Published online Nov 24, 2022. doi: 10.5306/wjco.v13.i11.929
Peer-review started: October 19, 2022
First decision: October 28, 2022
Revised: October 30, 2022
Accepted: November 6, 2022
Article in press: November 6, 2022
Published online: November 24, 2022
Processing time: 32 Days and 18.3 Hours
Gut microbiome (GM) composition and diversity have recently been studied as a biomarker of response to immune checkpoint blockade therapy (ICB) and of ICB-related colitis.
To conduct a systematic review on the role of GM composition and diversity in predicting response and colitis in patients with melanoma treated with ICB.
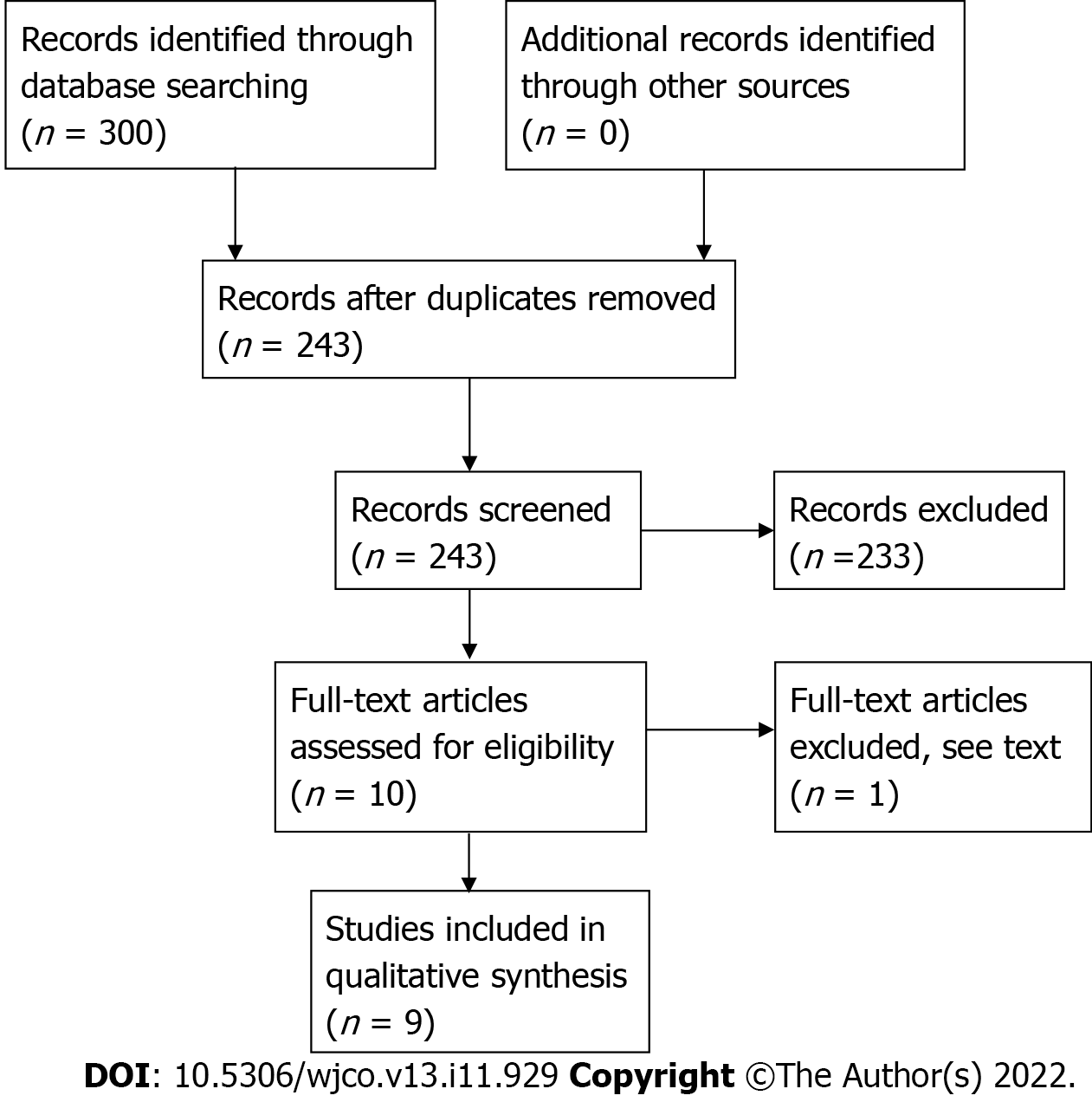
The review protocol was registered in PROSPERO: CRD42021228018. From a total of 300 studies, nine studies met inclusion criteria. Two studies were phase I clinical trials, while the remainder were prospective observational studies. All but one study has moderate risk of bias. In addition, we conducted a relevant search by Reference Citation Analysis (RCA) (https://www.referencecitationanalysis.com).
Fecal samples enriched in Firmicutes phylum were associated with good response to ICB, whereas the Bacteroidales family was associated with poor response to ICB. Samples with greater GM diversity were associated with more favorable response to ICB [hazard ratio (HR) = 3.57, 95% confidence interval = 1.02-12.52, P < 0.05]. Fecal samples with a higher abundance in Firmicutes were more susceptible to ICB-related colitis (P < 0.01) whereas samples enriched in Bacteroidetes were more resistant to ICB-related colitis (P < 0.05). Overall, there was limited concordance in the organisms in the GM identified to be associated with response to ICB, and studies evaluating GM diversity showed conflicting results.
This highlights the need for further prospective studies to confirm whether the GM could be used as a biomarker and potential intervention to modulate ICB response in melanoma patients.
Core Tip: Since the introduction of immune checkpoint inhibitors as part of standard of care for melanoma patients, there has been a growing interest in identifying biomarkers of response and immune related adverse events. Amongst these biomarkers, the composition of the gut microbiome has been one of the most intriguing discoveries. Our aim was to ascertain the current published evidence on the gut microbiome diversity and composition as a biomarker of response to immunotherapy. We demonstrated high variability in the results and limited concordance on the organisms identified. We highlight the conflicting aspects of these reports as well as their few commonalities.
- Citation: Oey O, Liu YY, Sunjaya AF, Simadibrata DM, Khattak MA, Gray E. Gut microbiota diversity and composition in predicting immunotherapy response and immunotherapy-related colitis in melanoma patients: A systematic review. World J Clin Oncol 2022; 13(11): 929-942
- URL: https://www.wjgnet.com/2218-4333/full/v13/i11/929.htm
- DOI: https://dx.doi.org/10.5306/wjco.v13.i11.929
Melanoma is the most lethal form of skin cancer accounting for 73% of skin-cancer related mortality and over 50000 deaths worldwide annually[1,2]. Survival for metastatic melanoma has significantly improved since the introduction of immunotherapy and targeted therapy with a 5-year survival rate of up to 50%[3-5]. Currently, the standard first-line therapy for metastatic melanoma include BRAF-targeted therapies and immune checkpoint blockade (ICB) consisting either anti-programmed death (PD)-1 monotherapy or combination of anti-PD-1 as well as anti-cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) therapy[6]. Despite the considerable benefit of ICB, 40%-60% of melanoma patients do not experience objective responses to the therapy[7-9]. Thus, tremendous efforts are now focused on identifying novel biomarkers which could accurately predict the subset of patients who would benefit from ICB[10-14]. These biomarkers include tumor mutational burden, cytokines, circulating tumor DNA, human leukocyte antigen, gut microbiota (GM) diversity and composition, among many others[15].
The GM is a community of 100 trillion microorganisms of more than 1000 species mainly bacteria but also, archaea, viruses and fungi which colonize the human intestines[16]. The relationship which exists between GM and the host is a mutualistic relationship where one benefits the other[16]. In return for the nutrients derived from the host, the GM performs numerous critical functions such as fermentation of dietary fiber into short-chain fatty acids; synthesis of vitamins; protection against pathologic gut microbes; and induction and regulation of the immune system[17,18]. The gut microbial balance is pivotal in the optimal functioning of all of these roles and thus any discrepancy in this delicate equilibrium could produce a state of dysbiosis which has been associated with many pathologies including cancer[19]. In the context of cancer, preclinical studies have demonstrated that some GM subpopulations have pro-tumorigenic effects, whereas others have tumor-suppressive effects[20-22]. Additionally, the GM has also been shown to modulate response to chemotherapy and immunotherapy[23-25]. This could be linked to the role of GM in metabolizing anti-cancer compounds and regulating the host’s immune response[16]. Thus, GM has been studied intensely as a potential biomarker of response to ICB[12,26-31]. This is particularly relevant for melanoma, where ICB has become standard of care given its demonstrated pronounced effectiveness.
Studies investigating GM composition and/or diversity in patients with melanoma have identified distinct GM composition in responders to ICB compared to non-responders, offering hope of a novel biomarker for predicting response to ICB[12,26-32]. Additionally, studies exploring whether certain GM composition and diversity could be predictive of ICB-related colitis - one of the major factors of ICB treatment cessation and thus failure to derive full benefit of ICB - have also been conducted[27,33]. This systematic review will be the first to compile the existing data regarding the role of GM composition and diversity in predicting response to ICB and ICB-related colitis specifically in patients with melanoma. Notably previous reviews have combined multiple cancers.
This review was conducted following the preferred reporting Items for systematic reviews and meta-analyses guidelines[34]. The review protocol was submitted to the international prospective register of systemic reviews (PROSPERO Registration number: CRD42021228018).
In this comprehensive literature search, original studies exploring the variation in GM community in fecal samples of melanoma patients who responded and did not respond to immunotherapy, experienced colitis and did not experience colitis were identified. Medline and Embase were searched for eligible papers published prior to December 2021 using the following search terms: (fecal OR gut) AND (microbiota OR microbiome) AND (melanoma) AND (immunotherapy OR checkpoint OR nivolumab OR ipilimumab OR pembrolizumab). OpenGrey and the Grey Literature Report were also searched for eligible unpublished papers and grey literature. The following keywords and its synonyms will be used for our search strategy: “fecal microbiota”, “melanoma”, “immunotherapy”.
Duplicate and irrelevant publication types such as symposium agendas were removed from the initial search results. Titles and abstracts of relevant publications were screened independently by Oliver O and Simadibrata DM based on inclusion and exclusion criteria stated below. Subsequently, reference lists within each relevant publication were examined for further pertinent studies. The full texts of these publications were then reviewed.
Inclusion criteria for the systematic review included randomized controlled trials (RCTs), original cohort, case-control studies published in a peer-reviewed journal exploring GM diversity and composition in fecal samples from melanoma patients treated with ICB which can be anti-PD-1 and/or anti-PD-L1 and/or anti-CTLA-4. Studies included should assess treatment outcome and/or ICB-related colitis incidence following treatment with ICB. Treatment outcomes should be determined by RECIST criteria and/or progression free survival (PFS) and/or overall survival (OS) and ICB-related colitis confirmed by colonoscopy.
Only studies which utilized fecal samples obtained from human subjects receiving ICB were included. Studies which assessed treatment response to immunotherapy in animal models were excluded. Two reviewers (Oliver O, Liu YY) independently screened and read the full text of the included articles for eligibility.
Two investigators (Oliver O, Liu YY) independently reviewed the eligible studies and extracted data from each study. Extracted variables included title, first author, year of publication, number of participants, type of immunotherapy received, GM analysis method, and study outcomes (GM composition and diversity in responders/non-responders and ICB-related colitis/non-ICB-related colitis patients). Extracted GM composition data included a list of the GM at the level of phyla, class, order, family, genus and species, whereas extracted GM diversity extracted included alpha diversity or the Shannon index. Any discrepancies found by the investigators on data extraction were resolved by consensus. In addition, we conducted a relevant search by Reference Citation Analysis (RCA) (https://www.referencecitationanalysis.com).
Non-randomized studies, including cohort studies, case-control studies and single-arm clinical trial that were included in this systematic review were independently evaluated by Oliver O and Simadibrata DM for any risk of bias using the Risk of Bias in Non-randomized Studies of interventions (ROBINS-I) assessment tool, a tool which assesses seven items: confounding, selection, intervention classification, deviation from intervention, missing data, measurement of outcome and selection of reported result. Each item was assessed according to the ROBINS-I guideline, where each bias domain can be classified as either low, moderate, serious or critical risk of bias, or no information mentioned.
The initial search from Medline, Embase, OpenGrey and Grey Literature retrieved 300 studies. After deduplication, the studies were screened by reviewing their abstracts and 10 articles selected for full assessment (Figure 1). One study by Vétizou et al[35] was excluded because while the fecal samples were obtained from patients treated with anti-CTLA-4, treatment response to ICB was assessed in an in-vivo mice model of melanoma following fecal transplantation rather than humans.
From the nine included studies, two studies were phase I clinical trials, while the remainder were prospective observational studies[26,28]. Unfortunately, no RCTs were available to date. According to the ROBINS-I assessment tool, all but one study was shown to have moderate risk of bias (Table 1). The study by Matson et al[29] had a serious risk of bias as there was a lack of clarity regarding the definition of intervention used.
| Ref. | Confounding | Selection | Intervention classification | Deviation from intervention | Missing data | Measurement of outcome | Selection of reported result | Overall |
| Dubin et al[33], 2016 | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Chaput et al[27], 2017 | Moderate | Low | Low | Moderate | Low | Low | Moderate | Moderate |
| Gopalakrishnan et al[12], 2018 | Moderate | Low | No information | Low | Low | Low | Low | Moderate |
| Matson et al[29], 2018 | Moderate | No information | Serious | No information | No information | Low | Moderate | Serious |
| Peters et al[30], 2019 | Moderate | Low | No information | No information | Low | Low | Low | Moderate |
| Baruch et al[26], 2020 | Moderate | Low | Low | Low | Moderate | Low | Low | Moderate |
| Wind et al[31], 2020 | Moderate | No information | No information | No information | No information | Low | Low | Moderate |
| Davar et al[28], 2021 | Moderate | Moderate | Low | Low | No information | Low | Low | Moderate |
| Andrews et al[36], 2021 | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
Eight studies assessed the role of GM composition and/or diversity and response to ICB in melanoma patients (Table 2). Seven studies compared the GM between responders and non-responders to ICB, and two studies analyzed the GM in patients undergoing fecal microbiota transplant (FMT).
| Ref. | Year | Therapy | Method | Sample size/ time point | Dominant microbes | Microbial diversity |
| Chaput et al[27], 2017 | 2017 | Anti-CTLA-4 | 16S rRNA gene sequencing of fecal samples | 26 before tx | Responders: Faecalibacterium and Firmicutes | N/A |
| Matson et al[29], 2018 | 2018 | Anti-PD-1 or anti-CTLA-4 | 16S rRNA gene and shotgun metagenome sequencing of fecal samples; qPCR on selected bacteria | 42 before tx | Responders: Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium Non-responders: Ruminococcus obeum and Roseburia intestinalis | N/A |
| Gopalakrishnan et al[12], 2018 | 2018 | Anti-PD-1 | 16S rRNA gene and shotgun metagenome sequencing of fecal samples | 43 before tx | Responders: Clostridiales, in particular Faecalibacterium Non-responders: Bacteroidales, in particular Bacteroides thetaiotaomicron; as well as Escherichia coli, and Anaerotruncus colihominis | Higher alpha diversity in patients with longer PFS |
| Peters et al[30], 2019 | 2019 | Anti-PD-1 or anti-CTLA-4 | 16S rRNA gene and shotgun metagenome sequencing of fecal samples | 27 before tx | Responders: Faecalibacterium, Parabacteroides, and Faecalibacterium prausnitzii Non-responders: Bacteroides and Biophilia | Higher microbial community richness and diversity was associated with longer PFS |
| Wind et al[31], 2020 | 2020 | Anti-PD-1 or anti-CTLA-4 | Shotgun metagenome sequencing of fecal samples | 25 before tx | Responders: Ruminococcus gnavus, Streptococcus parasanguinis, and Bacteroides massiliensis. Non-responders: Bifidobacterium longum and Peptostreptococcaceae | No significant difference in alpha-diversity between responder and non responders |
| Baruch et al[26], 2020 | 2020 | Anti-PD-1 refractory | 16S rRNA gene and shotgun metagenome sequencing of fecal samples | 10 anti-PD-1 refractory patients | FMT donors (responders): Lachnospiraceae, Veillonellaceae, and Ruminococcaceae Post FMT Responders: Enterococcaceae, Enterococcus, and Streptococcus australis Non-responders: Veillonella atypica | No significant difference in GM composition prior to FMT, but significant difference post-FMT between responders and non-responders Lower microbial richness in the donor of responding recipients |
| Davar et al[28].2021 | 2021 | Anti-PD-1 refractory | Shotgun metagenomic sequencing of fecal samples | 15 anti-PD-1 refractory patients, before FMT | Responders: Firmicutes (Lachnospiraceae and Ruminococcaceae families) and Actinobacteria (Bifidobacteriaceae and Coriobacteriaceae families) | Higher GM diversity of donors who were complete responders compared to donors who were partial responders No significant difference in GM diversity between donors and recipients prior to FMT |
| Andrews et al[36], 2021 | 2021 | Combined ICB - Anti-PD-1 and anti-CTLA-4 | 16S rRNA gene and shotgun metagenome sequencing of fecal samples | 38 | Responders: Bacteroides stercoris, Parabacteroides distasonis, Fournierella massiliensis. Non-responders: Klebsiella aerogenes and Lactobacillus rogosae | No significant difference in GM diversity between responders and non-responders |
The study by Chaput et al[27] assessing fecal GM composition of 26 metastatic melanoma patients prior to and post commencing anti-CTLA-4 therapy revealed that GM composition varied according to response. Patients showing long term response to therapy (nine out of 26 patients) were found with fecal samples with significantly higher Faecalibacterium percentages (P = 0.0092) while patients with poor clinical benefit had higher proportions of Bacteroides (P = 0.034). When patients were grouped based on their microbiota composition, those with high prevalence of Faecalibacterium and other Firmicutes had a longer PFS (P = 0.0039) and to a lesser extent longer OS (P = 0.051) relative to patients whose fecal samples were abundant with Bacteroides. Additionally, these patient groups were noted to derive long-term clinical benefit compared to the latter (67% vs 0%; P = 0.0017)[28].
In an analysis of stool samples from 42 metastatic melanoma patients prior to treatment with anti-PD-1 (n = 38) and anti-CTLA-4 (n = 4) therapy, Matson et al[29] showed a significant difference in GM composition between responders (16 patients) and non-responders (26 patients) (P < 0.01). In responders, eight microbial species namely, Enterococcus faecium, Collinsella aerofaciens, Bifidobacterium adolescentis, Klebsiella pneumoniae, Veillonella parvula, Parabacteroides merdae, Lactobacillus sp. and Bifidobacterium longum were found to be more abundant in responders than in non-responders[29]. In non-responders, two microbial species, specifically, Ruminococcus obeum and Roseburia intestinalis were more abundant[29]. To further assess the applicability of GM composition as a biomarker of response to ICB, they explored the correlation between the ratio of total numbers of potentially “beneficial” and “nonbeneficial” operational taxonomic units (OTUs), and change in tumor size, as assessed by the RECIST[29]. Patients with an OTU ratio of greater than 1.5 demonstrated clinical response to ICB[29].
In another study by Gopalakrishnan et al[12], fecal samples of 43 metastatic melanoma patients prior to treatment with anti-PD-1 therapy were analyzed. In responders (30 patients), analysis of fecal samples revealed abundance of GM from Ruminococcaceae family of the Clostridiales order, whereas in non-responders (13 patients), abundance of GM from the Bacteroidales order was noted[12]. Further analyses demonstrated that Faecalibacterium genus was notably enriched in fecal samples from responders and Bacteroides thetaiotaomicron, Escherichia coli, and Anaerotruncus colihominis were enriched in non-responders[12]. In addition, to investigate durability of response, patients were stratified based on their fecal composition of Faecalibacterium genus and Bacteroidales order and correlated to their PFS[12]. Results demonstrated that patients with Faecalibacterium-enriched fecal samples have longer PFS than those with low abundance (P = 0.03) and patients with Bacteroidales-enriched fecal samples have shorter PFS than those with low abundance (P = 0.05). Beyond specific microbial taxa, GM diversity, as assessed by Simpson's reciprocal index, was higher in responders compared to non-responders (P < 0.01)[12]. Moreover, high GM diversity was significantly associated with anti-PD-1 therapy response, when compared to patient groups of intermediate diversity [hazard ratio (HR) = 3.60, 95% confidence interval (CI): 1.02-12.74, P < 0.05) and low diversity (HR = 3.57, 95%CI: 1.02-12.52, P < 0.05). Other important predictors of therapy response include abundance of Faecalibacterium (HR = 2.92, 95%CI: 1.08-7.89) and Bacteroidales (HR = 0.39, 95%CI: 0.15-1.03) in the fecal microbiome[12].
Peters et al[30] examined the correlation between GM taxa and PFS in pre-treatment fecal samples of 27 metastatic melanoma patients receiving anti-PD-L1 and/or anti-CTLA-4. GM which was associated with shorter PFS included genera Bacteroides and Bilophila, and species Bacteroides ovatus, Blautia pro
Similarly, Wind et al[31] analyzed fecal samples from 25 metastatic melanoma patients - 12 responders, 13 non-responders - prior to start of treatment with anti-PD-1 or anti-CTLA-4. Analysis revealed that the fecal samples of responders were mainly enriched in Ruminococcus gnavus, Escherichia coli, Eubacterium biforme, Phascolarctobacterium succinatutens and Streptococcus salivarius, whereas samples from non-responders were abundant in Bifidobacterium longum, Prevotella copri, Coprococus sp, Eggerthella unclassified and Eubacterium ramulus[31]. When correlated with survival, fecal samples of participants enriched in Bacteroides massiliensis and Streptococcus parasanguinis were associated with longer PFS (HR: 3.79, 95%CI: 1.06-13.52 P = 0.04) and OS (HR: 5.05, 95%CI: 1.33-19.21, P = 0.017) respectively, whereas those who were carriers of Peptostreptococcaceae were associated with shorter PFS (HR: 0.18, 95%CI: 0.05-0.62, P = 0.007) and OS (HR: 0.12, 95%CI: 0.01-0.96, P = 0.046)[31]. In terms of GM diversity, as assessed by Shannon index, no significant difference between responders and non-responders was noted[31].
The study by Andrews et al[36] analyzed gut microbiome samples from a subset of 77 metastatic melanoma patients - 27 responders, 11 non-responders - who underwent combined ICB. There was no significant association in Firmicutes phyla and Clostridiales order and response to ICB (P = 0.39 and P = 0.38, respectively) and no significant difference in alpha diversity between responders and non-responders to ICB[36]. Fecal samples from responders were mainly enriched with Bacteroides stercoris, Parabacteroides distasonis and Fournierella massiliensis (P = 0.03, P = 0.04 and P = 0.008, respectively) while fecal samples from non-responders were abundant in Klebsiella aerogenes and Lactobacillus rogosae (P = 0.04 and P = 0.02, respectively)[36].
In a first clinical trial of its kind (phase 1), Baruch et al[26] demonstrated that FMT from anti-PD-1 treated metastatic melanoma patients who were complete responders (2 donors), triggered response to anti-PD-1 therapy in metastatic melanoma patients who were refractory to at least one line of anti-PD-1 therapy. Out of 10 patients included in the trial, 3 patients demonstrated objective responses with 1 achieving complete response and 2 patients achieving partial response[26]. Notably, the PFS milestone of 6 mo was reached in all responders[26]. Upon analysis of pre-treatment fecal samples of donors, donor of the responding recipients had a lower microbial richness than the other donor of the non-responding patients[26]. There was no significant difference on the GM composition prior to FMT of recipients who responded compared to those who did not respond[26]. Metagenome sequencing found that recipients post FMT have higher proportions of Veillonellaceae family and a lower relative abundance of Bifidobacterium bifidum. Donors were found with high amounts of Lachnospiraceae, Veillonellaceae, and Ruminococcaceae. Comparison of a small subset of non-responders with responders, found statistically significant higher abundance of Enterococcaceae, Enterococcus, and Streptococcus australis, and a lower relative abundance of Veillonella atypica. However clear deductions on specific GM taxa cannot be made, as there were non-responders and pre-treatment fecal samples with similar dynamics. it is crucial to note that this trial was primarily designed to assess safety of FMT and not statistically powered to assess efficacy[26].
In a separate trial, Davar et al[28] showed that fecal microbial transplant (FMT) from metastatic melanoma patients (7 donors) who had complete (4 donors) or partial response (3 donors) to anti-PD-1 therapy helped overcome resistance in anti-PD-1 treatment-refractory metastatic melanoma patients (15 patients). Following FMT and anti-PD-1 therapy, 6 out of 15 patients achieved clinical benefit, with 3 patients achieving objective responses and 3 patients experiencing stable disease lasting more than 12 mo[28]. Analysis of stools after FMT revealed that samples from responders were abundant in the phyla, Firmicutes (Lachnospiraceae and Ruminococcaceae) and Actinobacteria (Bifidobacteriaceae and Coriobacteriaceae) and had decreased proportions in phylum Bacteroidetes[28]. In terms of GM diversity assessed with inverse Simpson index, GM diversity of donors who were complete responders were more diverse than donors who were partial responders. There was no significant difference in GM diversity between donors and recipients prior to FMT[28].
To date only three studies have reported on the correlation between pre-treatment GM composition and/or diversity and ICB-related colitis (Table 3).
| Ref. | Year | Therapy | Method | Sample size/ timepoint | Dominant microbes | Microbial diversity |
| Dubin et al[33], 2015 | 2015 | Anti-CTLA-4 immunotherapy | 16S rRNA gene and shotgun metagenome sequencing of fecal samples | 34 | Colitis-resistant: Bacteroidetes | No significant difference in microbial richness and diversity |
| Chaput et al[27], 2017 | 2017 | Anti-CTLA-4 immunotherapy | 16S rRNA gene sequencing of fecal samples | 26 | Colitis-resistant: Bacteroidetes; Colitis-prone: Firmicutes | Decreased bacterial diversity was associated with colitis |
| Andrews et al[36], 2021 | Combined ICB - Anti-PD-1 and anti-CTLA-4 | 16S rRNA gene and shotgun metagenome sequencing of fecal samples | 38 | Colitis resistant: Firmicutes; Colitis prone: Bacteriodetes | No significant difference in alpha diversity |
Firstly, a prospective study by Dubin et al[33] explored the link between GM composition, and subsequent colitis development in 34 metastatic melanoma patients treated with ipilimumab, showed that the Bacteroidetes phylum was more abundant (P = < 0.05) in fecal samples of the 24 patients who did not develop ipilimumab-induced colitis compared to those who did. Further analysis revealed that within the Bacteroidetes phylum, the population of Bacteroidaceae, Rikenellaceae and Barnesiellaceae was significantly more abundant in the former than the latter (P < 0.01, P < 0.05 and P < 0.05 respectively)[33]. However, there was no significant difference in microbial richness and diversity, as assessed by Shannon and inverse Simpson indices, between those who developed ipilimumab-induced colitis relative to those who did not[33].
In a similar study by Chaput et al[27], analysis of fecal samples of metastatic melanoma patients receiving ipilimumab demonstrated high proportions of Firmicutes in patients who developed ipilimumab-induced colitis (P = 0.009). In contrast, fecal samples of those that did not develop colitis were mainly enriched with Bacteroidetes (P = 0.011)[27]. Accordingly, patients with the former GM composition also tend to have a shorter colitis-free cumulative incidence compared with patients with the latter composition[27]. Several OTUs known to be predictive to colitis such as F. prausnitzii L2-6, butyrate producing bacterium L2-21 and G. formicilis ATCC 27749 were associated with longer OS[27].
Finally, Andrews et al[36], analyzed gut microbiome samples in metastatic melanoma patients undergoing combined ICB and their link to ileitis and colitis events. No significant difference in alpha diversity was observed between those that did and did not develop colitis[36]. Fecal samples of patients developing colitis were enriched in Bacteroides intestinalis and Intestinibacter bartlettii (P = 0.009 and P = 0.009, respectively) while those that did not were abundant in Anaerotignum lactatifermentans and Dorea formicigenerans (P = 0.016 and P = 0.06, respectively)[36]. For both B. intestinalis and D. formicigenerans, associations with their risk of colitis were still maintained after adjustment using a logistic regression model [OR = 4.54 (95%CI = 1.06-24.7) and OR = 0.35 (95%CI = 0.082-1.35), respectively][36].
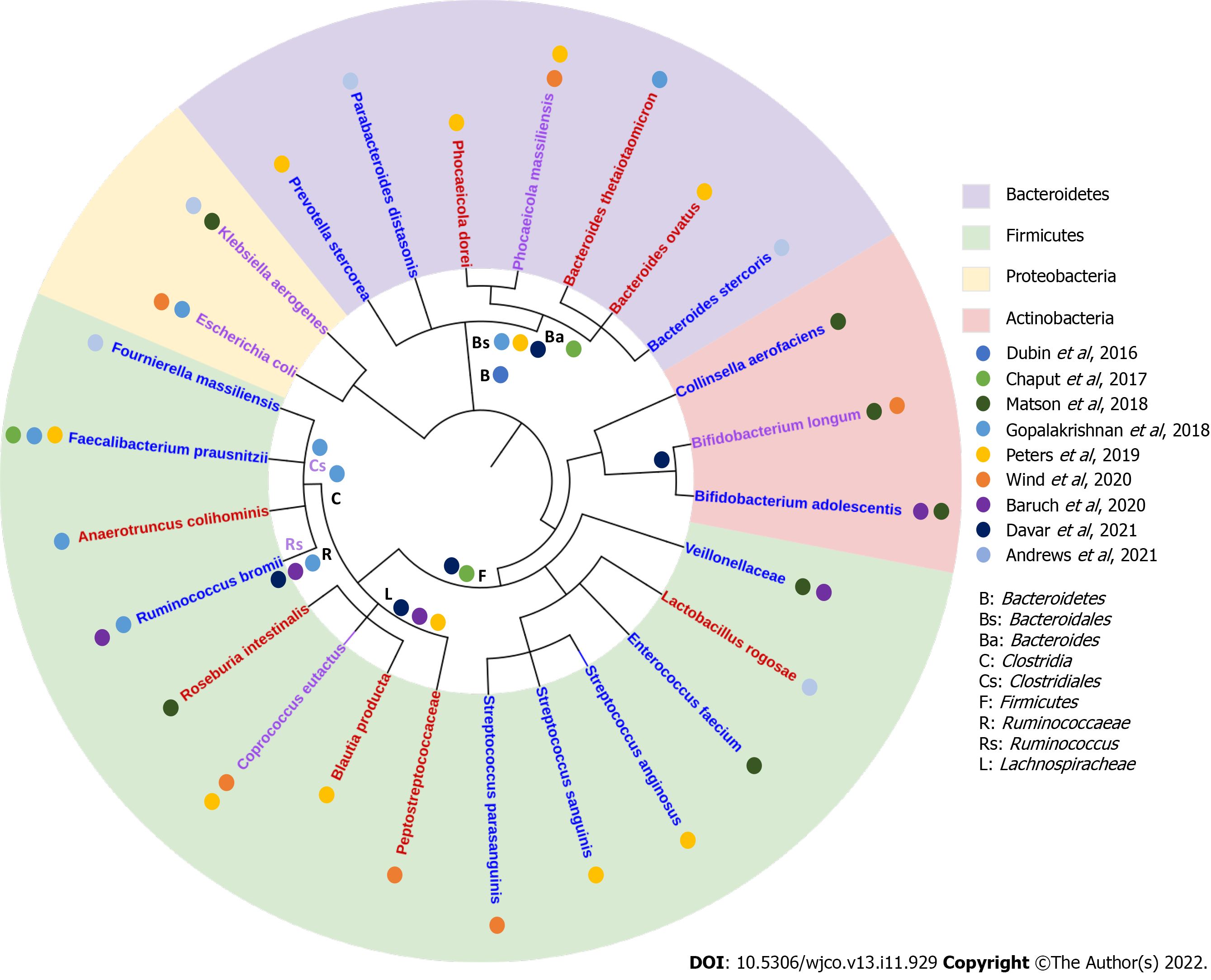
Our review of current reports assessing the GM composition relative to response to ICB, indicated high variability in the results and limited concordance on the organisms identified (Figure 2). Amongst the few commonalities, we found that fecal samples enriched in organisms from the Firmicutes phylum (Lachnospiraceae and Ruminococcaceae family) especially the Faecalibacterium genus were associated with ICB responders in 4 of 9 studies[12,27,28,31], while Bacteroidetes phylum was found in higher pro
In fact, our analysis mainly identified inconsistencies in the GM composition reported to be associated with response to ICB. For instance, Bifidobacterium longum was found to be abundant in responders in the study by Wind et al[31], but found to be enriched in non-responders in the study by Matson et al[29]. Some species from the Firmicutes family were found in both responders and non-responders such as Roseburia intestinalis[29]. Similarly, species from the Bacteroidales order were found in both responders and non-responders, such as Bacteroides massiliensis[31]. The overlap in GM composition in responders and non-responders may suggest that the functional capacity of the GM may be more important than individual GM family/order/species in determining response to ICB[30].
In contrast to individual species or taxas, GM diversity have been heralded to a marker of good health[37]. Here four studies - Gopalakrishnan et al[12], Peters et al[30], Wind et al[31] and, Andrews et al[36] - assessed its potential to predict ICB responsiveness. The two first studies demonstrated that higher GM diversity in the responder group compared to non-responder arm[12,30]. However, the other two studies, Wind et al[31] and Andrews et al[36], found no differences in GM diversity between both groups. Nevertheless, in other cancer types such as renal cell carcinoma and non-small cell lung cancer, greater GM diversity has also been associated with improved responses to anti-PD-1 therapy[37-39].
Study results showing associations with GM diversity were consistent with previous studies which showed that greater GM diversity is prevalent in healthy state across multiple diseases, plausibly suggesting that a greater GM diversity produces the optimal immune environment needed for normal physiological functioning[40-42]. One major reason is the promotion of a favorable immune phenotype, as evidenced by the positive correlation between Shannon diversity index and several CD8+ T cell and NK cell signatures, required to produce a robust anti-tumoral response[38].
Previous studies have demonstrated that GM from Firmicutes family and Bacteroidales order play a significant role in mediating the response to immunotherapy in melanoma patients[12,27,29]. For instance, abundance of Firmicutes was associated with increased frequencies of CD4+, CD8+ T cells, CD 45+ myeloid and lymphoid tumor-infiltrating cells and preserved cytokine response to anti-PD-1 therapy[12]. Additionally, abundance of Firmicutes was linked with decreased frequency of intestinal and systemic regulatory T cells (Tregs) and B7+ T cells, cells responsible for limiting immune response robustness[27]. This resulted in increased antigen presentation and effector T cell function in both the periphery and tumor microenvironment[12,27,29]. However, other GM such as Bacteroidales were unfavorable in terms of anti-tumoral response in that its abundance was associated with higher frequencies of Tregs and myeloid-derived suppressor cells and a blunted cytokine response[12]. These findings combined demonstrated that certain GM play a crucial role in mediating systemic and antitumor immune responses which have clear implications on efficacy on ICB therapy in metastatic melanoma patients.
Notably, GM has also been shown to potentially serve as not just a predictor of ICB therapy response, but also for boosting response to ICB therapy. FMT on anti-PD-1 treatment-refractory metastatic melanoma patients produced a complete response to anti-PD-1 therapy in one-third (9 out of 25 patients) of the otherwise therapy refractory patients[26,28].
Another aspect of the GM analyzed here, was its association with ICB-related colitis. The three studies included in this review demonstrated that GM which was abundant in ICB-related colitis-prone patients was enriched in responders to ICB (Firmicutes) while GM which was abundant in ICB-related colitis-resistant patients was enriched in non-responders to ICB (Bacteroidetes). This is consistent with the understanding that a more effective anti-tumoral response will produce greater off-target effects. The Bacteroidetes phyla has been linked with low-grade systemic inflammation, which could explain the observation that Bacteroidetes phyla was abundant in ICB-related colitis-resistant patients[27]. In line with this observation is the finding that level of Bacteroidetes is lower in inflammatory bowel disease - an autoimmune condition which produces chronic inflammation of the digestive tract - patients relative to healthy patients[43]. Conversely, Firmicutes phyla, especially F. prausnitzii has been associated with induction of Tregs which express high levels of CTLA-4, fueling speculation that it may cause sequestration of Tregs within the intestine[44]. Since Tregs express high levels of CTLA-4, their actions are inhibited, thereby limiting self-tolerance and promoting the development of colitis. These findings reiterate that GM has an immunomodulatory role, giving them the potential to be utilized as biomarkers of ICB-related colitis, in addition to response to ICB.
Our systematic review has several strengths. Firstly, unlike previous reviews which combined studies in various cancer types, this review focused solely on the effect of GM composition and diversity only in patients with melanoma. Secondly, we conducted a comprehensive search for RCTs and observational studies, performed a risk-of-bas assessment and studied clinically important outcomes - clinical response and ICB-related colitis - an adverse event reported in up to 25% of patients treated with ICB[45]. Thirdly, we only included studies which assessed response to immunotherapy in humans, not animals. However, several limitations exist in our systematic review. Studies which we included used distinct approaches when segregating patients into the responder and non-responder groups, using different response criteria to evaluate treatment response in patients. Additionally, there were differences in methods of stool collection and analysis of GM composition and diversity. For example, Chaput et al[46] collected multiple stool samples every 3 wk of ICB, while other studies such as Dubin et al[33] and Matson et al[29] collected stool samples only prior to initiation of ICB. Furthermore, only 4 studies considered confounding factors such as variation in diet and antibiotic use[27,29-31]. Therefore, inter-study comparison of the GM composition and diversity in responders vs non-responders and those who experienced colitis vs non-colitis should be addressed with caution. Furthermore, included studies only enrolled a small number of patients, which could explain inconsistent results between studies.
In conclusion, GM composition and diversity holds some potential as a biomarker of response and toxicity to ICB in melanoma. Larger prospective studies with standardized experimental protocol ought to be conducted to elucidate whether distinct GM signatures are required for robust response to different ICB regimens. Additionally, more studies correlating metagenomic and metatranscriptomic data of GM to outcomes of melanoma patients on immunotherapy ought to be performed as the functional capacity may be more important rather than individual GM family/order/species. In addition, we eagerly await the outcome of multiple large-scale RCTs involving FMT in the context of ICB-refractory melanoma such as NCT04577729 and NCT04988841 (PICASSO) (ClinicalTrails.gov).We foresee that together with other promising biomarkers, GM composition and diversity will be integrated into a multiparameter model to accurately predict which subset of melanoma patients are likely to respond to ICB[10,11,47].
Survival for metastatic melanoma has significantly improved since the introduction of immune checkpoint blockade (ICB) therapy. However, despite their considerable efficacy, 40%-60% of melanoma patients do not experience objective responses to the therapy. Additionally, some patients experience ICB-related colitis as a consequence of ICB therapy, preventing them from deriving the full benefit of ICB therapy. Recent studies have demonstrated that the gut microbiome (GM) may affect tumor immunity by regulating the host immune system and tumor micro-environment, thus suggesting that GM may affect response to ICB therapy and susceptibility of ICB-related colitis.
The GM has shown great potential as a biomarker of response to ICB therapy in melanoma patients. Previous studies investigating GM composition and/or diversity in patients with melanoma have identified distinct GM composition and diversity in responders to ICB compared to non-responders, as well as those more susceptible to ICB-related colitis than those who are not.
To be the first to compile the existing data regarding the role of GM composition and diversity in predicting response to ICB and ICB-related colitis specifically in patients with melanoma.
Comprehensive literature search was done in various platforms using the following search terms: (fecal OR gut) AND (microbiota OR microbiome) AND (melanoma) AND (immunotherapy OR checkpoint OR nivolumab OR ipilimumab OR pembrolizumab). From a total of 300 studies, nine studies met inclusion criteria. Two studies were phase I clinical trials, while the remainder were prospective observational studies. All but one study has moderate risk of bias. Data from these studies including but not limited to, number of participants, type of immunotherapy received, GM analysis method, and GM composition and diversity were collected and interpreted.
Fecal samples enriched in Firmicutes phylum were associated with good response to ICB therapy, however they were associated with increased susceptibility to ICB-related colitis. Fecal samples enriched in Bacteroidales family were associated with poor response to ICB. Samples with greater GM diversity were associated with more favorable response to ICB. Fecal samples enriched in Bacteroidetes were associated with decreased incidence of ICB-related colitis. Overall, there was limited concordance in the organisms in the GM identified to be associated with response to ICB, and studies evaluating GM diversity showed conflicting results.
GM composition and diversity holds some potential as a biomarker of response and toxicity to ICB in melanoma. Further prospective studies, including several RCTs that are underway, are needed to confirm whether the GM could be used as a biomarker and potential intervention to modulate ICB response in melanoma patients.
With other promising biomarkers, GM composition and diversity holds potential to be integrated into a multiparameter model to accurately predict which subset of melanoma patients are likely to respond to ICB.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: American Society of Clinical Oncology; American Association for Cancer Research.
Specialty type: Oncology
Country/Territory of origin: Australia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: MI SC, China; Wen XL, China S-Editor: Wang LL L-Editor: A P-Editor: Wang LL
| 1. | Gershenwald JE, Guy GP Jr. Stemming the Rising Incidence of Melanoma: Calling Prevention to Action. J Natl Cancer Inst. 2016;108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 63507] [Article Influence: 15876.8] [Reference Citation Analysis (174)] |
| 3. | Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph R, Weber JS, Dronca R, Mitchell TC, Patnaik A, Zarour HM, Joshua AM, Zhao Q, Jensen E, Ahsan S, Ibrahim N, Ribas A. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann Oncol. 2019;30:582-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 579] [Cited by in RCA: 671] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 4. | Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, Ferrucci PF, Smylie M, Hogg D, Hill A, Márquez-Rodas I, Haanen J, Guidoboni M, Maio M, Schöffski P, Carlino MS, Lebbé C, McArthur G, Ascierto PA, Daniels GA, Long GV, Bastholt L, Rizzo JI, Balogh A, Moshyk A, Hodi FS, Wolchok JD. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med. 2019;381:1535-1546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1826] [Cited by in RCA: 2606] [Article Influence: 434.3] [Reference Citation Analysis (0)] |
| 5. | Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko E, Chiarion Sileni V, Schachter J, Garbe C, Bondarenko I, Gogas H, Mandalá M, Haanen JBAG, Lebbé C, Mackiewicz A, Rutkowski P, Nathan PD, Ribas A, Davies MA, Flaherty KT, Burgess P, Tan M, Gasal E, Voi M, Schadendorf D, Long GV. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N Engl J Med. 2019;381:626-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 676] [Cited by in RCA: 936] [Article Influence: 156.0] [Reference Citation Analysis (0)] |
| 6. | Domingues B, Lopes JM, Soares P, Pópulo H. Melanoma treatment in review. Immunotargets Ther. 2018;7:35-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 309] [Cited by in RCA: 449] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 7. | Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10799] [Cited by in RCA: 11738] [Article Influence: 782.5] [Reference Citation Analysis (0)] |
| 8. | Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, Ferrucci PF, Hill A, Wagstaff J, Carlino MS, Haanen JB, Maio M, Marquez-Rodas I, McArthur GA, Ascierto PA, Long GV, Callahan MK, Postow MA, Grossmann K, Sznol M, Dreno B, Bastholt L, Yang A, Rollin LM, Horak C, Hodi FS, Wolchok JD. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6070] [Cited by in RCA: 6169] [Article Influence: 616.9] [Reference Citation Analysis (0)] |
| 9. | Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J, Lorigan P, Neyns B, Blank CU, Hamid O, Mateus C, Shapira-Frommer R, Kosh M, Zhou H, Ibrahim N, Ebbinghaus S, Ribas A; KEYNOTE-006 investigators. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med. 2015;372:2521-2532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4026] [Cited by in RCA: 4459] [Article Influence: 445.9] [Reference Citation Analysis (1)] |
| 10. | Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44-56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1568] [Cited by in RCA: 1853] [Article Influence: 308.8] [Reference Citation Analysis (0)] |
| 11. | Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, Kuo F, Kendall SM, Requena D, Riaz N, Greenbaum B, Carroll J, Garon E, Hyman DM, Zehir A, Solit D, Berger M, Zhou R, Rizvi NA, Chan TA. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 768] [Cited by in RCA: 807] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 12. | Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2999] [Cited by in RCA: 3249] [Article Influence: 464.1] [Reference Citation Analysis (0)] |
| 13. | Pavlick AC, Fecher L, Ascierto PA, Sullivan RJ. Frontline Therapy for BRAF-Mutated Metastatic Melanoma: How Do You Choose, and Is There One Correct Answer? Am Soc Clin Oncol Educ Book. 2019;39:564-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 14. | Marsavela G, Lee J, Calapre L, Wong SQ, Pereira MR, McEvoy AC, Reid AL, Robinson C, Warburton L, Abed A, Khattak MA, Meniawy TM, Dawson SJ, Sandhu S, Carlino MS, Menzies AM, Scolyer RA, Long GV, Amanuel B, Millward M, Ziman MR, Rizos H, Gray ES. Circulating Tumor DNA Predicts Outcome from First-, but not Second-line Treatment and Identifies Melanoma Patients Who May Benefit from Combination Immunotherapy. Clin Cancer Res. 2020;26:5926-5933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 15. | Morad G, Helmink BA, Sharma P, Wargo JA. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell. 2021;184:5309-5337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 957] [Article Influence: 239.3] [Reference Citation Analysis (0)] |
| 16. | Lepage P, Leclerc MC, Joossens M, Mondot S, Blottière HM, Raes J, Ehrlich D, Doré J. A metagenomic insight into our gut's microbiome. Gut. 2013;62:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 243] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 17. | Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2488] [Cited by in RCA: 3377] [Article Influence: 307.0] [Reference Citation Analysis (1)] |
| 18. | Morowitz MJ, Carlisle EM, Alverdy JC. Contributions of intestinal bacteria to nutrition and metabolism in the critically ill. Surg Clin North Am. 2011;91:771-785, viii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 128] [Article Influence: 9.1] [Reference Citation Analysis (1)] |
| 19. | Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16:406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 196] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 20. | Jan G, Belzacq AS, Haouzi D, Rouault A, Métivier D, Kroemer G, Brenner C. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 2002;9:179-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 250] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 21. | Lara-Tejero M, Galán JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290:354-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 402] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 22. | Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, Palmer DC, Boni A, Muranski P, Yu Z, Gattinoni L, Antony PA, Rosenberg SA, Restifo NP. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117:2197-2204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 475] [Cited by in RCA: 443] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 23. | Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, Schlitzer A, Ginhoux F, Apetoh L, Chachaty E, Woerther PL, Eberl G, Bérard M, Ecobichon C, Clermont D, Bizet C, Gaboriau-Routhiau V, Cerf-Bensussan N, Opolon P, Yessaad N, Vivier E, Ryffel B, Elson CO, Doré J, Kroemer G, Lepage P, Boneca IG, Ghiringhelli F, Zitvogel L. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1147] [Cited by in RCA: 1531] [Article Influence: 127.6] [Reference Citation Analysis (0)] |
| 24. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-563.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 1444] [Article Influence: 180.5] [Reference Citation Analysis (0)] |
| 25. | Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, Jiang W, Cai S, Zhao P, Song R, Li P, Qin N, Fang W. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7:193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 363] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 26. | Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, Rotin D, Anafi L, Avivi C, Melnichenko J, Steinberg-Silman Y, Mamtani R, Harati H, Asher N, Shapira-Frommer R, Brosh-Nissimov T, Eshet Y, Ben-Simon S, Ziv O, Khan MAW, Amit M, Ajami NJ, Barshack I, Schachter J, Wargo JA, Koren O, Markel G, Boursi B. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 963] [Article Influence: 192.6] [Reference Citation Analysis (0)] |
| 27. | Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M, Vaysse T, Marthey L, Eggermont A, Asvatourian V, Lanoy E, Mateus C, Robert C, Carbonnel F. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 585] [Cited by in RCA: 909] [Article Influence: 129.9] [Reference Citation Analysis (0)] |
| 28. | Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, Deblasio RN, Menna C, Ding Q, Pagliano O, Zidi B, Zhang S, Badger JH, Vetizou M, Cole AM, Fernandes MR, Prescott S, Costa RGF, Balaji AK, Morgun A, Vujkovic-Cvijin I, Wang H, Borhani AA, Schwartz MB, Dubner HM, Ernst SJ, Rose A, Najjar YG, Belkaid Y, Kirkwood JM, Trinchieri G, Zarour HM. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 843] [Cited by in RCA: 997] [Article Influence: 249.3] [Reference Citation Analysis (0)] |
| 29. | Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1390] [Cited by in RCA: 2092] [Article Influence: 298.9] [Reference Citation Analysis (1)] |
| 30. | Peters BA, Wilson M, Moran U, Pavlick A, Izsak A, Wechter T, Weber JS, Osman I, Ahn J. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med. 2019;11:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 165] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 31. | Wind TT, Gacesa R, Vich Vila A, de Haan JJ, Jalving M, Weersma RK, Hospers GAP. Gut microbial species and metabolic pathways associated with response to treatment with immune checkpoint inhibitors in metastatic melanoma. Melanoma Res. 2020;30:235-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398:1002-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 681] [Article Influence: 170.3] [Reference Citation Analysis (0)] |
| 33. | Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, No D, Gobourne A, Littmann E, Huttenhower C, Pamer EG, Wolchok JD. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 560] [Cited by in RCA: 762] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 34. | Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62:e1-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 7600] [Article Influence: 475.0] [Reference Citation Analysis (1)] |
| 35. | Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Bérard M, Nigou J, Opolon P, Eggermont A, Woerther PL, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1834] [Cited by in RCA: 2514] [Article Influence: 251.4] [Reference Citation Analysis (0)] |
| 36. | Andrews MC, Duong CPM, Gopalakrishnan V, Iebba V, Chen WS, Derosa L, Khan MAW, Cogdill AP, White MG, Wong MC, Ferrere G, Fluckiger A, Roberti MP, Opolon P, Alou MT, Yonekura S, Roh W, Spencer CN, Curbelo IF, Vence L, Reuben A, Johnson S, Arora R, Morad G, Lastrapes M, Baruch EN, Little L, Gumbs C, Cooper ZA, Prieto PA, Wani K, Lazar AJ, Tetzlaff MT, Hudgens CW, Callahan MK, Adamow M, Postow MA, Ariyan CE, Gaudreau PO, Nezi L, Raoult D, Mihalcioiu C, Elkrief A, Pezo RC, Haydu LE, Simon JM, Tawbi HA, McQuade J, Hwu P, Hwu WJ, Amaria RN, Burton EM, Woodman SE, Watowich S, Diab A, Patel SP, Glitza IC, Wong MK, Zhao L, Zhang J, Ajami NJ, Petrosino J, Jenq RR, Davies MA, Gershenwald JE, Futreal PA, Sharma P, Allison JP, Routy B, Zitvogel L, Wargo JA. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat Med. 2021;27:1432-1441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 270] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 37. | Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 839] [Cited by in RCA: 1231] [Article Influence: 175.9] [Reference Citation Analysis (0)] |
| 38. | Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, Zheng H, Yao C, Wang Y, Lu S. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J Thorac Oncol. 2019;14:1378-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 370] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 39. | Salgia NJ, Bergerot PG, Maia MC, Dizman N, Hsu J, Gillece JD, Folkerts M, Reining L, Trent J, Highlander SK, Pal SK. Stool Microbiome Profiling of Patients with Metastatic Renal Cell Carcinoma Receiving Anti-PD-1 Immune Checkpoint Inhibitors. Eur Urol. 2020;78:498-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 140] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 40. | Mosca A, Leclerc M, Hugot JP. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front Microbiol. 2016;7:455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 387] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 41. | Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6397] [Cited by in RCA: 5647] [Article Influence: 352.9] [Reference Citation Analysis (1)] |
| 42. | Vincent C, Stephens DA, Loo VG, Edens TJ, Behr MA, Dewar K, Manges AR. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome. 2013;1:18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 43. | Verma R, Verma AK, Ahuja V, Paul J. Real-time analysis of mucosal flora in patients with inflammatory bowel disease in India. J Clin Microbiol. 2010;48:4279-4282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 44. | Romano E, Kusio-Kobialka M, Foukas PG, Baumgaertner P, Meyer C, Ballabeni P, Michielin O, Weide B, Romero P, Speiser DE. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc Natl Acad Sci U S A. 2015;112:6140-6145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 478] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 45. | Som A, Mandaliya R, Alsaadi D, Farshidpour M, Charabaty A, Malhotra N, Mattar MC. Immune checkpoint inhibitor-induced colitis: A comprehensive review. World J Clin Cases. 2019;7:405-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 201] [Cited by in RCA: 189] [Article Influence: 31.5] [Reference Citation Analysis (5)] |
| 46. | Nel Van Zyl K, Whitelaw AC, Newton-Foot M. The effect of storage conditions on microbial communities in stool. PLoS One. 2020;15:e0227486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Pedersen JG, Madsen AT, Gammelgaard KR, Aggerholm-Pedersen N, Sørensen BS, Øllegaard TH, Jakobsen MR. Inflammatory Cytokines and ctDNA Are Biomarkers for Progression in Advanced-Stage Melanoma Patients Receiving Checkpoint Inhibitors. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |