Published online Feb 15, 2016. doi: 10.4291/wjgp.v7.i1.138
Peer-review started: July 2, 2015
First decision: September 22, 2015
Revised: October 10, 2015
Accepted: December 17, 2015
Article in press: December 18, 2015
Published online: February 15, 2016
Processing time: 216 Days and 7.8 Hours
AIM: To investigate the intestinal functions of the NKCC1 Na+-K+-2Cl cotransporter (SLC12a2 gene), differential mRNA expression changes in NKCC1-null intestine were analyzed.
METHODS: Microarray analysis of mRNA from intestines of adult wild-type mice and gene-targeted NKCC1-null mice (n = 6 of each genotype) was performed to identify patterns of differential gene expression changes. Differential expression patterns were further examined by Gene Ontology analysis using the online Gorilla program, and expression changes of selected genes were verified using northern blot analysis and quantitative real time-polymerase chain reaction. Histological staining and immunofluorescence were performed to identify cell types in which upregulated pancreatic digestive enzymes were expressed.
RESULTS: Genes typically associated with pancreatic function were upregulated. These included lipase, amylase, elastase, and serine proteases indicative of pancreatic exocrine function, as well as insulin and regenerating islet genes, representative of endocrine function. Northern blot analysis and immunohistochemistry showed that differential expression of exocrine pancreas mRNAs was specific to the duodenum and localized to a subset of goblet cells. In addition, a major pattern of changes involving differential expression of olfactory receptors that function in chemical sensing, as well as other chemosensing G-protein coupled receptors, was observed. These changes in chemosensory receptor expression may be related to the failure of intestinal function and dependency on parenteral nutrition observed in humans with SLC12a2 mutations.
CONCLUSION: The results suggest that loss of NKCC1 affects not only secretion, but also goblet cell function and chemosensing of intestinal contents via G-protein coupled chemosensory receptors.
Core tip: The NKCC1 Na+-K+-2Cl- cotransporter is a major mechanism of Cl- uptake in support of secretion. To investigate its intestinal functions we analyzed mRNA expression changes in NKCC1-null intestines. Differentially expressed genes included digestive enzymes and a large number of olfactory and other G-protein coupled receptors that function in chemical sensing. This suggests that loss of NKCC1 affects not only secretion, but also digestion and chemosensing of components of the intestinal contents. The results likely have relevance to recent evidence showing that mutations in the human NKCC1 gene cause unexplained food intolerance and failure of intestinal function requiring parenteral nutrition.
- Citation: Bradford EM, Vairamani K, Shull GE. Differential expression of pancreatic protein and chemosensing receptor mRNAs in NKCC1-null intestine. World J Gastrointest Pathophysiol 2016; 7(1): 138-149
- URL: https://www.wjgnet.com/2150-5330/full/v7/i1/138.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v7.i1.138
The NKCC1 Na+-K+-2Cl- cotransporter, encoded by the SLC12A2 gene, is expressed in all mammalian tissues and is particularly prominent in epithelial cells of the gastrointestinal tract[1]. Detailed immunohistochemical studies show that it is localized to basolateral membranes of intestinal secretory epithelia[1], where it mediates uptake of much of the Cl- that is then secreted through the apical cystic fibrosis transmembrane conductance regulator (CFTR). In addition to secretory epithelia, NKCC1 is expressed in all segments of the gastrointestinal tract and throughout the crypt and villus epithelium, including both absorptive cells and goblet cells[1]. Thus, loss or inhibition of NKCC1 could have wide-ranging effects on gastrointestinal function. In fact, a National Institutes of Health web site (http://www.genome.gov/27551936) on Rare or Undiagnosed Diseases reports that humans with SLC12A2 mutations have “exercise intolerance, dilated cardiomyopathy, and episodic hypoglycemia progressing to unexplained failure of intestinal function/food tolerance with subsequent parenteral nutrition dependency”. The basis for the severe intestinal phenotype in humans is not known, but since it does not mimic the Cystic Fibrosis intestinal phenotype, it is unlikely to be due entirely to the secretory defect.
When anion secretion is impaired, the intestinal lumen can become desiccated, which leads to constipation, obstructions, and intussusception. Mice lacking CFTR usually develop lethal intestinal obstructions by 6 wk of age and require treatment with an osmotic laxative for normal survival[2], but the intestinal phenotype of Nkcc1-/- mice is much less severe[3]. In the original study, a small percentage of the Nkcc1-/- mice on a mixed genetic background developed intestinal impactions[3]. However, Nkcc1-/- mice on the FVB/N background (Swiss mice carrying the Fv1b sensitizing allele) that were used for the current study do not develop impactions, and the intestinal tract appears histologically normal.
Loss of NKCC1 in mice causes a defect in intestinal anion secretion as measured by reductions in the bumetanide-sensitive component in short circuit currents in duodenum, jejunum, and cecum[3-5]. However, compensatory pathways for basolateral Cl- uptake have been identified in duodenum and colon[5,6], providing an explanation of why the loss of NKCC1 does not cause a profound defect in anion secretion in the intestinal tract. To gain a better understanding of the functions of NKCC1 and the molecular consequences of NKCC1-deficiency in the intestine, we performed microarray analyses of mRNA from Nkcc1-/- small intestine. The observed alterations in the expression of mRNAs encoding digestive enzymes and receptors that mediate chemical sensing could be involved in intestinal pathologies resulting from the inhibition or loss of NKCC1.
Nkcc1-/- mice were generated as previously described[3] and inbred onto the FVB/N background for at least 10 generations. FVB/N are Swiss mice, derived from the HFSF/N strain, which carry the Fv1b (Friend Leukemia Virus strain B) sensitizing allele. These Nkcc1-/- mice are available from the Jackson Labs Mutant Mouse Regional Resource Center and can be identified by searching (http://www.jax.org/mmrrc/) with the gene symbol Slc12a2 (strain name: FVB.Cg-Slc12a2tm1Ges). Wild-type (WT) FVB/N mice were originally obtained from Jackson Labs and bred in house. All experimental pairs were derived by breeding of NKCC1 heterozygous breeding pairs, were between 8 and 12 wk of age, and were age- and gender-matched. Mouse numbers (n) are indicated in methods or figure legends. Mice were maintained in a specific pathogen-free barrier facility, with access to food and water ad libitum, and all experiments were approved by the University of Cincinnati Animal Care and Use Committee.
Mice were anesthetized by intraperitoneal injection of Avertin (2.5% solution of Tribromethanol) at 500 mg/kg, followed by cervical dislocation after the mice were anesthetized. Intestinal segments were removed and flushed with ice-cold phosphate-buffered saline. Some samples were frozen in liquid nitrogen and used for extraction of total RNA using Tri-Reagent (Molecular Research Center) and others were processed for immunolocalization studies as described in detail below.
Northern blot analysis was performed using 10 mg of total RNA per sample, and blots were hybridized with 32P-labeled cDNA as previously described[7]. Primer sequences are given in Table 1. Quantification of band intensity, including background subtraction and normalization to the ribosomal L32 subunit mRNA, was done using ImageQuant software (Amersham Biosciences).
| Name | Sequence |
| Pdx1 | F: ACCAAAGCTCACGCGTGGAAA |
| R: TGATGTGTCTCTCGGTCAAGTT | |
| Insulin 1 | F: TTGCCCTCTGGGAGCCCAAA |
| R: CAGATGCTGGTGCAGCACTG | |
| Insulin 2 | F: TCTTCCTCTGGGAGTCCCAC |
| R: CAGATGCTGGTGCAGCACTG | |
| Glut2 | F: CATTGCTGGAAGAAGCGTATCAG |
| R: GAGACCTTCTGCTCAGTCGACG | |
| Amylase | F: TTAACGATAATAAATGTAATGGAGAA |
| R: CCCTGCTACTCCAATGTCAA | |
| Trypsin | F: TGAGCAGTTTGTCAATTCTGCC |
| R: GCATGATGTCGTTGTTCAGGG | |
| Elastase | F: GTAGTTGCAGCCCAGAGAGG |
| R: TCTGCTATGTCACAGGCTGG | |
| Gp2 | F: TCCCTGCCAGAATCACACGGT |
| R: GGAGGTCCTGCCTACAGACACA | |
| L32 | F: GATCTAGCGGCCGCCATCTGTTTACGGCATCATG |
| R: TAGATCGCGGCCGCCGCTCCCATAACCGATGTTGG |
For quantitative real-time polymerase chain reaction (PCR), 4 μg of total RNA was reversed transcribed using oligo d(T) and SuperScript II reverse transcriptase (Invitrogen). The cDNA was diluted 5-fold in water. Primer sequences are given in Table 1. For each primer set, optimization using serially-diluted standards and melting curve analysis was performed. Real-time PCR was run on a DNA Engine Opticon II (MJ Research) thermalcycler using iQ SYBR Green (BioRad). For each gene, samples were run at least in duplicate, and mean values were normalized to the expression of L32. Standard curves were prepared for each gene on every run. All pairwise comparisons were done using a student’s t-test; P-values of < 0.05 were considered significant.
Intestinal segments were fixed in 10% neutral buffered formalin overnight and embedded in paraffin. Goblet cells were identified with hemotoxylin and eosin (H and E) or Alcian Blue/Periodic Acid-Schiff (AB/PAS) staining as described previously[8]. For immunofluorescence, sections (n = 4 WT and 3 NKCC1-null mutant) were deparaffinized in Citrisolve, rehydrated through successive ethanol washes, and boiled for 5 min in a sodium citrate buffer. Sections were incubated with primary antibodies for 12 h at room temperature, washed, and incubated with either another primary antibody or the appropriate secondary antibodies for 12 h at room temperature. Polyclonal antibodies against amylase (Sigma), trypsin (Abcam), elastase (Abcam), and intestinal trefoil factor 3 (TFF3) (ITF-Santa Cruz Biotechnology) were used at a 1:100 dilution; all secondary antibodies (Alexa Fluor, Molecular Probes) were used at a 1:200 dilution, and DAPI was included in the mounting medium (Vector Laboratories) for staining of nuclei. For localization studies using both fluorescence and AB/PAS, immunohistochemistry was done first and the slides were photographed. The slides were then washed, AB/PAS staining was performed, and the same areas were imaged. For colocalization studies, images were merged in Adobe Photoshop.
Microarray analyses were performed by the University of Cincinnati Genomics and Microarray Laboratory core facility exactly as described previously[8] using the Duke University Murine Operon v.3.0 spotted Array, which consisted of the Operon mouse oligonucleotide library, version 3.0, comprised of 70-mer oligonucleotides representing mouse genes. For analysis of NKCC1-null small intestine, total RNA was isolated from the entire small intestine of 8-wk old WT and NKCC1-null mutant (KO) mice (n = 6 of each genotype) on the FVB/N background, and cDNA was generated and labeled with Cy3 or Cy5 fluorophores and hybridized with the microarrays. The fold-changes and statistical significance for the microarray analysis were calculated exactly as described previously[8]. Briefly, Cy3 and Cy5 labeling was alternated between WT and KO samples (dye-flip controls) to control for the influence of fluorophore labeling on probe hybridization. Values are represented as fold-changes (ratio of fluorescence intensity values), with positive values indicating upregulation and negative values indication downregulation in KO relative to WT. The complete set of microarray data as well a list of the significantly changed genes and more detailed methods have been deposited in the National Center for Biotechnology Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are available through GEO accession number GSE19117. The statistical analysis of the data has been reviewed by Dr. Mario Medvedovic, Director of the Division of Biostatistics and Bioinformatics, Department of Environmental Health, University of Cincinnati.
Gene Onlology analysis was performed using the online GOrilla program[9] (Gene Ontology enRIchment anaLysis and visuaLizAtion tool) (http://cbl-gorilla.cs.technion.ac.il/). Two analysis options were used: (1) Single Rank List, with the entire gene set ranked according to P values; and (2) Two Unranked Lists, in which a target list of genes with a specific range of P values is compared against the list of all genes analyzed (13551 total). The Single Rank option avoids setting an arbitrary cutoff of P values and was useful for identifying GO categories that were highly enriched at the top end of the significance range. In the Two List analysis, significance and enrichment was calculated based on the number of genes in each GO category that appear in the target list and background list. Enrichment (N, B, n, b) is defined as: N is the total number of genes, B is the total number of genes associated with a specific GO term, n is the number of genes in the top of the user’s input list or in the target set when appropriate, b is the number of genes in the intersection; Enrichment = (b/n)/(B/N). The program calculates statistical probabilities using the hypergeometric distribution[9].
RNA Seq mRNA expression data for human duodenum and pancreas was obtained from the European Bioinformatics Institute (EBI) Expression Atlas (http://www.ebi.ac.uk/gxa/home). This data base allowed an evaluation of the relative expression levels of the various pancreatic genes that were upregulated in the duodenum of Nkcc1-/- mice. The data were available through baseline experiments, Uhlen’s lab, in which 32 human tissues are represented.
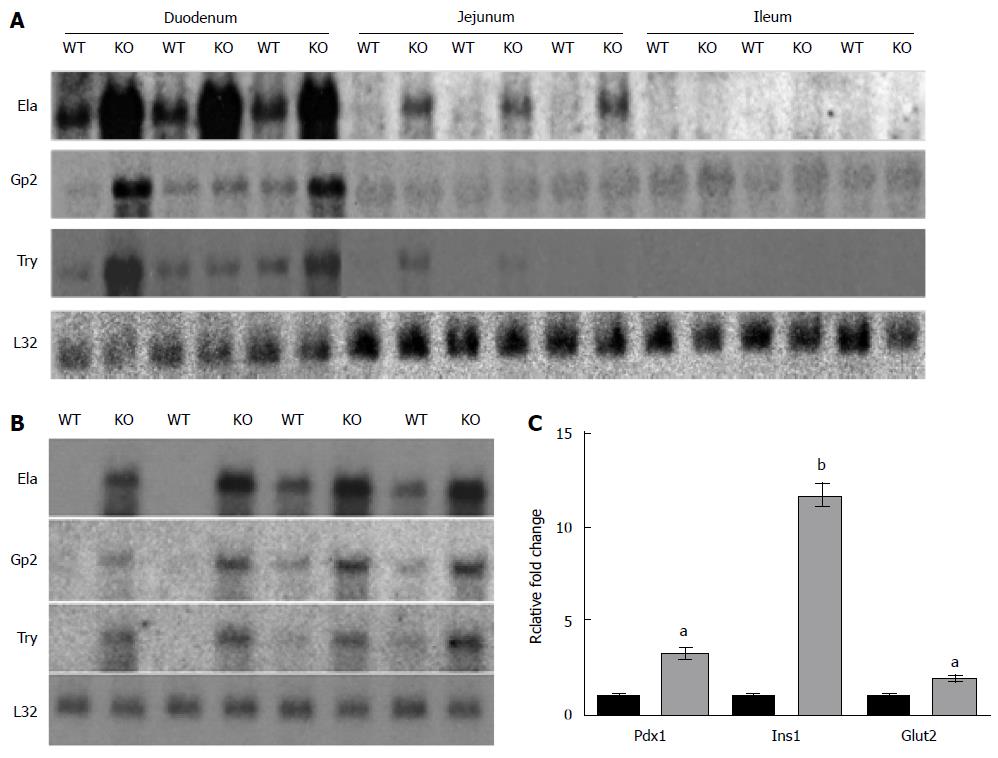
Microarray analysis was performed using small intestine RNA from the Nkcc1-/- and WT mice. Among the top 30 genes exhibiting differential expression in the Nkcc1-null intestine, all were upregulated, with induction ranging from 3.5-fold to 15.8-fold (Table 2). A pattern of genes typically associated with the exocrine pancreas, namely the digestive enzymes and components of the zymogen granules, was immediately apparent. Cpa1 is a pancreatic carboxypeptidase that was upregulated 15.8-fold. Elastases 1, 2, and 3B were all up-regulated and exhibited high fluorescence intensities. Several serine proteases, including trypsins 1, 2, 4, and 10, as well as chymotrypsins A, B and C, were also up-regulated. Amylase, another digestive enzyme produced by the pancreas, was increased 8.7-fold. Besides digestive enzymes, a number of the highly upregulated genes encode proteins that are associated with zymogen granules (see Discussion). These included glycoprotein 2 (Gp2, 3.9-fold), pancreatic protein disulfide isomerase (Pdia2, 4.5-fold), and syncollin (Sycn, 3.0-fold; P < 0.001, data not shown).
| Gene symbol | Description | Average intensity | Fold change | P value |
| Cpa1 | Carboxypeptidase A1, pancreatic | 1811 | 15.86 | 0.00001 |
| Elane | Elastase 2 | 2118 | 12.05 | 0.00001 |
| Glb1l3 | Galactosidase, beta 1 like 3 | 234 | 10.27 | 0.0001 |
| Try4 | Trypsin 4 | 2251 | 9.91 | 0.00001 |
| Rnase1 | Ribonuclease 1, pancreatic RNase A | 592 | 9.28 | 0.00001 |
| Cpb1 | Carboxypeptidase B1 | 866 | 9.15 | 0.00001 |
| Amy2a5 | Amylase 2, pancreatic | 691 | 8.75 | 0.00001 |
| Try10 | Trypsin 10 | 2306 | 8.33 | 0.00001 |
| Ctrc | Chymotrypsin C (caldecrin) | 397 | 7.79 | 0.0003 |
| Ctrb1 | Chymotrypsinogen B1 | 3360 | 7.66 | 0.00001 |
| Pnlip | Pancreatic lipase | 1938 | 7.24 | 0.00001 |
| Prss1 | Protease, serine 1 (trypsin 1) | 1481 | 6.93 | 0.00001 |
| Cel | Bile salt activated lipase, pancreatic | 356 | 6.73 | 0.00001 |
| Cela3b | Elastase 3B, pancreatic | 1576 | 6.69 | 0.00001 |
| Prss2 | Protease, serine 2 (trypsin II) | 716 | 6.52 | 0.0001 |
| Pnliprp1 | Pancreatic lipase related protein 1 | 574 | 5.83 | 0.0001 |
| Muc6 | Mucin 6, gastric | 152 | 5.78 | 0.0053 |
| Tff2 | Trefoil factor 2 | 1391 | 5.77 | 0.0200 |
| Cela1 | Elastase 1, pancreatic | 549 | 4.84 | 0.0002 |
| V1rh10 | Vomeronasal 1 receptor, H10 | 107 | 4.66 | 0.0008 |
| Calb3 | Calbindin D9K | 609 | 4.64 | 0.00001 |
| Reg1 | Lithostathine 1 (pancreatic stone protein 1) | 5648 | 4.56 | 0.0008 |
| Pdia2 | Protein disulfide isomerase, pancreatic | 292 | 4.47 | 0.0004 |
| Akp3 | Alkaline phosphatase 3, intestine | 366 | 4.02 | 0.00001 |
| Gp2 | Glycoprotein 2 (zymogen granule membrane) | 377 | 3.92 | 0.0001 |
| Cep350 | Centrosomal protein 350 | 141 | 3.92 | 0.0003 |
| Bmper | BMP-binding endothelial regulator | 318 | 3.78 | 0.0011 |
| Olfr846 | Olfactory receptor 846 | 432 | 3.66 | 0.0001 |
| Reg2 | Lithostathine 2 (pancreatic stone protein 2) | 253 | 3.58 | 0.0383 |
| Ctrl | Chymotrypsin like; chymotrypsin A | 1431 | 3.49 | 0.0030 |
In addition to enzymes typical of the exocrine pancreas, genes commonly expressed in the endocrine pancreas were also upregulated. These included Reg1 and Reg2 (regenerating islet or pancreatic stone proteins)[10], which were upregulated 4.6- and 3.6-fold (Table 2), insulin 1 (Ins1, 1.92-fold; P < 0.003), and pancreatic-duodenal homeobox 1 (Pdx1; 2.53-fold, P < 0.002), which is heavily involved in regulation of β-cell function[11]. Glut2, which is the most abundant facilitative glucose transporter in the intestine[12], was also upregulated (1.65-fold, P < 0.007).
Northern blot analysis and quantitative RT-PCR of several genes validated the results of the microarray analysis (Figure 1). The expression of elastase (4.7-fold), amylase (1.9-fold), glycoprotein 2 (3.4-fold), and trypsin (6.0-fold) were all significantly up-regulated in the duodenum (Figure 1A and B). The Northern blot data indicated that the up-regulation of these genes observed in microarray analyses of the Nkcc1-/- small intestine occurs primarily in the duodenum. It should be noted that expression of these genes in the duodenum appeared to be robust and did not require long exposure times to be detected, even in the WT samples. RT-PCR confirmed upregulation of Pdx1, Ins1, and Glut2 in the Nkcc1-/- small intestine (Figure 1C).
In previous studies the stomach, intestine, liver, and pancreas of Nkcc1-/- mice appeared histologically normal[3], and no apparent differences in the morphology of the gastrointestinal tract was observed in the current study. Based on the results of the microarray analysis and histology, it is evident that the basic population of cells in the intestine of the Nkcc1-/- mice is not significantly altered. Expression of the Paneth cell markers matrix metalloproteinase 7 and lysozyme were not significantly changed, and neither were the expression of the major goblet cell markers mucin 2 and TFF3. The expression of sucrase-isomaltase (alpha glucosidase), associated with terminally-differentiated enterocytes, was also not significantly different. Collectively, these data support the histological assessment that the basic structure and cell populations in the gut of the mutant mice are not significantly changed.
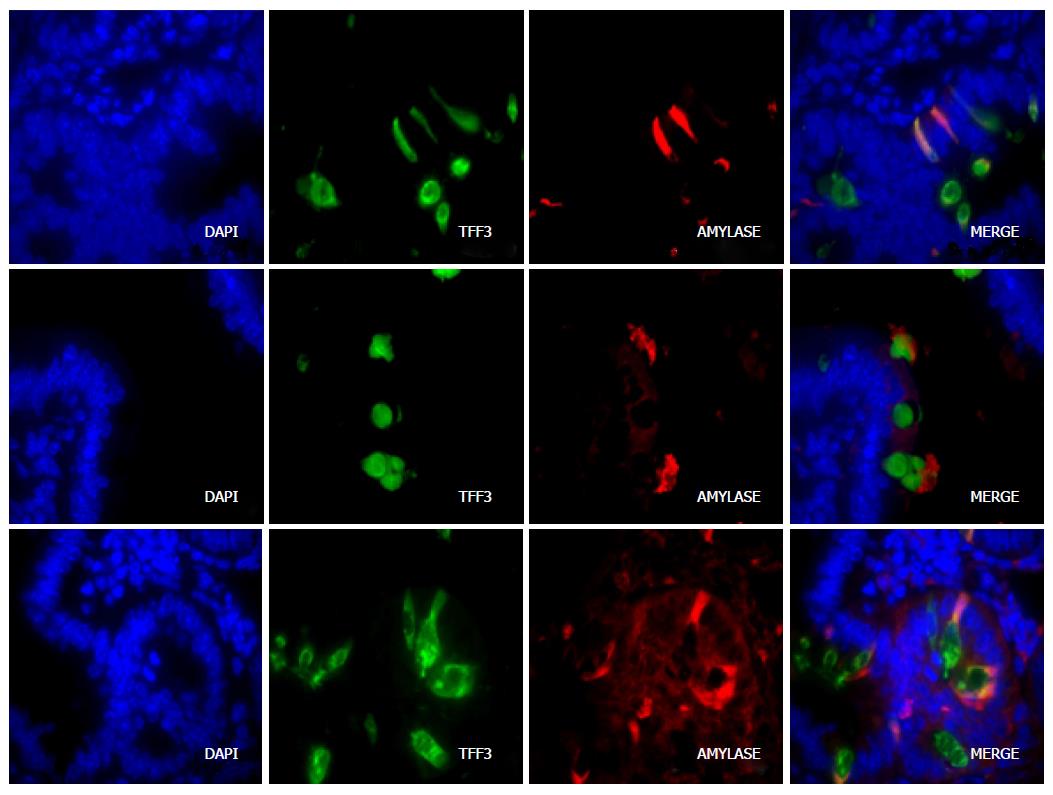
Immunofluorescence of amylase, trypsin, and elastase identified cytoplasmic staining of a subset of cells in the crypts and along the villi. Morphologically, these cells were clearly goblet cells. Co-immunofluorescence experiments using an antibody against intestinal TFF3, which is a goblet-cell specific marker, demonstrated that amylase, elastase and trypsin co-localize with Tff3 in goblet cells (Figure 2). Although mucus is known to bind antibodies non-specifically, in sections containing only the primary or secondary antibody, the goblet cells excluded all background fluorescence, indicating that the staining for each antibody was specific.
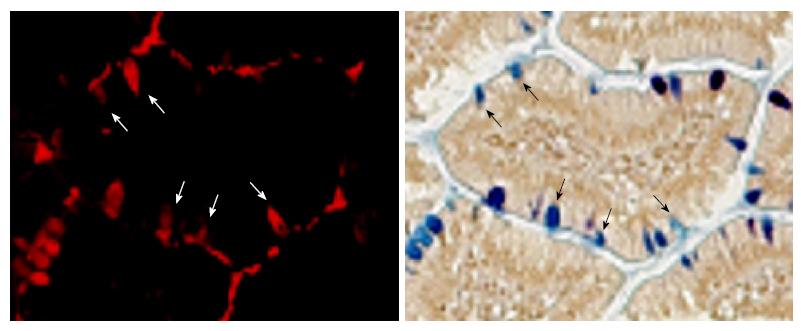
Interestingly, all amylase-positive cells contained TFF3; however, not all TFF3-positive cells were amylase-positive. This indicates that only a subset of goblet cells produce pancreatic-type peptidases and auxiliary zymogen granule proteins. Immunolocalization of amylase in cells that were also stained with AB/PAS provided further evidence that expression was occurring in a subset of goblet cells, as there were AB/PAS-positive goblet cells that lacked amylase (Figure 3). In some cases, it was possible to identify an amylase or elastase-positive cloud or cap over the lumenal border of the cells, and above the mucus, suggesting that these proteins are secreted from a separate compartment, or from separate granules, than TFF3-labeled mucins.
The differentially expressed genes were subjected to Gene Ontology analysis using the online GOrilla program[9], and significant GO categories were identified (Table 3). Using the single rank option (see Methods), the GO categories included those dealing with digestion and closely related protease and triglyceride lipase activities, which were highly enriched in the top end of the significance range. Sensory perception and the closely related olfactory receptor/G-protein coupled receptor activities were also enriched using the single rank option; these categories included genes spanning a much greater significance range; this was particularly apparent for olfactory receptor activities. Similar results were obtained using the two list option and a significance cutoff of P < 0.025, which analyzed the top 390 genes out of 13551 associated with known GO categories.
| GO category | P-value | Enrichment | (N, B, n, b) |
| Single ranked list | |||
| GO:0008236 Serine-type peptidase activity | 3.38E-14 | 35.6 | (13551, 135, 31, 11) |
| GO:0008233 Peptidase activity | 7.66E-11 | 12.6 | (13551, 451, 31, 13) |
| GO:0007586 Digestion | 1.82E-10 | 209.8 | (13551, 19, 17, 5) |
| GO:0007600 Sensory perception | 3.45E-8 | 1.67 | (13551, 817, 1270, 128) |
| GO:0006508 Proteolysis | 4.01E-8 | 7.65 | (13551, 743, 31, 13) |
| GO:0004984 Olfactory receptor activity | 7.11E-8 | 1.82 | (13551, 552, 1270, 94) |
| GO:0004806 Triglyceride lipase activity | 7.01E-7 | 271.0 | (13551, 10, 15, 3) |
| GO:0004930 GPCR activity | 3.86E-5 | 2.41 | (13551, 512, 352, 32) |
| Two unranked lists (target and background) | |||
| GO:0008236 Serine-type peptidase activity | 6.03E-8 | 4.63 | (13551, 135, 390, 18) |
| GO:0007586 Digestion | 5.79E-7 | 12.8 | (13551, 19, 390, 7) |
| GO:0004930 GPCR activity | 1.30E-5 | 2.24 | (13551, 512, 390, 33) |
| GO:0004984 Olfactory receptor activity | 5.86E-5 | 2.08 | (13551, 552, 390,3 3) |
| GO:0004806 Triglyceride lipase activity | 1.24E-4 | 13.9 | (13551, 10, 390, 4) |
| GO:0008233 Peptidase activity | 2.71E-4 | 2.08 | (13551, 451, 390, 27) |
The most statistically significant GO categories included genes encoding the pancreatic enzymes that were discussed above, along with genes for additional peptidases and lipases, most of which are directed at digestion of food in the intestinal tract (Table 4). Most of these genes were upregulated; the few that were modestly down-regulated were peptidases with either no apparent role in digestion (Otud4, Dpep3, and Hpn) or that were expressed at very low levels for a digestive enzyme, such as pepsinogen 5.
| Gene symbol | Description | Average intensity | Fold change | P-value |
| Cpa1 | Carboxypeptidase A1, pancreatic | 1811 | 15.86 | 0.00001 |
| Elane | Elastase 2 | 2118 | 12.05 | 0.00001 |
| Try4 | Trypsin 4 | 2251 | 9.91 | 0.00001 |
| Cpb1 | Carboxypeptidase B1 (tissue) | 866 | 9.15 | 0.00001 |
| Try10 | Trypsin 10 | 2306 | 8.33 | 0.00001 |
| Ctrc | Chymotrypsin C (caldecrin) | 397 | 7.79 | 0.0003 |
| Ctrb1 | Chymotrypsinogen B | 3360 | 7.66 | 0.00001 |
| Pnlip | Pancreatic lipase | 1938 | 7.24 | 0.00001 |
| Prss1 | Protease, serine 1 (trypsin 1) | 1481 | 6.93 | 0.00001 |
| Cel | Carboxyester lipase, pancreatic | 356 | 6.73 | 0.00001 |
| Cela3b | Elastase 3B, pancreatic | 1576 | 6.69 | 0.00001 |
| Prss2 | Trypsin II, anionic | 716 | 6.52 | 0.0001 |
| Pnliprp1 | Pancreatic lipase related protein 1 | 574 | 5.83 | 0.0001 |
| Cela1 | Elastase 1, pancreatic | 549 | 4.84 | 0.0002 |
| 2210010C04Rik | RIKEN cDNA 2210010C04 gene | 125 | 3.89 | 0.0005 |
| Ctrl | Chymotrypsin like; chymotrypsin A | 1431 | 3.49 | 0.0030 |
| Klk1 | Kallikrein 1 | 278 | 1.82 | 0.0180 |
| Klk1b26 | Kallikrein 1-related petidase b26 | 988 | 1.80 | 0.0030 |
| Pnliprp2 | Pancreatic lipase related protein 2 | 833 | 1.74 | 0.0181 |
| Klk1b21 | Kallikrein 21 | 386 | 1.66 | 0.0036 |
| Cyp7b1 | Cytochrome P450 7B1 | 100 | 1.53 | 0.0229 |
| Prcp | Prolyl carboxypeptidase (angiotensinase C) | 566 | 1.51 | 0.0047 |
| Amz1 | Archaelysin family metallopeptidase 1 | 887 | 1.35 | 0.0121 |
| Otud4 | OTU domain containing 4 | 524 | -1.34 | 0.0221 |
| Pga5 | Pepsinogen 5, group I | 168 | -1.42 | 0.0198 |
| Dpep3 | Dipeptidase 3 | 301 | -1.48 | 0.0197 |
| Hpn | Serine protease hepsin | 206 | -1.99 | 0.0211 |
The most intriguing changes involved altered expression of receptors involved in chemical sensing (Table 5). These included some G-protein coupled receptors (GPCRs) that are known to be involved in chemical sensing in the intestinal tract, such as Gpbar1[13] and Ffar2[14], as well as a large number of G-protein receptors of the olfactory receptor family and several of the vomeronasal receptor family. As discussed below, this suggests that the loss of NKCC1 affects chemosensing in the intestinal tract, and also suggests that olfactory and vomeronasal receptors play a major role in this process.
| Gene symbol | Description | Average intensity | Fold change | P-value |
| V1rh10 | Vomeronasal 1 receptor, H10 | 107 | 4.66 | 0.0008 |
| Olfr846 | Olfactory receptor 846 | 432 | 3.66 | 0.0001 |
| Olfr491 | Olfactory receptor 491 | 79 | 3.18 | 0.0051 |
| Olfr419 | Olfactory receptor 419 | 477 | 3.12 | 0.0004 |
| Olfr827 | Olfactory receptor 827 | 122 | 2.65 | 0.0001 |
| Olfr992 | Olfactory receptor 992 | 300 | 2.53 | 0.0005 |
| Olfr506 | Olfactory receptor 506 | 143 | 2.48 | 0.0049 |
| Olfr812 | Olfactory receptor 812 | 53 | 2.37 | 0.0048 |
| Gpr63 | G protein-coupled receptor 63 | 42 | 2.08 | 0.0023 |
| Olfr1080 | Olfactory receptor 1080 | 66 | 1.90 | 0.0107 |
| V1rh1 | Vomeronasal 1 receptor, H2 | 60 | 1.88 | 0.0071 |
| Olfr344 | Olfactory receptor 344 | 50 | 1.87 | 0.0057 |
| Olfr1340 | Olfactory receptor 1340 | 79 | 1.83 | 0.0123 |
| Olfr378 | Olfactory receptor 378 | 67 | 1.81 | 0.0072 |
| Olfr643 | Olfactory receptor 643 | 77 | 1.72 | 0.0080 |
| Olfr1000 | Olfactory receptor 996 | 475 | 1.72 | 0.0127 |
| Olfr1134 | Olfactory receptor 1134 | 139 | 1.71 | 0.0101 |
| Olfr906 | Olfactory receptor 906 | 134 | 1.68 | 0.0038 |
| Olfr736 | Olfactory receptor 736 | 92 | 1.67 | 0.0166 |
| Olfr433 | Olfactory receptor 433 | 189 | 1.65 | 0.0039 |
| Olfr78 | Olfactory receptor 78 | 125 | 1.62 | 0.0087 |
| Olfr90 | Olfactory receptor 90 | 106 | 1.56 | 0.0235 |
| Prokr1 | Prokineticin receptor 1 | 196 | 1.56 | 0.0096 |
| Olfr1494 | Olfactory receptor 1494 | 457 | 1.54 | 0.0180 |
| Olfr1462 | Olfactory receptor 1462 | 260 | 1.52 | 0.0183 |
| Olfr444 | Olfactory receptor 444 | 252 | 1.52 | 0.0154 |
| Nmur1 | Neuromedin U receptor 1 | 196 | 1.51 | 0.0177 |
| Olfr1366 | Olfactory receptor 1366 | 89 | 1.50 | 0.0207 |
| Olfr804 | Olfactory receptor 804 | 404 | 1.48 | 0.0175 |
| Gpbar1 | G protein-coupled bile acid receptor 1 | 129 | 1.47 | 0.0216 |
| Gprc5a | G protein-coupled receptor, C5A | 936 | 1.46 | 0.0142 |
| Olfr740 | Olfactory receptor 739 | 172 | 1.43 | 0.0164 |
| Olfr1156 | Olfactory receptor 152 | 156 | 1.43 | 0.0211 |
| Rxfp4 | Relaxin family peptide receptor 4 | 397 | 1.37 | 0.0132 |
| Igf2r | Insulin-like growth factor 2 receptor | 1089 | -1.39 | 0.0158 |
| Olfr641 | Olfactory receptor 641 | 652 | -1.39 | 0.0211 |
| Olfr48 | Olfactory receptor 140 | 99 | -1.48 | 0.0143 |
| Vmn1r210 | Vomeronasal 1 receptor 210 | 192 | -1.54 | 0.0127 |
| Grm2 | Glutamate receptor, metabotropic 2 | 179 | -1.63 | 0.0028 |
| Vmn1r5 | Vomeronasal 1 receptor 5 | 124 | -1.64 | 0.0071 |
| Ffar2 | Free fatty acid receptor 2 | 203 | -1.85 | 0.0092 |
The results of this study show that the loss of NKCC1 in small intestine leads to upregulation of digestive enzymes, many of which are typical of the exocrine pancreas, and to differential expression of a large number of genes encoding chemosensing receptors. As noted in the Introduction, loss-of-function mutations in the SLC12A2 (NKCC1) gene lead to unexplained failure of intestinal function/food tolerance (http://www.genome.gov/27551936). Although it is possible that impaired secretion contributes to this phenotype, considerable compensation for loss of NKCC1-mediated basolateral Cl--uptake occurs in the intestine[5,6] and Cystic Fibrosis patients and mice exhibit a more profound secretory defect than NKCC1-null mice. This suggests that factors other than impaired secretion account for the severe intestinal phenotype in humans with Nkcc1 mutations.
The most striking result of the microarray analyses, although perhaps of limited clinical importance (discussed below), was the increased expression of digestive enzymes that are normally expressed at high levels in the pancreas and the restriction of their expression to a subset of goblet cells. Because it is critical to prevent premature release of digestive enzymes within the pancreas, many digestive enzymes are produced and packaged as inactive forms in zymogen granules, which release the enzymes upon secretion into the duodenum. Several genes that encode proteins involved in zymogen granule function (Gp2, Sync, and Pdia2), were upregulated. Glycoprotein 2 is the most abundant protein on zymogen granules and is thought to regulate exocytosis at the apical pole of the granule[15]. Syncollin is found on the luminal surface of zymogen granules, and syncollin-deficient mice exhibit impaired exocytosis of granules from pancreatic acinar cells[16]. Pancreatic protein disulfide isomerase plays a major role in forming the disulfide linkages of secretory proteins and has been localized to zymogen granule membranes[17]. These expression patterns are consistent with increased synthesis and packaging of zymogen granule proteins in the Nkcc1-/- intestine.
During embryonic development, the endoderm gives rise to the pancreas, liver, and the epithelial lining of the gastrointestinal tract. This requires the expression of various transcription factors, as well as interactions between the epithelium and surrounding mesenchyme[18,19]. It has become clear that the relative plasticity of the pancreas, liver, and intestine is high, and that certain populations of cells within these organs can undergo transdifferentiation. Although this initially seemed to be an interesting possibility, it is unlikely that transdifferentiation is occurring in the Nkcc1-/- small intestine. Several transcription factors, including Pdx1 and pancreas transcription factor 1 (Ptf1a) have been shown to mediate plasticity between the pancreas and small intestine. Expression of Pdx1 is a key regulatory step in the development of both the pancreas and the duodenum[20], and inactivation of Ptf1a drives pancreatic precursor cells toward intestinal cell fates[21]. Although expression of pancreatic genes was upregulated, Pdx1 was only modestly increased and expression of Ptf1a was not changed (data not shown). Furthermore, when we examined relative expression levels of the pancreatic enzymes in human pancreas and duodenum that are available through the EBI Expression Atlas (see methods) it was clear that the pancreatic enzymes are expressed at several orders of magnitude higher levels in pancreas than in duodenum. Thus, the apparently robust expression of pancreatic enzymes in the Nkcc1-/- intestine is likely to be far below the levels of expression occurring in the pancreas. Similarly, although insulin (Ins1) was mildly induced in Nkcc1-/- intestine, the EBI expression Atlas data indicates that insulin expression in human pancreas is far greater than that in duodenum (relative transcript levels 551 in pancreas vs 0.3 in duodenum). Although we obtained an antibody against human insulin, which readily and specifically identified pancreatic islet cells in the mouse (data not shown), we were unable to detect insulin in Nkcc1-/- intestine, consistent with exceedingly low levels of expression.
The reason for the increased expression of pancreatic enzymes in goblet cells is unclear. Histological analyses gave no evidence for an increase in goblet cells in Nkcc1-/- intestine, and the expression levels of Tff3 (intestinal trefoil factor) and Muc2 (mucin 2), which are expressed at very high levels in goblet cells, were not significantly changed. However, NKCC1 has been shown to be expressed at high levels on basolateral membranes of goblet cells[1], and there is evidence that it affects mucous secretion[22]. Thus, it is possible that the absence of NKCC1 specifically in goblet cells has a direct effect on the expression of pancreatic enzymes. Whether this is due to subtle effects of NKCC1-deficiency on the differentiation of goblet cells is unclear.
Despite the changes in mRNA expression patterns between the WT and Nkcc1-/- intestine, previous studies of two different NKCC1 knockouts revealed no significant histological changes between the two genotypes in the adult intestine[3,4]. Furthermore, detailed morphometric analyses of the intestinal tracts of 8-wk-old WT and Nkcc1-/- mice, the same age as used in the current study, revealed no significant differences in cell structure[3]. We cannot, of course, rule out subtle changes in cell differentiation resulting from changes during early development or during regular turnover of the epithelium. However, the normal epithelial cell structure and relatively modest changes in expression of two of the major transcription factors involved in development of the pancreas and intestine, argue against defects in development and/or epithelial cell differentiation as the basis of the observed expression changes. Also, as discussed below, NKCC1 serves a direct role in the process of chemical sensing in olfactory neurons, suggesting that the changes in expression of chemosensing receptors is a direct response to the absence NKCC1 rather than being secondary to developmental defects.
Although less dramatic than the changes in pancreatic genes, the significant changes in expression of a large number of G-protein coupled chemosensing receptors suggest that the loss of NKCC1 may cause major impairments in the sensing of nutrients and other constituents of the intestinal contents. It is clear from many studies that GPCRs play a major role in intestinal chemosensing[23]. In fact, some of the GPCRs identified as differentially expressed in our microarray analyses have been shown to play important roles in chemosensing in the gut. Gpbar1 (1.47-fold increase) is expressed in enteroendocrine L-cells, where it senses bile acids and transduces a signal that leads to secretion of glucagon-like peptide 1 (GLP-1) and peptide tyrosine-tyrosine, thereby regulating glucose homeostasis[13,24]. Ffar2 (-1.85-fold decrease) is expressed in enteroendocrine cells and serves as a sensor of short-chain fatty acids[14]. Gpr63 (2.1-fold increase), which was expressed at relatively low levels, has been identified as a receptor for several lipids, including N-Arachidonylethanolamine[25] and both sphingosine 1-phosphate and dioleoylphasphatidic acid[26]. Grm2 (-1.63-fold decrease) is one of the metabotropic glutamate receptors that is involved in nutrient sensing through taste receptors[27]. Several of the differentially expressed GPCRs serve as receptors for peptide hormones that affect gut function. These include Rxfp4 (1.37-fold increase), which is the receptor for the orexigenic (appetite-stimulating) insulin-like peptide 5[28], and Nmur1 (1.51-fold increase), which is a receptor for neuromedin U and functions in glucose and appetite homeostasis[29].
The GPCRs that are known to play major roles in chemical sensing include taste receptors, olfactory receptors, and volmeronasal receptors. Taste receptors are known to function in the intestinal tract and to play major roles in nutrient sensing and subsequent signaling events[27,30-32]. There is less information about the role of olfactory receptors in chemosensing in the intestinal tract, but recent evidence indicates that olfactory receptors are expressed on colonic epithelial tissues, where they may detect bacterial metabolites in the luminal contents[33]. In addition, specific olfactory receptors have been shown to be differentially expressed in the duodenum of obesity-prone rats fed a high-fat diet[34]. The authors speculated that these receptors may be involved in sensing and responding to dietary fat. Specific olfactory receptors have also been identified in human enterochromaffin cells and when stimulated can lead to the release of serotonin[35,36]. Vomeronasal receptors also function in chemosensing[37], but as far as we are aware there is no previous evidence for their expression and function in the intestinal tract. Nevertheless, V1rh10 was the most highly upregulated gene among the G-protein coupled receptors (4.66-fold), indicating that it’s expression is responsive to the loss of NKCC1.
Previous studies have shown that NKCC1 plays an important role in signaling through olfactory receptors. Odorant-induced Cl- currents, which are required for signaling, were reduced in olfactory neurons of Nkcc1-/- mice[38], indicating that NKCC1 is an important mechanism for Cl--uptake in olfactory neurons. However, later studies provided strong evidence for additional Cl--loading mechanisms that can provide some compensation for the loss of NKCC1[39,40], and physiological studies indicated that WT and Nkcc1-/- mice have similar sensitivities to several specific odorants[41]. In more recent studies, however, loss of NKCC1 caused defects in the sensitivity to a complex mixture of odorants, and RNA Seq analysis of WT and Nkcc1-/- olfactory epithelium revealed significant differential expression changes involving a large number of olfactory receptors[42]. Thus, loss of NKCC1 leads to differential expression changes involving chemosensing receptors in both the olfactory epithelium[42] and in the small intestine, as indicated in the current study.
In summary, the microarray analysis of Nkcc1-/- small intestine indicates that the loss of NKCC1 leads both to increased expression of pancreatic enzymes in a subset of goblet cells and to differential expression of chemosensing GPCRs, including a large number of olfactory receptors. Because a number of GPCRs with well-established roles in chemosensing in the intestine were among the receptors that were changed, it seems likely that the differential expression of olfactory receptors is also indicative of altered sensing of components of the luminal contents. This finding raises the possibility that the episodic hypoglycemia and failure of intestinal function/food tolerance observed in human patients with NKCC1 (SLC12A2 gene) mutations may be due to chronic deficiencies in their ability to properly sense and respond to nutrients and other constituents of the intestinal lumen. Finally, it should also be noted that loop diuretics that inhibit NKCC1 have been associated with an increased incidence of intestinal dysfunction, including constipation[43] and indications of a poor outcome following ischemic colitis[44]. It is therefore possible that inhibition of NKCC1 by these common therapeutic agents could lead to intestinal dysfunction similar to that observed in NKCC1 genetic deficiencies. Whether this is due to developmental abnormalities involving one or more cell types in the intestinal epithelium and/or is a more direct response to impaired chemosensing is unclear.
In the intestine, the basolateral NKCC1 Na+-K+-2Cl- cotransporter is the major mechanism of chloride uptake in support of secretion; however, it is not restricted to secretory epithelia and may serve other functions as well. To gain insights regarding novel functions of NKCC1 in the intestine, mRNA expression patterns in wild-type and NKCC1-null intestine were analyzed.
This study was designed to determine changes in gene expression occurring in the intestinal tract of mice lacking NKCC1. Analysis of those patterns provided evidence that NKCC1 plays an important role in chemical sensing in the intestinal tract, a relatively new area of investigation.
This study demonstrates for the first time that mRNA levels for a large number of olfactory receptors and other G-protein coupled receptors involved in chemical sensing in the intestinal tract are altered in response to the loss of NKCC1.
The effects of NKCC1 ablation on the expression of large numbers of chemosensory receptors raises the possibility that the failure of intestinal function in response to genetic mutations in the human NKCC1 gene and side effects of loop diuretic treatment in the intestine could be due in part to impaired chemical sensing.
The Slc12a2 gene, encoding the NKCC1 Na+-K+-2Cl- cotransporter, was genetically ablated in the mouse model used in this study. The corresponding human gene is SLC12A2; mutations in human SLC12A2 gene have been shown to cause failure of intestinal function.
This study aimed to analyze the mRNA expression changes in the intestine of NKCC1-/- mice. The methods of this study, methods were properly used to identify cell types, and results were also presented clearly.
P- Reviewer: Koh TW, Wang YN S- Editor: Ji FF L- Editor: A E- Editor: Jiao XK
| 1. | Jakab RL, Collaco AM, Ameen NA. Physiological relevance of cell-specific distribution patterns of CFTR, NKCC1, NBCe1, and NHE3 along the crypt-villus axis in the intestine. Am J Physiol Gastrointest Liver Physiol. 2011;300:G82-G98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 2. | Clarke LL, Gawenis LR, Franklin CL, Harline MC. Increased survival of CFTR knockout mice with an oral osmotic laxative. Lab Anim Sci. 1996;46:612-618. [PubMed] |
| 3. | Flagella M, Clarke LL, Miller ML, Erway LC, Giannella RA, Andringa A, Gawenis LR, Kramer J, Duffy JJ, Doetschman T. Mice lacking the basolateral Na-K-2Cl cotransporter have impaired epithelial chloride secretion and are profoundly deaf. J Biol Chem. 1999;274:26946-26955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 297] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 4. | Grubb BR, Lee E, Pace AJ, Koller BH, Boucher RC. Intestinal ion transport in NKCC1-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2000;279:G707-G718. [PubMed] |
| 5. | Walker NM, Flagella M, Gawenis LR, Shull GE, Clarke LL. An alternate pathway of cAMP-stimulated Cl secretion across the NKCC1-null murine duodenum. Gastroenterology. 2002;123:531-541. [PubMed] |
| 6. | Gawenis LR, Bradford EM, Alper SL, Prasad V, Shull GE. AE2 Cl-/HCO3- exchanger is required for normal cAMP-stimulated anion secretion in murine proximal colon. Am J Physiol Gastrointest Liver Physiol. 2010;298:G493-G503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Woo AL, Gildea LA, Tack LM, Miller ML, Spicer Z, Millhorn DE, Finkelman FD, Hassett DJ, Shull GE. In vivo evidence for interferon-gamma-mediated homeostatic mechanisms in small intestine of the NHE3 Na+/H+ exchanger knockout model of congenital diarrhea. J Biol Chem. 2002;277:49036-49046. [PubMed] |
| 8. | Bradford EM, Sartor MA, Gawenis LR, Clarke LL, Shull GE. Reduced NHE3-mediated Na+ absorption increases survival and decreases the incidence of intestinal obstructions in cystic fibrosis mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G886-G898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 9. | Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2442] [Cited by in RCA: 2520] [Article Influence: 157.5] [Reference Citation Analysis (0)] |
| 10. | Qiu L, List EO, Kopchick JJ. Differentially expressed proteins in the pancreas of diet-induced diabetic mice. Mol Cell Proteomics. 2005;4:1311-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 75] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Kaneto H, Matsuoka TA, Miyatsuka T, Kawamori D, Katakami N, Yamasaki Y, Matsuhisa M. PDX-1 functions as a master factor in the pancreas. Front Biosci. 2008;13:6406-6420. [PubMed] |
| 12. | Thorens B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia. 2015;58:221-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 493] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 13. | Ullmer C, Alvarez Sanchez R, Sprecher U, Raab S, Mattei P, Dehmlow H, Sewing S, Iglesias A, Beauchamp J, Conde-Knape K. Systemic bile acid sensing by G protein-coupled bile acid receptor 1 (GPBAR1) promotes PYY and GLP-1 release. Br J Pharmacol. 2013;169:671-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 14. | Nøhr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, Sichlau RM, Grunddal KV, Poulsen SS, Han S. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154:3552-3564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 428] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 15. | Parker EM, Zaman MM, Freedman SD. GP2, a GPI-anchored protein in the apical plasma membrane of the pancreatic acinar cell, co-immunoprecipitates with src kinases and caveolin. Pancreas. 2000;21:219-225. [PubMed] |
| 16. | Wäsle B, Turvey M, Larina O, Thorn P, Skepper J, Morton AJ, Edwardson JM. Syncollin is required for efficient zymogen granule exocytosis. Biochem J. 2005;385:721-727. [PubMed] |
| 17. | Bruneau N, Lombardo D, Levy E, Bendayan M. Roles of molecular chaperones in pancreatic secretion and their involvement in intestinal absorption. Microsc Res Tech. 2000;49:329-345. [PubMed] |
| 18. | Montgomery RK, Mulberg AE, Grand RJ. Development of the human gastrointestinal tract: twenty years of progress. Gastroenterology. 1999;116:702-731. [PubMed] |
| 19. | Stainier DY. No organ left behind: tales of gut development and evolution. Science. 2005;307:1902-1904. [PubMed] |
| 20. | Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983-995. [PubMed] |
| 21. | Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128-134. [PubMed] |
| 22. | Dolganov GM, Woodruff PG, Novikov AA, Zhang Y, Ferrando RE, Szubin R, Fahy JV. A novel method of gene transcript profiling in airway biopsy homogenates reveals increased expression of a Na+-K+-Cl- cotransporter (NKCC1) in asthmatic subjects. Genome Res. 2001;11:1473-1483. [PubMed] |
| 23. | Reimann F, Tolhurst G, Gribble FM. G-protein-coupled receptors in intestinal chemosensation. Cell Metab. 2012;15:421-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 24. | Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1223] [Cited by in RCA: 1393] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 25. | Yin H, Chu A, Li W, Wang B, Shelton F, Otero F, Nguyen DG, Caldwell JS, Chen YA. Lipid G protein-coupled receptor ligand identification using beta-arrestin PathHunter assay. J Biol Chem. 2009;284:12328-12338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 252] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 26. | Niedernberg A, Tunaru S, Blaukat A, Ardati A, Kostenis E. Sphingosine 1-phosphate and dioleoylphosphatidic acid are low affinity agonists for the orphan receptor GPR63. Cell Signal. 2003;15:435-446. [PubMed] |
| 27. | Xiao W, Feng Y, Holst JJ, Hartmann B, Yang H, Teitelbaum DH. Glutamate prevents intestinal atrophy via luminal nutrient sensing in a mouse model of total parenteral nutrition. FASEB J. 2014;28:2073-2087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Grosse J, Heffron H, Burling K, Akhter Hossain M, Habib AM, Rogers GJ, Richards P, Larder R, Rimmington D, Adriaenssens AA. Insulin-like peptide 5 is an orexigenic gastrointestinal hormone. Proc Natl Acad Sci USA. 2014;111:11133-11138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 29. | Dalbøge LS, Pedersen SL, Secher T, Holst B, Vrang N, Jelsing J. Neuromedin U inhibits food intake partly by inhibiting gastric emptying. Peptides. 2015;69:56-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Mace OJ, Lister N, Morgan E, Shepherd E, Affleck J, Helliwell P, Bronk JR, Kellett GL, Meredith D, Boyd R. An energy supply network of nutrient absorption coordinated by calcium and T1R taste receptors in rat small intestine. J Physiol. 2009;587:195-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 123] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 31. | Wang JH, Inoue T, Higashiyama M, Guth PH, Engel E, Kaunitz JD, Akiba Y. Umami receptor activation increases duodenal bicarbonate secretion via glucagon-like peptide-2 release in rats. J Pharmacol Exp Ther. 2011;339:464-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 32. | Rønnestad I, Akiba Y, Kaji I, Kaunitz JD. Duodenal luminal nutrient sensing. Curr Opin Pharmacol. 2014;19:67-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Kaji I, Karaki S, Kuwahara A. Taste sensing in the colon. Curr Pharm Des. 2014;20:2766-2774. [PubMed] |
| 34. | Primeaux SD, Braymer HD, Bray GA. High fat diet differentially regulates the expression of olfactory receptors in the duodenum of obesity-prone and obesity-resistant rats. Dig Dis Sci. 2013;58:72-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 35. | Braun T, Voland P, Kunz L, Prinz C, Gratzl M. Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology. 2007;132:1890-1901. [PubMed] |
| 36. | Kidd M, Modlin IM, Gustafsson BI, Drozdov I, Hauso O, Pfragner R. Luminal regulation of normal and neoplastic human EC cell serotonin release is mediated by bile salts, amines, tastants, and olfactants. Am J Physiol Gastrointest Liver Physiol. 2008;295:G260-G272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 171] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 37. | Chamero P, Leinders-Zufall T, Zufall F. From genes to social communication: molecular sensing by the vomeronasal organ. Trends Neurosci. 2012;35:597-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 38. | Reisert J, Lai J, Yau KW, Bradley J. Mechanism of the excitatory Cl- response in mouse olfactory receptor neurons. Neuron. 2005;45:553-561. [PubMed] |
| 39. | Nickell WT, Kleene NK, Gesteland RC, Kleene SJ. Neuronal chloride accumulation in olfactory epithelium of mice lacking NKCC1. J Neurophysiol. 2006;95:2003-2006. [PubMed] |
| 40. | Nickell WT, Kleene NK, Kleene SJ. Mechanisms of neuronal chloride accumulation in intact mouse olfactory epithelium. J Physiol. 2007;583:1005-1020. [PubMed] |
| 41. | Smith DW, Thach S, Marshall EL, Mendoza MG, Kleene SJ. Mice lacking NKCC1 have normal olfactory sensitivity. Physiol Behav. 2008;93:44-49. [PubMed] |
| 42. | Haering C, Kanageswaran N, Bouvain P, Scholz P, Altmüller J, Becker C, Gisselmann G, Wäring-Bischof J, Hatt H. Ion transporter NKCC1, modulator of neurogenesis in murine olfactory neurons. J Biol Chem. 2015;290:9767-9779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 43. | Fosnes GS, Lydersen S, Farup PG. Constipation and diarrhoea - common adverse drug reactions? A cross sectional study in the general population. BMC Clin Pharmacol. 2011;11:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 44. | Mosli M, Parfitt J, Gregor J. Retrospective analysis of disease association and outcome in histologically confirmed ischemic colitis. J Dig Dis. 2013;14:238-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |