-October 2008 | NA | 378 | 48.5 | G1: 15, G2: 127, G3: 56, Unknown: 178; NA | G1/G2: 72%; I/II: NA | NA | 195 (50.1); score 4; MAB5382 | OS; DFS | (U) OS: 1.05 (0.81-1.36), 0.714 | Estimate | 73.8 |
| DFS: 1.01 (0.81-1.25), 0.963 | ||||||||||||
| Huang et al, [46], 2014, Taiwan | 1991-2001 | NA | 96 | 46.5 | NA | NA; NA | NA | 29 (30.2); score ≥ 3; NA | OS; DFS | (M): OS: 1.61 (0.63-4.13), 0.322 | Reported | NA |
| DFS: 2.18 (0.93-5.11), 0.073 | ||||||||||||
| Koo and Jung[49], 2010, South Korea | January 2000-December 2001 | NA | 182 of 224 | 48.8 | NA; I: 32, II: 123, III: 69 | G1/G2: NA; I/II: 69% | Luminal A: 115, luminal B: 20, HER2 +: 29, TNBC: 60 | 33 (18.1); ≥ 1%; EP1215Y | OS; RFS | (U): OS: 2.94 (1.98-4.37), < 0.0001 | Survival curve | 89.6 |
| RFS: 2.78 (1.42-5.46), 0.003 | ||||||||||||
| Ramírez-Tortosa et al [20], 2022, Spain | NA | Prospective | 88 of 95 | 20 | G1: 20, G2: 37, G3: 34, unknown: 4 | G1/G2: 63%; I/II: 68% | Luminal A: 31, luminal B: 28, HER2 +: 20, TNBC: 13, Missing: 3 | 35 (39.8); ≥ 5%; NA | OS; DFS | OS: (M) 1.31 (0.81-2.13), 0.295; DFS: (U) 1.70 (1.12-2.57), 0.013; (M) 2.5 (1.0-6.2), 0.047 | DFS: U: Survival curve OS: Estimate | 88.8 |
| Stage: II: 63, III: 29, unknown: 3 | ||||||||||||
| Sato-Tadano et al[15], 2013, China | 2004-2008 | NA | 118 | 57 | NA; I: 68, II: 31, III: 19 | G1/G2: NA; I/II: 84% | Luminal A: 23, luminal B: 15, HER2 +: 4, TNBC: 10 | 62 (52.5); score ≥ 3; NA | BCSS; DFS | (U) BCSS: 1.12 (0.78-1.61), 0.54; DFS: 1.07 (0.74-1.53), 0.72 | Estimate | 57 |
| Trastour et al[30], 2007, France | 1993 | Retrospective | 132 | 62 | G1: 57, G2: 44, G3: 10 | G1/G2: 91%; I/II: NA | NA | 59 (44.7); > 1% | OS, DFS | (U) OS: 1.84 (1.31-2.5), 0.0005; DFS: 1.64 (1.28-2.1), 0.0001 | U: Estimate | 138 |
| (M) OS: 1.25 (0.89-1.76), 0.2 | M: DFS reported | |||||||||||
| NA | Antiserum 2087 | DFS: 4.2 [2.1-8.5], < 0.001 | M: OS estimate | |||||||||
| Schoppmann et al[40], 2006, Austria | NA | Prospective | 119 | 50.9 | G1: 9, G2: 54, G3: 56 | G1/G2: 53%; I/II: NA | NA | 30 (25.2); score 3; H1α67 | OS; DFS | OS: (U) 1.60 (1.05-2.41), 0.027; DFS: (U) 1.59 (1.05-2.40), 0.029; (M) 1.61 (1.06-2.43), 0.025 | Estimate | 110 |
| NA | ||||||||||||
| Yan et al[36], 2009, Australia | NA | NA | 125 | NA | G1: 7, G2: 30, G3: 67, Unknow: 19; NA | G1/G2: 36%; I/II: NA | Luminal: 49, HER2 +: 6, TNBC: 37 | 55 (44.0); score 1; NA | DFS | (U) 3.25 (1.01-10.51), 0.049; | Survival curve | 64 |
| Nie et al[51], 2018, China | January 2013-December 2014 | NA | 220 | 46 | NA; II: 54, III: 166 | G1/G2: NA; I/II: 25% | Luminal A: 8, luminal B: 127, HER2 +: 50, TNBC: 35 | 150 (68.2); score ≥ 1; NA | DFS | (M) 4.17 (1.01–17.17), 0.048 | Reported | 220 |
| Chen et al[47], 2007, Taiwan | 1988-2002 | Retrospective | 104 | NA | G1: 33, G2: 43, G3: 28; NA | G1/G2: 73%; I/II: NA | NA | 47 (45.2); score 3; H1α67 | DFS | (U): 3.82 (2.14-6.84), < 0.0001 | Reported | > 120 |
| Cui and Jiang[45], 2019, China | January 2012-December 2015 | Retrospective | 87 of 126 | 50.4 | G1: 5, G2: 48, G3: 34 | G1/G2: 70%; I/II: 84% | TNBC | 36 (41.4); score ≥ 1; ab51608 | DFS | (U): 2.03 (1.36–3.51), < 0.001; (M): 2.22 (1.47–3.77), < 0.001 | Reported | 24 |
| I: 19, II: 54, III: 14 | ||||||||||||
| Generali et al[35], 2006, Italy | January 1997-December 2001 | Prospective | 187 | NA | G2: 95; G3: 135; missing: 3 | G1/G2: 41%; I/II: NA | NA | 138 (73.8); score ≥ 1; ESEE122 | DFS | (U) 1.83 (1.10-3.04), 0.02 | Survival curve | 53 |
| NA | ER +: 1.39 (1.00-1.92), 0.05 | |||||||||||
| Jin et al[44], 2016, South Korea | 2003-2006 | NA | 270 | NA | G2: 59, G3: 211; NA | G1/G2: 22%; I/II: NA | TNBC | 39 (14.4); ≥ 1%; NA | DFS | (U) 1.26 (1.04-1.52), 0.017 | U: Survival curve M: Reported | NA |
| (M) 2.62 (1.33-5.15), 0.05 | ||||||||||||
| Kuijper et al[14], 2005, Netherlands | NA | NA | 37 | NA | NA | NA; NA | NA | 15 (40.5); ≥ 1%; NA | DFS | (U) 4.39 (1.14-16.99), 0.032 | Survival curve | NA |
| Vleugel et al[28], 2005, Netherlands | NA | NA | 200 | NA | G1: 61, G2: 78, G3: 61; NA | G1/G2: 70%; I/II: NA | NA | 88 (44.0); ≥ 1%; NA | RFS | (M) 2.23 (1.18-4.21), 0.01 | Reported | 105 |
| Shi et al[50], 2017, China | September 2004-September 2008 | Retrospective | 60 | 53 | NA; I: 20, II: 28, III: 12 | G1/G2: NA; I/II: 80% | NA | 20 (33.3); ≥ 5%; ab85886 | DFS | (U): 4.76 (2.17, 10.44), < 0.001 | Survival curve | 60 |
| Tan et al[16], 2007, United Kingdom | NA | NA | 295 | 57 | G1: 37, G2: 66, G3: 50; NA | G1/G2: 67%; I/II: NA | NA | 125 (42.4); score ≥ 2; ESEE122 | DFS | (U) 1.60 (1.02-2.42), 0.04 | Reported | 105 |
| Shamis et al[21], 2022, United Kingdom | 1995-1998 | NA | Cohort I: 289 of 373 | NA | NA, | NA; NA | ER + | 39 (13.5); NA; H1α67 | DFS | (U) 1.52 (1.08-2.13), 0.015 | Survival curve | > 12 |
| Marton et al[38], 2012, Croatia | 2001-2005 | NA | 31 | 61.7 | G1: 7, G2: 19, G3: 5; NA | G1/G2: 84%; I/II: NA | NA | 7 (22.6); score ≥ 1; NA | DFS | (U) 2.20 (0.95-5.11), 0.066 | Estimate | 144 |
| (M) 2.74 (1.18-6.36), 0.019 |
Forty studies reported 57 analyses, including 28 analyses (n = 5833) of the connection linking HIF-1α expression with OS in breast cancer patients and 29 analyses (n = 5691) of the connections linking HIF-1α expression to DFS among individuals with breast cancer. The most commonly reported cutoff values for HIF-1α expression were nuclear staining exceeding 1% (n = 7) and 5% (n = 5), as well as HIF-1α expression scores (adopting a scoring approach that merges stain intensity with positive cell percentage) exceeding 1 (n = 8) and 3 (n = 6). There were differences in the antibodies employed across the various studies, with 12 different antibodies involved. The most common was HIFα67 (n = 6)[29,32,33,40,43,47], followed by EP1215Y (n = 4)[17,31,49,52].
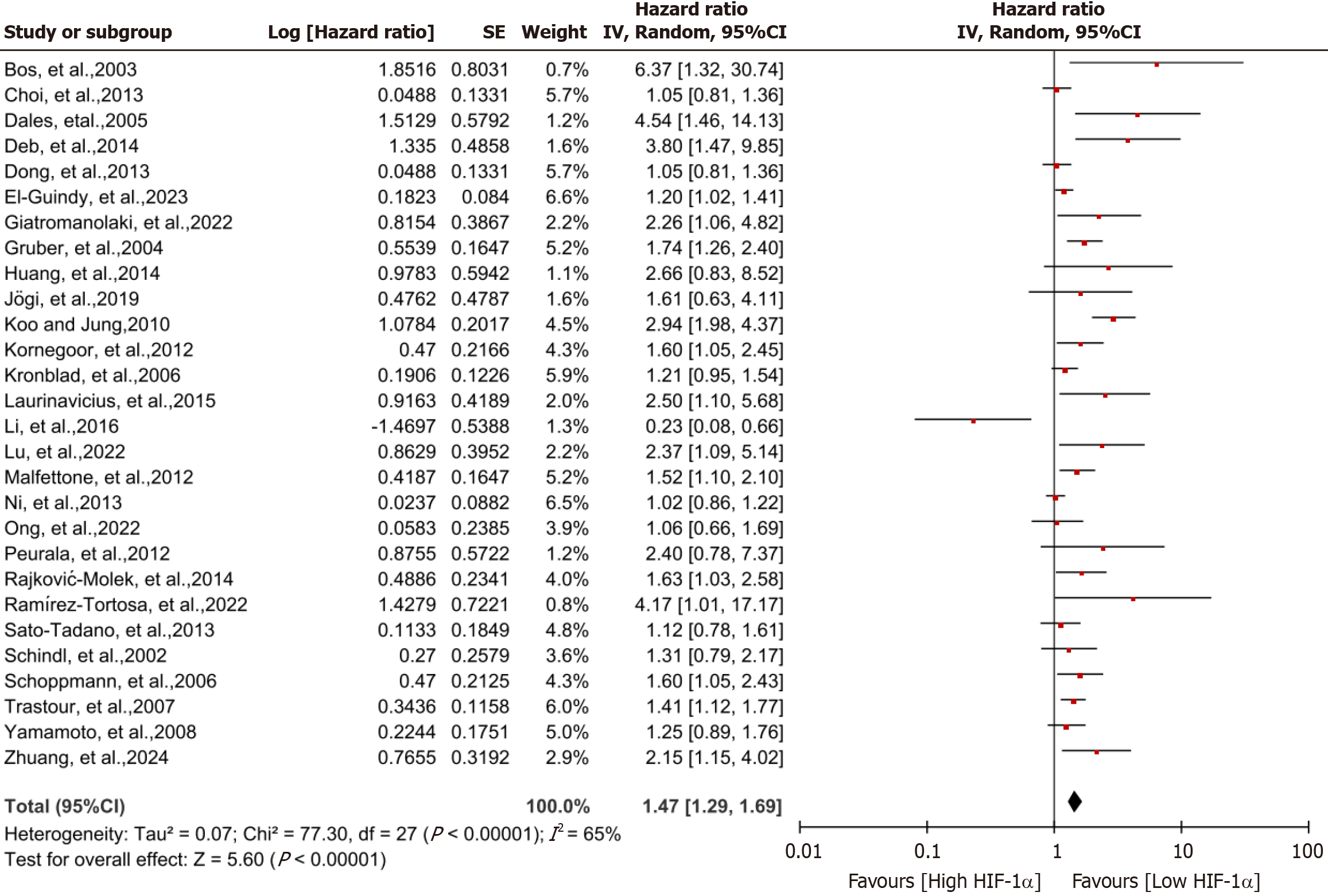
Effect of HIF-1α on OS of breast cancer patients: Figure 2 displays the forest plot of HIF-1α expression among indi
| Analysis | Number of studies | Pooled HR ratio (95%CI) | I2 statistic (%) | χ2 P value for heterogeneity | Analytical model | P value for overall effect |
| Primary analyses | ||||||
| OS | 28 | 1.47 (1.29-1.69) | 65 | < 0.00001 | REM | < 0.00001 |
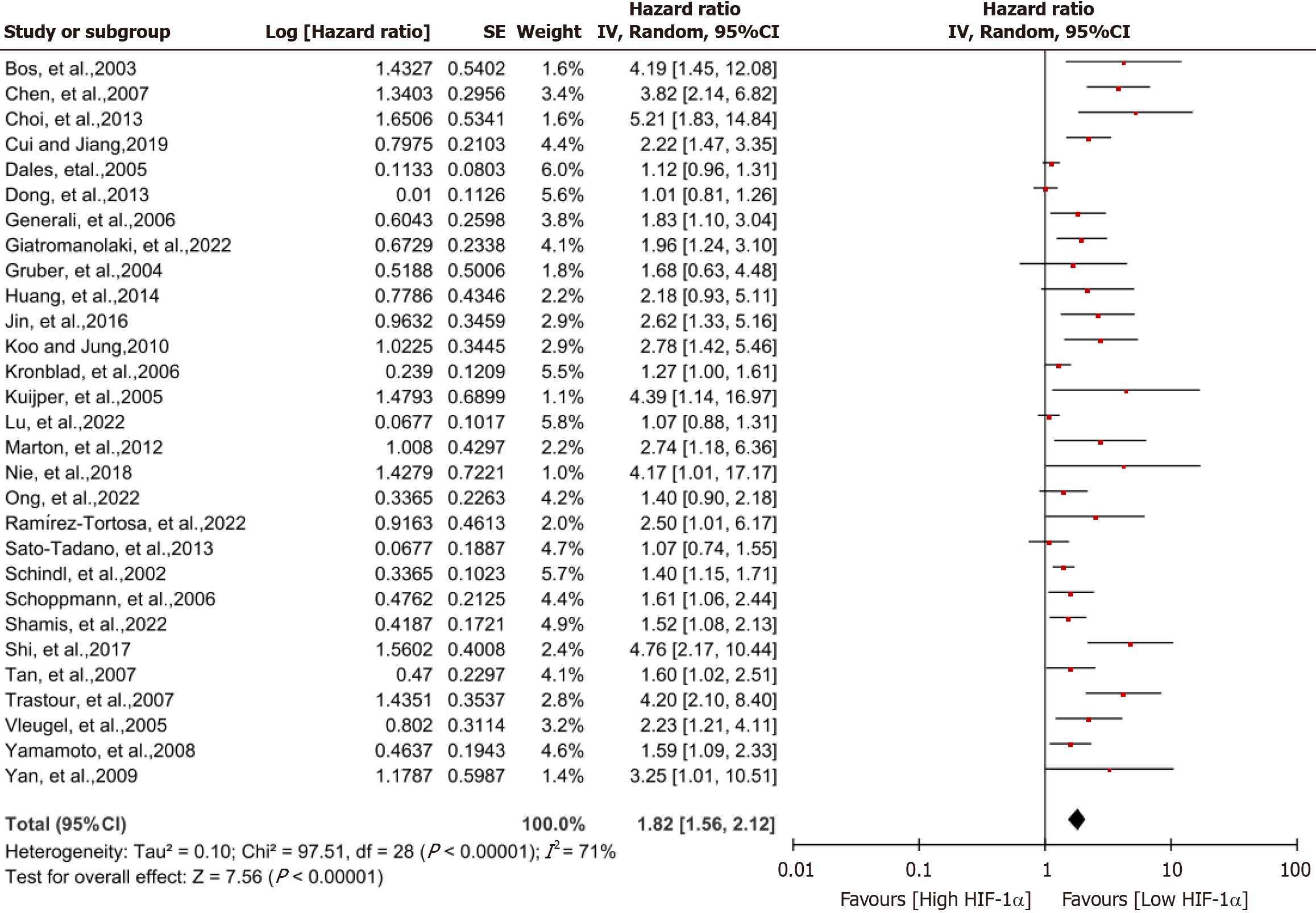
| DFS | 29 | 1.82 (1.56-2.12) | 71 | < 0.00001 | REM | < 0.00001 |
| Sensitivity analyses | ||||||
| OS | ||||||
| Exclusion of study with the largest effect size[42] | 27 | 1.51 (1.31-1.74) | 62 | < 0.0001 | REM | < 0.00001 |
| Sample size ≥ 200[12,19,26,33,34,37,38,52,53] | 9 | 1.20 (1.06-1.36) | 48 | 0.05 | FEM | 0.004 |
| NOS scoring ≥ 7[12,13,19,20,32,37,53] | 7 | 1.46 (1.08-1.97) | 62 | 0.02 | REM | 0.02 |
| DFS | ||||||
| Exclusion of study with the largest effect size[34] | 28 | 1.88 (1.60-2.22) | 69 | < 0.00001 | REM | < 0.00001 |
| Sample size ≥ 200[16,19,21,28,33,34,37,44,51-53] | 11 | 1.44 (1.21-1.71) | 63 | 0.003 | REM | < 0.0001 |
| NOS scoring ≥ 7[16,19-21,32,36,37,45,53] | 9 | 1.57 (1.24-1.99) | 63 | 0.005 | REM | 0.0002 |
Effects of HIF-1α on recurrence-free and metastatic survival of breast cancer patients: Figure 4 presents a forest plot describing the correlation linking HIF-1α expression to DFS in breast cancer patients. High HIF-1α expression was related to a greater risk of recurrence and metastasis, resulting in a shorter DFS (HR = 1.82; 95%CI: 1.56-2.12, I2 = 71%; P < 0.00001). This finding substantiates the idea that high HIF-1α expression serves as a prognostic factor influencing recurrence and metastasis in breast cancer (Table 2).
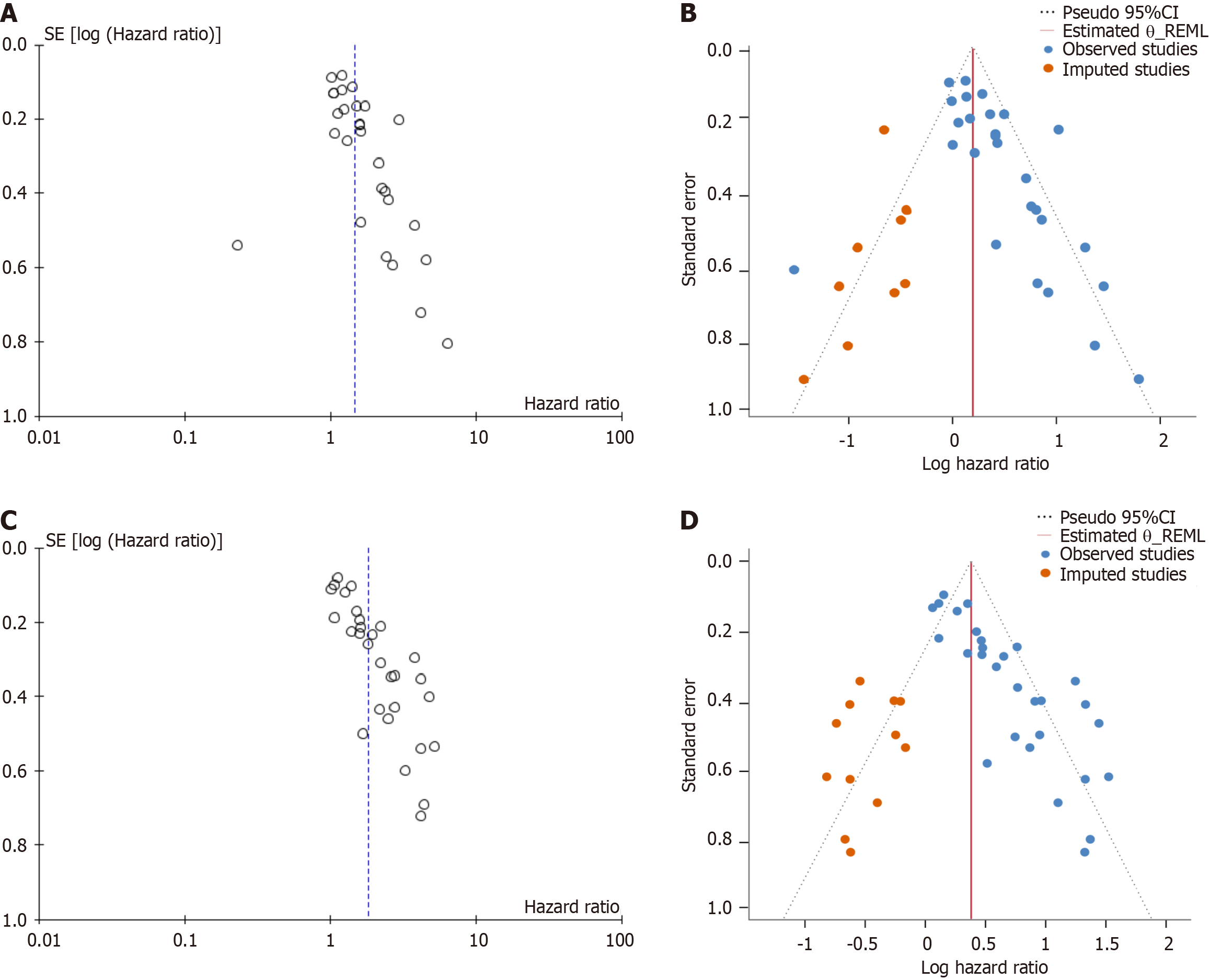
A leave-one-out sensitivity analysis was employed. Excluding the study that has the greatest effect size[34], and this did not change the influence of HIF-1α on DFS (HR = 1.88; 95%CI: 1.60-2.22; I2 = 69%; P < 0.00001). Sensitivity analyses restricted to large sample sizes or high-quality studies alone also did not alter the overall HR (Table 2). The funnel plot exhibited asymmetry, with the calculated P value (P = 0.001) from Egger’s asymmetry test falling below 0.05, highlighting possible selection bias in the systematic review of DFS interval studies (Figure 3C). Furthermore, the trim-and-fill method analysis revealed that twelve missing studies were input to account for potential bias in the DFS analysis (Figure 3D). However, after adjusting for publication bias, the significant overall meta-analysis effect size suggests the presence of limited or non-significant publication bias in the DFS studies.
Subgroup analysis of the effect of HIF-1α on survival of breast cancer patients: Table 3 and Table 4 present the detailed outcomes of the subgroup analysis, suggesting that age, study design, publication year, HIF-1α expression cutoff value, tumor stage, and tumor grade do not influence the connection linking HIF-1α expression to OS or DFS in breast cancer patients. In other words, HIF-1α expression served as a prognostic factor in various subgroups (P > 0.05) (Tables 3 and 4).
| Stratified analysis | Number of studies | Pooled HR ratio (95%CI) | I2 statistic (%) | χ2 P value for heterogeneity | Analytical model | χ2 P value for subgroup differences |
| Age (median) | ||||||
| < 53[17,18,20,33,39,40,46,48,49,52,53] | 11 | 1.35 (1.21-1.51) | 76 | < 0.00001 | REM | - |
| ≥ 53[12,13,15,19,27,29,30,32,34,38,54] | 11 | 1.52 (1.33-1.74) | 45 | 0.05 | FEM | 0.18 |
| Location | ||||||
| Asia[15,18,19,22,42,43,46,48,49,52,53] | 11 | 1.55 (1.39-1.73) | 31 | 0.11 | FEM | - |
| Europe[12,13,20,26,27,29-34,37-40,54] | 16 | 1.58 (1.39-1.79) | 29 | 0.14 | FEM | |
| Africa[17] | 1 | 1.20 (1.02-1.41) | NA | - | - | 0.002 |
| Antibody | ||||||
| H1α67[29,32,33,40,43] | 5 | 1.52 (1.26-1.83) | 27 | 0.24 | FEM | - |
| EP1215Y[17,31,49,52] | 4 | 1.42 (1.22-1.64) | 87 | < 0.0001 | REM | - |
| ab51608[18,22] | 2 | 2.23 (1.37-3.64) | 0 | 0.85 | FEM | - |
| H206[34,39] | 2 | 1.65 (1.21-2.25) | 70 | 0.07 | REM | - |
| NB100- 123H2[37] | 1 | 1.21 (0.95-1.54) | NA | - | - | - |
| NB100- 131[38] | 1 | 1.63 (1.03-2.58) | NA | - | - | - |
| BD610959[26] | 1 | 1.61 (0.63-4.11) | NA | - | - | - |
| Ab82832[48] | 1 | 0.23 (0.08-0.66) | NA | - | - | - |
| Antiserum 2087[30] | 1 | 1.41 (1.12-1.77) | NA | - | - | - |
| MAB5382[53] | 1 | 1.05 (0.81-1.36) | NA | - | - | - |
| ESEE122[27] | 1 | 2.26 (1.06-4.82) | NA | - | - | 0.16 |
| Cut-off value | ||||||
| Percentage ≥ 1%[26,30,37,39,49] | 5 | 1.49 (1.30-1.71) | 72 | 0.006 | REM | - |
| Percentage ≥ 5%[13,17,20,27,32,34,38,43,52] | 9 | 1.36 (1.20-1.55) | 59 | 0.01 | REM | - |
| Scoring ≥ 1 [18,19,29,31,42,54] | 6 | 1.21 (1.05-1.39) | 69 | 0.007 | REM | - |
| Scoring ≥ 3[12,15,22,33,40,46,48,53] | 8 | 1.25 (1.06-1.47) | 70 | 0.002 | REM | 0.71 |
| Study design | ||||||
| Retrospective[12,19,30,34,38] | 5 | 1.48 (1.23-1.77) | 59 | 0.04 | REM | - |
| Prospective[17,20,27,33,37,40] | 6 | 1.27 (1.13-1.44) | 26 | 0.24 | FEM | 0.18 |
| Publish date | ||||||
| Before 2013[13,15,29,30,32-34,37,39,40,42,43,49,52-54] | 16 | 1.32 (1.22-1.44) | 68 | < 0.0001 | REM | - |
| After 2013[12,17-20,22,26,27,31,38,46,48] | 12 | 1.35 (1.19-1.54) | 64 | 0.001 | REM | 0.53 |
| Grading | ||||||
| G1/G2 < 65%[13,17,19,20,29,32,37,39,40,54] | 10 | 1.35 (1.21-1.50) | 40 | 0.09 | FEM | - |
| G1/G2 ≥ 65%[22,30,31,38,42,43,48,52,53] | 9 | 1.19 (1.07-1.33) | 71 | 0.0005 | REM | 0.54 |
| Staging | ||||||
| I/II < 70%[18,20,38,42,48,49] | 6 | 1.26 (1.09-1.45) | 87 | < 0.0001 | REM | - |
| I/II ≥ 70%[15,22,26,43] | 4 | 1.32 (1.03-1.69) | 11 | 0.34 | REM | 0.69 |
| Molecular typing | ||||||
| TNBC[17,19] | 2 | 1.18 (1.01-1.38) | 0 | 0.62 | FEM | - |
| Gender | ||||||
| Male[12,13] | 2 | 1.85 (1.25-2.72) | 62 | 0.1 | REM | - |
| Female[15,17-20,22,26,27,29-34,37-40,42,43,46,48,49,52-54] | 26 | 1.29 (1.21-1.38) | 65 | < 0.00001 | REM | 0.08 |
| Stratified analysis | Number of studies | Pooled HR ratio (95%CI) | I2 statistic (%) | χ2 P value for heterogeneity | Analytical model | χ2 P value for subgroup differences |
| Age median | ||||||
| ≤ 53[18,20,33,40,45,46,49,51-53] | 10 | 1.32 (1.18-1.47) | 74 | < 0.001 | REM | - |
| >53[15,16,19,27,29,30,32,34,41,50] | 10 | 1.35 (1.20-1.53) | 76 | < 0.001 | REM | 0.74 |
| Location | ||||||
| Asia[15,18,19,43-47,49-53] | 13 | 1.37 (1.23-1.53) | 80 | < 0.001 | REM | - |
| Europe[14,16,20,21,27-30,32-37,40,41] | 16 | 1.73 (1.45-2.07) | 60 | 0.001 | REM | 0.51 |
| Antibody | ||||||
| H1α67[29,32,33,40,43,47] | 6 | 1.61 (1.38-1.87) | 63 | 0.02 | REM | - |
| EP1215Y[49,52] | 2 | 3.34 (1.90-5.90) | 0 | 0.32 | FEM | - |
| ESEE122[16,27,35] | 3 | 1.79 (1.36-2.34) | 0 | 0.82 | FEM | - |
| ab51608[18,45] | 2 | 1.23 (1.03-1.47) | 90 | 0.002 | REM | - |
| H206[34] | 1 | 1.12 (0.96-1.31) | NA | - | - | - |
| MAB5382[53] | 1 | 1.01 (0.81-1.26) | NA | - | - | - |
| NB100- 123H2[37] | 1 | 1.27 (1.00-1.61) | NA | - | - | - |
| ab85886[50] | 1 | 4.76 (2.17-10.44) | NA | - | - | - |
| Antiserum 2087[30] | 1 | 4.20 (2.10-8.40) | NA | - | - | 0.0004 |
| Cut-off value | ||||||
| Percentage ≥ 1%[14,28,30,37,44,49] | 6 | 1.70 (1.40-2.05) | 73 | 0.002 | REM | - |
| Percentage ≥ 5%[20,27,32,34,43,50,52] | 7 | 1.36 (1.19-1.55) | 81 | < 0.001 | REM | - |
| Scoring ≥ 1[16,18,19,29,35,36,41,45,51] | 9 | 1.40 (1.21-1.61) | 60 | 0.01 | REM | - |
| Scoring ≥ 3[15,33,40,46,47,53] | 6 | 1.31 (1.15-1.48) | 78 | 0.0005 | REM | 0.25 |
| Study design | ||||||
| Retrospective[19,30,34,45,47,50] | 6 | 1.42 (1.25-1.62) | 88 | < 0.01 | REM | - |
| Prospective[20,27,33,35,37,40] | 6 | 1.46 (1.28-1.66) | 8 | 0.37 | FEM | 0.79 |
| Recruitment period | ||||||
| Before 2013[14-16,21,28-30,32-37,40,41,43,47,49,52,53] | 19 | 1.37 (1.26-1.48) | 73 | < 0.001 | REM | - |
| After 2013[18-21,27,44-46,50,51] | 10 | 1.29 (1.15-1.44) | 71 | 0.0003 | REM | 0.63 |
| Grading | ||||||
| G1/G2 < 65%[19,20,29,32,35-37,40,44,45] | 10 | 1.63 (1.40-1.89) | 37 | 0.12 | FEM | - |
| G1/G2 ≥ 65%[16,28,30,41,43,47,52,53] | 8 | 2.25 (1.47-3.45) | 82 | < 0.001 | REM | 0.35 |
| Staging | ||||||
| I/II < 70%[18,20,49,51] | 4 | 1.22 (1.01-1.47) | 76 | 0.005 | REM | - |
| I/II ≥ 70%[15,43,45,50] | 4 | 1.65 (1.33-2.05) | 79 | 0.003 | REM | 0.85 |
| Molecular typing | ||||||
| TNBC[19,44,45] | 3 | 1.79 (1.03-3.11) | 72 | 0.03 | REM | - |
The meta-analysis results from four studies[17,19,44,45] on triple-negative breast cancer verified that HIF-1α expression remains a prognostic factor for these patients. Analysis of two studies[17,19] focusing on OS revealed that the OS of patients with high HIF-1α expression is shorter than that of patients with low expression (HR = 1.18; 95%CI: 1.01-1.38, I2 = 0%; P = 0.03). Three studies[19,44,45] focusing on DFS indicated that patients exhibiting high HIF-1α levels have significantly shorter DFS compared to those with low expression (HR = 1.79; 95%CI: 1.03-3.11, I2 = 72%; P = 0.03) (Tables 3 and 4).
Furthermore, the antibody that was used to detect HIF-1α expression can influence the relationship between HIF-1α expression and breast cancer prognosis. The impact of HIF-1α expression detected with the EP1215Y antibody on DFS was greater than that of HIF-1α expression detected with other antibodies (P = 0.0004) (Table 4). However, the effect of the antibody that was used to detect HIF-1α expression on OS was not significant (P = 0.16) (Table 3). Additionally, the region where the study population was located had a significant effect on the correlation linking HIF-la expression with OS in patients with breast cancer (P = 0.002) (Table 3). It is worth noting that two studies[12,13] on male breast cancer revealed that HIF-1α expression remained a prognostic factor for patients, with a shorter OS observed in patients with high HIF-1α expression (HR = 1.85; 95%CI: 1.25-2.72, I2 = 62%, P = 0.002) (Table 3).
Identifying the risk of disease recurrence and death among breast cancer patients is crucial for guiding the monitoring of recurrence and risk of death and for selecting adjuvant therapies. However, accurately identifying these risks for individual patients remains challenging[55]. HIF-1α serves as the master transcriptional driver enabling cellular adaptation under hypoxia, and it influences the initiation and progression of breast cancer through various mechanisms. HIF-1α can regulate breast cancer invasion, metastasis, and immune evasion. Numerous clinical prognostic studies have confirmed that HIF-1α expression can affect the prognosis of breast cancer patients[10]. This systematic review and meta-analysis comprehensively searched and systematically analyzed 40 published studies examining the correlation linking HIF-1α expression with the risk of recurrence and death in breast cancer patients. These findings indicate that compared to patients with low HIF-1α expression, those with high HIF-1α expression not only have an increased risk of recurrence but also a significantly elevated risk of death. Notably, this systematic review combined with meta-analysis additionally validated the notable prognostic significance of HIF-1α expression within triple-negative breast cancer, which has the worst prognosis. Thus, this study represents the most up-to-date and comprehensive evaluation of the prognostic role of HIF-1α expression in breast cancer patients, and it is believed that this study will provide valuable evidence for the prediction of breast cancer prognosis.
Hypoxia can regulate breast cancer invasion, metastasis, and immune evasion through multiple mechanisms, thereby influencing breast cancer progression and prognosis. First, under hypoxic conditions, cancer cells overexpress HIF-1α, upregulate the transcription of angiogenesis-related factors such as vascular endothelial growth factor (VEGF) and platelet-derived growth factor, and promote breast cancer progression by regulating angiogenesis[56,57]. Second, hypoxia can induce the transcription of drug resistance-related genes, such as MDR1, MRP1, and BRCP, leading to chemotherapy resistance and enabling breast cancer cells to evade the effects of cytotoxic drugs, resulting in metastasis[58,59]. Furthermore, HIF-1α can induce epithelial-mesenchymal transition and establish an immunosuppressive tumor microenvironment; during these processes, cancer cell intercellular contacts are disrupted, and these cells acquire motility, accelerating tumor metastasis[60]. Metastasis significantly increases the risk of mortality. The results of this systematic review and meta-analysis revealed that patients of high HIF-1α expression breast cancer are more prone to recurrence and metastasis than those with low expression, suggesting that upregulated HIF-1α expression may affect breast cancer recurrence and metastasis by regulating angiogenesis and drug resistance, thereby influencing the prognosis of breast cancer. Therefore, targeting HIF-1α or VEGF may help facilitate vascular network stabilization in neoplastic tissues, enhance chemotherapeutic agent penetration, mitigate oxygen-deprived tumor niches and immune-evasive milieus, counteract therapeutic resistance mechanisms, and potentially reverse the prognosis of breast cancer recurrence and metastasis[61]. Research has demonstrated that HIF-1α is involved in multiple signaling pathways, such as the HIF-1, Wnt, and VEGF signaling pathways, and regulates the proliferation and invasion of triple-negative breast cancer cells by recruiting coactivator associated arginine methyltransferase 1[62]. Notably, the findings of this systematic review indicate that triple-negative breast cancer patients with high HIF-1α expression have shorter OS and DFS, suggesting a role of HIF-1α in promoting tumor effects, specifically in triple-negative breast cancer recurrence and metastasis. However, subgroup analysis for other molecular subtypes was not possible due to the lack of studies analyzing the prognostic implications for HIF-1α expression in other molecular subtypes of breast cancer separately. Nevertheless, based on the prognostic significance of HIF-1α in the overall population of breast cancer patients, it can be inferred that HIF-1α also has a prognostic impact on other molecular subtypes of breast cancer.
The results of this meta-analysis suggest that breast cancer patients exhibiting high levels of HIF-1α have a higher mortality rate compared to those with lower expression levels. Among the different age subgroups, although the mo
Furthermore, the choice of antibodies that are used to detect HIF-1α expression can affect the ability of HIF-1α expression to predict DFS in breast cancer patients. Therefore, it is recommended to refer to the evidence from this systematic review when selecting antibodies for the detection of HIF-1α expression in studies examining its prognostic role in breast cancer to ensure the selection of antibodies with more stable prognostic significance. Additionally, evidence from prospective cohort studies on the predictive significance of HIF-1α expression for breast cancer prognosis is more consistent, suggesting that rigorous and well-designed study protocols can ensure the consistency and accuracy of prognostic research results. This finding also supports the evidence from subgroup analyses of prospective cohort studies, which are more compelling. Sensitivity analyses performed using the leave-one-out method and by omitting small or low-quality studies did not reveal any significant influence of individual studies or specific types of studies on the overall meta-analysis results, confirming the stability of the findings of this meta-analysis.
Although we comprehensively evaluated the correlation linking HIF-1α expression in breast cancer with patient prognosis, this systematic review and meta-analysis have some limitations. First, there are differences in the cutoff values used in the included studies, so the determination of high HIF-1α expression may vary significantly across studies. This may affect the true correlation that is linking HIF-1α expression levels to breast cancer prognosis, resulting in substantial heterogeneity among studies. Second, due to the lack of detailed data for individual molecular subtypes of breast cancer in the included studies, accurately predicting the impact of different molecular subtypes on the established linkage HIF-1α overexpression and patient prognosis was not possible. Furthermore, restricting the analysis to studies published in Chinese or English may lead to language bias. Thus, considering these factors, we recommend that the results of this meta-analysis be interpreted with caution. Moreover, to improve the quality of prognostic tumor marker reports, we recommend following the Reporting Recommendations for Tumor Marker Prognostic Studies guidelines when new related research is conducted.
In summary, this meta-analysis confirms that HIF-1α expression can influence the prognosis of patients with breast cancer. Higher HIF-1α expression is associated with shorter DFS and OS. Hypoxia, which is a significant characteristic of cancer, plays a crucial role in the initiation and progression of breast cancer. Our study revealed that HIF-1α, which is a key indicator of hypoxia in the tumor microenvironment, serves as a prognostic marker for adverse outcomes in breast cancer patients. These findings offer valuable insights for evaluating the biological behavior of breast cancer and guiding decisions on adjuvant treatment. Nevertheless, although the overall meta-analysis suggests that HIF-1α expression affects the prognosis of all breast cancer patients, due to the limited molecular subtype information in the included studies, we can only precisely assess the risk of recurrence, metastasis, and death in a subgroup of triple-negative breast cancer patients exhibiting high HIF-1α expression. Hence, designing more rigorous studies to determine the prognostic si
The authors wish to acknowledge Dr. Ping ZG, Professor of College of Public Health, Zhengzhou University, for his help in guiding and reviewing the statistical methods of this study.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 7677] [Article Influence: 7677.0] [Reference Citation Analysis (2)] |
| 2. | Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2279] [Cited by in RCA: 4524] [Article Influence: 4524.0] [Reference Citation Analysis (3)] |
| 3. | Saxena K, Jolly MK, Balamurugan K. Hypoxia, partial EMT and collective migration: Emerging culprits in metastasis. Transl Oncol. 2020;13:100845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 135] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 4. | Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1050] [Cited by in RCA: 1094] [Article Influence: 99.5] [Reference Citation Analysis (0)] |
| 5. | Liu X, Xie P, Hao N, Zhang M, Liu Y, Liu P, Semenza GL, He J, Zhang H. HIF-1-regulated expression of calreticulin promotes breast tumorigenesis and progression through Wnt/β-catenin pathway activation. Proc Natl Acad Sci U S A. 2021;118:e2109144118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 6. | Wang R, Godet I, Yang Y, Salman S, Lu H, Lyu Y, Zuo Q, Wang Y, Zhu Y, Chen C, He J, Gilkes DM, Semenza GL. Hypoxia-inducible factor-dependent ADAM12 expression mediates breast cancer invasion and metastasis. Proc Natl Acad Sci U S A. 2021;118:e2020490118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 7. | Wicks EE, Semenza GL. Hypoxia-inducible factors: cancer progression and clinical translation. J Clin Invest. 2022;132:e159839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 313] [Article Influence: 104.3] [Reference Citation Analysis (0)] |
| 8. | Zuo Q, Yang Y, Lyu Y, Yang C, Chen C, Salman S, Huang TY, Wicks EE, Jackson W 3rd, Datan E, Qin W, Semenza GL. Plexin-B3 expression stimulates MET signaling, breast cancer stem cell specification, and lung metastasis. Cell Rep. 2023;42:112164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 9. | Zhao Z, Mu H, Li Y, Liu Y, Zou J, Zhu Y. Clinicopathological and prognostic value of hypoxia-inducible factor-1α in breast cancer: a meta-analysis including 5177 patients. Clin Transl Oncol. 2020;22:1892-1906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Shamis SAK, McMillan DC, Edwards J. The relationship between hypoxia-inducible factor 1α (HIF-1α) and patient survival in breast cancer: Systematic review and meta-analysis. Crit Rev Oncol Hematol. 2021;159:103231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 11. | Wang W, He YF, Sun QK, Wang Y, Han XH, Peng DF, Yao YW, Ji CS, Hu B. Hypoxia-inducible factor 1α in breast cancer prognosis. Clin Chim Acta. 2014;428:32-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Deb S, Johansson I, Byrne D, Nilsson C; kConFab Investigators, Constable L, Fjällskog ML, Dobrovic A, Hedenfalk I, Fox SB. Nuclear HIF1A expression is strongly prognostic in sporadic but not familial male breast cancer. Mod Pathol. 2014;27:1223-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Kornegoor R, Verschuur-Maes AH, Buerger H, Hogenes MC, de Bruin PC, Oudejans JJ, Hinrichs B, van Diest PJ. Fibrotic focus and hypoxia in male breast cancer. Mod Pathol. 2012;25:1397-1404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Kuijper A, van der Groep P, van der Wall E, van Diest PJ. Expression of hypoxia-inducible factor 1 alpha and its downstream targets in fibroepithelial tumors of the breast. Breast Cancer Res. 2005;7:R808-R818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Sato-Tadano A, Suzuki T, Amari M, Takagi K, Miki Y, Tamaki K, Watanabe M, Ishida T, Sasano H, Ohuchi N. Hexokinase II in breast carcinoma: a potent prognostic factor associated with hypoxia-inducible factor-1α and Ki-67. Cancer Sci. 2013;104:1380-1388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Tan EY, Campo L, Han C, Turley H, Pezzella F, Gatter KC, Harris AL, Fox SB. Cytoplasmic location of factor-inhibiting hypoxia-inducible factor is associated with an enhanced hypoxic response and a shorter survival in invasive breast cancer. Breast Cancer Res. 2007;9:R89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | El-Guindy DM, Ibrahim FM, Ali DA, El-Horany HE, Sabry NM, Elkholy RA, Mansour W, Helal DS. Hypoxia-induced autophagy in triple negative breast cancer: association with prognostic variables, patients' survival and response to neoadjuvant chemotherapy. Virchows Arch. 2023;482:823-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 18. | Lu XL, Zhan R, Zhao GM, Qian ZH, Gong CC, Li YQ. Expression of CDK13 Was Associated with Prognosis and Expression of HIF-1α and beclin1 in Breast Cancer Patients. J Invest Surg. 2022;35:442-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Ong CHC, Lee DY, Lee B, Li H, Lim JCT, Lim JX, Yeong JPS, Lau HY, Thike AA, Tan PH, Iqbal J. Hypoxia-regulated carbonic anhydrase IX (CAIX) protein is an independent prognostic indicator in triple negative breast cancer. Breast Cancer Res. 2022;24:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 20. | Ramírez-Tortosa CL, Alonso-Calderón R, Gálvez-Navas JM, Pérez-Ramírez C, Quiles JL, Sánchez-Rovira P, Jiménez-Morales A, Ramírez-Tortosa M. Hypoxia-Inducible Factor-1 Alpha Expression Is Predictive of Pathological Complete Response in Patients with Breast Cancer Receiving Neoadjuvant Chemotherapy. Cancers (Basel). 2022;14:5393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 21. | Shamis SAK, Quinn J, Mallon EEA, Edwards J, McMillan DC. The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer. J Histochem Cytochem. 2022;70:479-494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 22. | Zhuang JY, Huang ZN, Weng ZJ, Liu MM, Huang XQ, He D, Shao CK, Dong M. Expression and clinical significance of hypoxia-induced long non-coding RNA TCONS_I2_00001955 in breast cancer. Breast Cancer. 2024;31:317-328. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 23. | Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34245] [Cited by in RCA: 40323] [Article Influence: 1440.1] [Reference Citation Analysis (2)] |
| 24. | Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4738] [Cited by in RCA: 4941] [Article Influence: 274.5] [Reference Citation Analysis (0)] |
| 25. | Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7948] [Cited by in RCA: 9044] [Article Influence: 361.8] [Reference Citation Analysis (0)] |
| 26. | Jögi A, Ehinger A, Hartman L, Alkner S. Expression of HIF-1α is related to a poor prognosis and tamoxifen resistance in contralateral breast cancer. PLoS One. 2019;14:e0226150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 27. | Giatromanolaki A, Gkegka AG, Pouliliou S, Biziota E, Kakolyris S, Koukourakis M. Hypoxia and anaerobic metabolism relate with immunologically cold breast cancer and poor prognosis. Breast Cancer Res Treat. 2022;194:13-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 28. | Vleugel MM, Greijer AE, Shvarts A, van der Groep P, van Berkel M, Aarbodem Y, van Tinteren H, Harris AL, van Diest PJ, van der Wall E. Differential prognostic impact of hypoxia induced and diffuse HIF-1alpha expression in invasive breast cancer. J Clin Pathol. 2005;58:172-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 180] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 29. | Gruber G, Greiner RH, Hlushchuk R, Aebersold DM, Altermatt HJ, Berclaz G, Djonov V. Hypoxia-inducible factor 1 alpha in high-risk breast cancer: an independent prognostic parameter? Breast Cancer Res. 2004;6:R191-R198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 94] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 30. | Trastour C, Benizri E, Ettore F, Ramaioli A, Chamorey E, Pouysségur J, Berra E. HIF-1alpha and CA IX staining in invasive breast carcinomas: prognosis and treatment outcome. Int J Cancer. 2007;120:1451-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 185] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 31. | Laurinavicius A, Green AR, Laurinaviciene A, Smailyte G, Ostapenko V, Meskauskas R, Ellis IO. Ki67/SATB1 ratio is an independent prognostic factor of overall survival in patients with early hormone receptor-positive invasive ductal breast carcinoma. Oncotarget. 2015;6:41134-41145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 32. | Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, Semenza GL, van Diest PJ, van der Wall E. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 393] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 33. | Schindl M, Schoppmann SF, Samonigg H, Hausmaninger H, Kwasny W, Gnant M, Jakesz R, Kubista E, Birner P, Oberhuber G; Austrian Breast and Colorectal Cancer Study Group. Overexpression of hypoxia-inducible factor 1alpha is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin Cancer Res. 2002;8:1831-1837. [PubMed] |
| 34. | Dales JP, Garcia S, Meunier-Carpentier S, Andrac-Meyer L, Haddad O, Lavaut MN, Allasia C, Bonnier P, Charpin C. Overexpression of hypoxia-inducible factor HIF-1alpha predicts early relapse in breast cancer: retrospective study in a series of 745 patients. Int J Cancer. 2005;116:734-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 194] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 35. | Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield S, Bersiga A, Allevi G, Milani M, Aguggini S, Gandolfi V, Dogliotti L, Bottini A, Harris AL, Fox SB. Hypoxia-inducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12:4562-4568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 204] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 36. | Yan M, Rayoo M, Takano EA; KConFab Investigators, Fox SB. BRCA1 tumours correlate with a HIF-1alpha phenotype and have a poor prognosis through modulation of hydroxylase enzyme profile expression. Br J Cancer. 2009;101:1168-1174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 37. | Kronblad A, Jirström K, Rydén L, Nordenskjöld B, Landberg G. Hypoxia inducible factor-1alpha is a prognostic marker in premenopausal patients with intermediate to highly differentiated breast cancer but not a predictive marker for tamoxifen response. Int J Cancer. 2006;118:2609-2616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Rajković-Molek K, Mustać E, Hadžisejdić I, Jonjić N. The prognostic importance of nuclear factor κB and hypoxia-inducible factor 1α in relation to the breast cancer subtype and the overall survival. Appl Immunohistochem Mol Morphol. 2014;22:464-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 39. | Malfettone A, Saponaro C, Paradiso A, Simone G, Mangia A. Peritumoral vascular invasion and NHERF1 expression define an immunophenotype of grade 2 invasive breast cancer associated with poor prognosis. BMC Cancer. 2012;12:106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 40. | Schoppmann SF, Fenzl A, Schindl M, Bachleitner-Hofmann T, Nagy K, Gnant M, Horvat R, Jakesz R, Birner P. Hypoxia inducible factor-1alpha correlates with VEGF-C expression and lymphangiogenesis in breast cancer. Breast Cancer Res Treat. 2006;99:135-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 41. | Marton I, Knezevic F, Ramic S, Milosevic M, Tomas D. Immunohistochemical expression and prognostic significance of HIF-1α and VEGF-C in neuroendocrine breast cancer. Anticancer Res. 2012;32:5227-5232. [PubMed] |
| 42. | Ni X, Zhao Y, Ma J, Xia T, Liu X, Ding Q, Zha X, Wang S. Hypoxia-induced factor-1 alpha upregulates vascular endothelial growth factor C to promote lymphangiogenesis and angiogenesis in breast cancer patients. J Biomed Res. 2013;27:478-485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Yamamoto Y, Ibusuki M, Okumura Y, Kawasoe T, Kai K, Iyama K, Iwase H. Hypoxia-inducible factor 1alpha is closely linked to an aggressive phenotype in breast cancer. Breast Cancer Res Treat. 2008;110:465-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 120] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 44. | Jin MS, Lee H, Park IA, Chung YR, Im SA, Lee KH, Moon HG, Han W, Kim K, Kim TY, Noh DY, Ryu HS. Overexpression of HIF1α and CAXI predicts poor outcome in early-stage triple negative breast cancer. Virchows Arch. 2016;469:183-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 45. | Cui J, Jiang H. Prediction of postoperative survival of triple-negative breast cancer based on nomogram model combined with expression of HIF-1α and c-myc. Medicine (Baltimore). 2019;98:e17370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 46. | Huang CJ, Lian SL, Hou MF, Chai CY, Yang YH, Lin SF, Chang HW. SNP 1772 C > T of HIF-1α gene associates with breast cancer risk in a Taiwanese population. Cancer Cell Int. 2014;14:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 47. | Chen HH, Su WC, Lin PW, Guo HR, Lee WY. Hypoxia-inducible factor-1alpha correlates with MET and metastasis in node-negative breast cancer. Breast Cancer Res Treat. 2007;103:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 48. | Li M, Xiao D, Zhang J, Qu H, Yang Y, Yan Y, Liu X, Wang J, Liu L, Wang J, Duan X. Expression of LPA2 is associated with poor prognosis in human breast cancer and regulates HIF-1α expression and breast cancer cell growth. Oncol Rep. 2016;36:3479-3487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 49. | Koo JS, Jung W. Alteration of REDD1-mediated mammalian target of rapamycin pathway and hypoxia-inducible factor-1α regulation in human breast cancer. Pathobiology. 2010;77:289-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 50. | Shi L, Zhao C, Pu H, Zhang Q. FBP1 expression is associated with basal-like breast carcinoma. Oncol Lett. 2017;13:3046-3056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 51. | Nie C, Lv H, Bie L, Hou H, Chen X. Hypoxia-inducible factor 1-alpha expression correlates with response to neoadjuvant chemotherapy in women with breast cancer. Medicine (Baltimore). 2018;97:e13551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 52. | Choi J, Jung WH, Koo JS. Metabolism-related proteins are differentially expressed according to the molecular subtype of invasive breast cancer defined by surrogate immunohistochemistry. Pathobiology. 2013;80:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 53. | Dong M, Wan XB, Yuan ZY, Wei L, Fan XJ, Wang TT, Lv YC, Li X, Chen ZH, Chen J, Lin Q, Wen JY, Ma XK, Liu Q, Wu XY. Low expression of Beclin 1 and elevated expression of HIF-1α refine distant metastasis risk and predict poor prognosis of ER-positive, HER2-negative breast cancer. Med Oncol. 2013;30:355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 54. | Peurala E, Koivunen P, Bloigu R, Haapasaari KM, Jukkola-Vuorinen A. Expressions of individual PHDs associate with good prognostic factors and increased proliferation in breast cancer patients. Breast Cancer Res Treat. 2012;133:179-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 55. | Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin Cancer Biol. 2018;52:56-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 279] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 56. | Semenza GL. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends Mol Med. 2012;18:534-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 176] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 57. | Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2349] [Cited by in RCA: 3794] [Article Influence: 474.3] [Reference Citation Analysis (0)] |
| 58. | Chen J, Ding Z, Peng Y, Pan F, Li J, Zou L, Zhang Y, Liang H. HIF-1α inhibition reverses multidrug resistance in colon cancer cells via downregulation of MDR1/P-glycoprotein. PLoS One. 2014;9:e98882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 59. | Li D, Xu D, Chen P, Xie J. Notch1 signaling modulates hypoxia-induced multidrug resistance in human laryngeal cancer cells. Mol Biol Rep. 2022;49:6235-6240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 60. | Ye LY, Chen W, Bai XL, Xu XY, Zhang Q, Xia XF, Sun X, Li GG, Hu QD, Fu QH, Liang TB. Hypoxia-Induced Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma Induces an Immunosuppressive Tumor Microenvironment to Promote Metastasis. Cancer Res. 2016;76:818-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 220] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 61. | Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74:665-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 591] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 62. | Feng D, Gao J, Liu R, Liu W, Gao T, Yang Y, Zhang D, Yang T, Yin X, Yu H, Huang W, Wang Y. CARM1 drives triple-negative breast cancer progression by coordinating with HIF1A. Protein Cell. 2024;15:744-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |