Published online Sep 22, 2021. doi: 10.4291/wjgp.v12.i5.84
Peer-review started: March 3, 2021
First decision: April 19, 2021
Revised: April 28, 2021
Accepted: August 11, 2021
Article in press: August 11, 2021
Published online: September 22, 2021
Processing time: 196 Days and 9 Hours
Cholangiocarcinoma (CCA) is a type of cancer with increasing prevalence around the world that originates from cholangiocytes, the epithelial cells of the bile duct. The tumor begins insidiously and is distinguished by high grade neoplasm, poor outcome, and high risk for recurrence. Liver transplantation has become broadly accepted as a treatment option for CCA. Liver transplantation is expected to play a crucial role as palliative and curative therapy for unresectable hilar CCA and intrahepatic CCA. The purpose of this study was to determine which cases with CCA should be subjected to liver transplantation instead of resection, although reported post-transplant recurrence rate averages approximately 20%. This review also aims to highlight the molecular current frontiers of CCA and directions of liver transplantation for CCA.
Core Tip: Currently, there are many controversial hypotheses concerning liver transplantation in cholangiocarcinoma (CCA) and risk factors and molecular pathogenesis of CCA, with a focus on primary sclerosing cholangitis. Here, we mainly review the current advances in classification and treatment of CCA.
- Citation: Safarpour AR, Askari H, Ejtehadi F, Azarnezhad A, Raeis-Abdollahi E, Tajbakhsh A, Abazari MF, Tarkesh F, Shamsaeefar A, Niknam R, Sivandzadeh GR, Lankarani KB, Ejtehadi F. Cholangiocarcinoma and liver transplantation: What we know so far? World J Gastrointest Pathophysiol 2021; 12(5): 84-105
- URL: https://www.wjgnet.com/2150-5330/full/v12/i5/84.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v12.i5.84
Cholangiocarcinomas (CCA), also known as bile duct cancer, constitute a diverse group of biliary epithelial tumors affecting the intrahepatic CCA (iCCA), perihilar CCA (pCCA), and distal bile duct CCA (dCCA)[1]. CCA is the second leading cause of liver malignancy after hepatocellular carcinoma (HCC), and the overall incidence and mortality rates of CCA have increased progressively worldwide in the last 4 decades[2]. Primary sclerosing cholangitis (PSC) as a chronic liver disease can increase the risk for CCA reaching approximately 10% or 398-fold vs with the general population[3,4]. CCA has remained the common cause of death at the global level among PSC patients, whereby 30% of all CCAs are recognized annually after diagnosing PSC[5,6]. CCA is generally considered to be one of the contraindications in relation to liver transplantation characterized by poor prognosis. CCA patients have a median survival of 2 years following diagnosis. The only potentially curative treatment chance is surgery, depending on the stages of disease[7]. It has been shown that neoadjuvant therapy with liver transplantation as a novel treatment exhibits better survival rates with fewer recurrence in comparison with conventional resection for localized, node-negative hilar cholangiocarcinoma (hCCA)[8]. CCAs are a highly aggressive epithelial malignancy, and many patients represent advanced stages of disease[9]. Early detection of CCA still remains a challenge owing to its ‘silent’ clinical feature (most patients in the initial stage at the time of diagnosis are asymptomatic) and difficult to reach anatomical sites[10]. It seems that the use of liver transplantation for the treatment of CCA can influence clinical outcomes in patients around the world. This class of tumor driving from the bile duct epithelial cells is clinically malignant, and its occurrence and prognosis are mostly associated with its anatomic location within the biliary tree and its chance to achieve complete resection with negative margins[11]. This review summarizes the risk factors and molecular pathogenesis of CCA, with a focus on PSC and liver transplantation along with advances in classification and treatment.
CCA may be originated from the different cell types of the biliary tract, including cholangiocytes, the epithelial cells lining of the biliary surface epithelium, the epithelial cells of the peribiliary glands, hepatic progenitor cells, or any other mature hepatocytes that have become malignant. In this regard, CCA could also be classified in terms of anatomical, histological, and molecular aspects[12].
According to anatomical location of the tumor, CCA will most commonly be classified into three sub-groups: (1) iCCA; (2) pCCA; and (3) dCCA (Table 1)[12,13]. Given the tumor location, iCCA typically arises from the intrahepatic biliary tract including segmental bile ducts to smaller branches of the intrahepatic biliary system. Thus, this subtype of CCA occurred in the periphery of the second-order bile ducts[13]. Also, iCCA represents approximately 20% of all CCA reported cases[14]. pCCA arises around the hepatic ducts and their junctions[15]. Finally, dCCA refers to the malignancy that occurs in the common bile duct, i.e. originated from Vater’s ampulla[14,16].
| Anatomical classification | Histomorphological classification | Molecular specification (gene alterations) | |
| iCCA | Small intrahepatic bile ducts iCCA | Mass forming tumors[17] | IDH1/2, FGFR2, EPHA2, BAP1[14,19] |
| Large intrahepatic bile ducts iCCA | Mass forming, periductal, or intraductal mucinous tumors[17,18] | EPHA2, BAP1, KRAS, TP53, GNAS, NRAS, MRAS, SMAD4[12,14,21] | |
| eCCA | Perihilar CCA | Intraductal mucinous tumors[12,17] | KRAS, TP53, GNAS, NRAS, MRAS, SMAD4, ARID1B, PRKACA, BRAF[14,19,24] |
| Distal CCA | Periductal mucinous tumors[12,17] | KRAS, TP53, GNAS, NRAS, MRAS, SMAD4, ARID1B, PRKACB, BRAF[14,19,24] | |
From the histological point of view, characteristics of pCCA and dCCA that can be considered as extrahepatic CCA (eCCA) subtypes are conventionally mucin-producing adenocarcinomas or papillary tumors. On the other hand, iCCAs are more heterogeneous than two other subtypes of CCA. Histological studies showed that the adenocarcinoma is formed by columnar to cuboidal epithelial cells in the tubular structures, acini formation, and micropapillary architecture with variable morphological aspects, which are the most common types of iCCA[12].
Moreover, it has been suggested that, according to the level or size of the displayed bile duct, iCCA is classified into two main histological subtypes. First, the small bile duct iCCA that presents as small-sized tubular or acinar adenocarcinoma. These tumors commonly originated from small intrahepatic bile ducts, progenitor cells, and mature hepatocytes[17]. In contrast, large bile ducts iCCA derive from large intrahepatic bile ducts and/or associated peribiliary glands. Moreover, depending on the origin of the large bile duct iCCA, the histological aspects of this subtype of iCCAs are partly similar to pCCA and dCCA. However, the gross examination is not sufficient for accurate tumor classification, and further histological, molecular, and clinical investigation is required[18].
First, it needs to be explained that, due to some differences in the characteristics of the existing studies, including different molecular detection methods and diversity in the selection of populations, there is still no consensus on the molecular characteristics of CCA classification[17]. However, it is possible to establish an acceptable relationship between the anatomical and molecular aspects of CCA subtypes. Integrative molecular analyses not only provided the functional information for CCA classification but also were used to understand the pathogenesis and signaling pathways underlying the CCA carcinogenesis and progression[14].
Mutation-based classification is the main approach of CCA molecular classification. For instance, the isoforms 1 and 2 of isocitrate dehydrogenase (IDH1 and IDH2) and NRAS mutations are the main molecular manifestation of iCCA, whereas eCCA typically showed TP53, KRAS, and BRAF mutations[14,19]. Also, it has been reported that IDH1/2 and BAP1 mutations and fibroblast growth factor receptor 2 (FGFR2) fusions are the main molecular characteristics of iCCA, while protein kinase CAMP-activated catalytic subunit alpha (PRKACA) and AT-rich interactive domain-containing protein 1B mutations are more common in eCCA. Besides, KRAS, GNAS, and TP53 mutations are shared between iCCA and eCCA[19]. Interestingly, FGFR2 pairs with PRKACA in iCCA, as well as PRKACB in eCCA[20]
On the other hand, previous molecular studies also have attempted to connect the morphological CCA subtypes with specific molecular-based patterns. In this regard, the large-duct type iCCAs have a specific molecular property such as high mutation frequency of oncogenes and tumor suppressor genes and lack other gene mutations that are typically seen in small-duct iCCA. It has been reported that KRAS and TP53 are two prominent genes with high mutation frequency in the large-duct type iCCAs as well as lack of IDH1/2 mutations and FGFR2-fusions, which are molecular characteristics of small-duct iCCA[21].
In addition to mutation and sequence alterations, epigenetic study based on the methylation profiles of CCA subtypes can be used for CCA classification. For example, CCA has been related to hypermethylation at the promoter of tumor suppressor genes, such as DAPK, P14 (ARF), and ASC[22]. Moreover, despite the different patterns of methylation in GC-rich regions (CpG islands) in the CCA subtypes-related genes, it has been revealed that there is an alteration in CpG methylation that belonged to WNT, transforming growth factor-β, phosphatidylinositol 3 kinase, mitogen-activated protein kinase, and NOTCH signaling pathways[14]. Furthermore, the results of various studies showed that molecular characteristics of CCA subtypes consisting of sequence and copy number alterations, gene expression, and DNA methylation can be categorized into different clusters, but the details of this issue are beyond the scope of this article[23].
In addition to the mentioned above, another recent large cohort of CCA suggested that according to whole-gene expression data, chromosomal aberrations, and signaling pathway activation, CCA can be divided into two molecular subgroups: (1) inflammation class; and (2) proliferation class, which accounts for 38% and 62% of CCA cases, respectively[24]. The inflammation class of CCA has been characterized by the activation of inflammatory response and overexpression of T helper 2)-related cytokines and down-regulation of Th1-related cytokines. Moreover, it has been reported that several oncogenic pathways were enriched in the proliferation class that is accompanied by activation of receptor tyrosine kinase pathways (i.e. epidermal growth factor, RAS, AKT, MET, angiogenesis-related vascular endothelial growth factor, and platelet-derived growth factor) and Kirsten rat sarcoma viral oncogene homolog mutations[14,24].
Despite all of the before-mentioned data about molecular CCA classification, many other studies provide more useful information about molecular characteristics of CCA subtypes, such as the information derived from the noncoding RNA alteration, proteomics, and radiogenomic studies, which should be discussed in a separate article focusing on molecular classification of CCA[24].
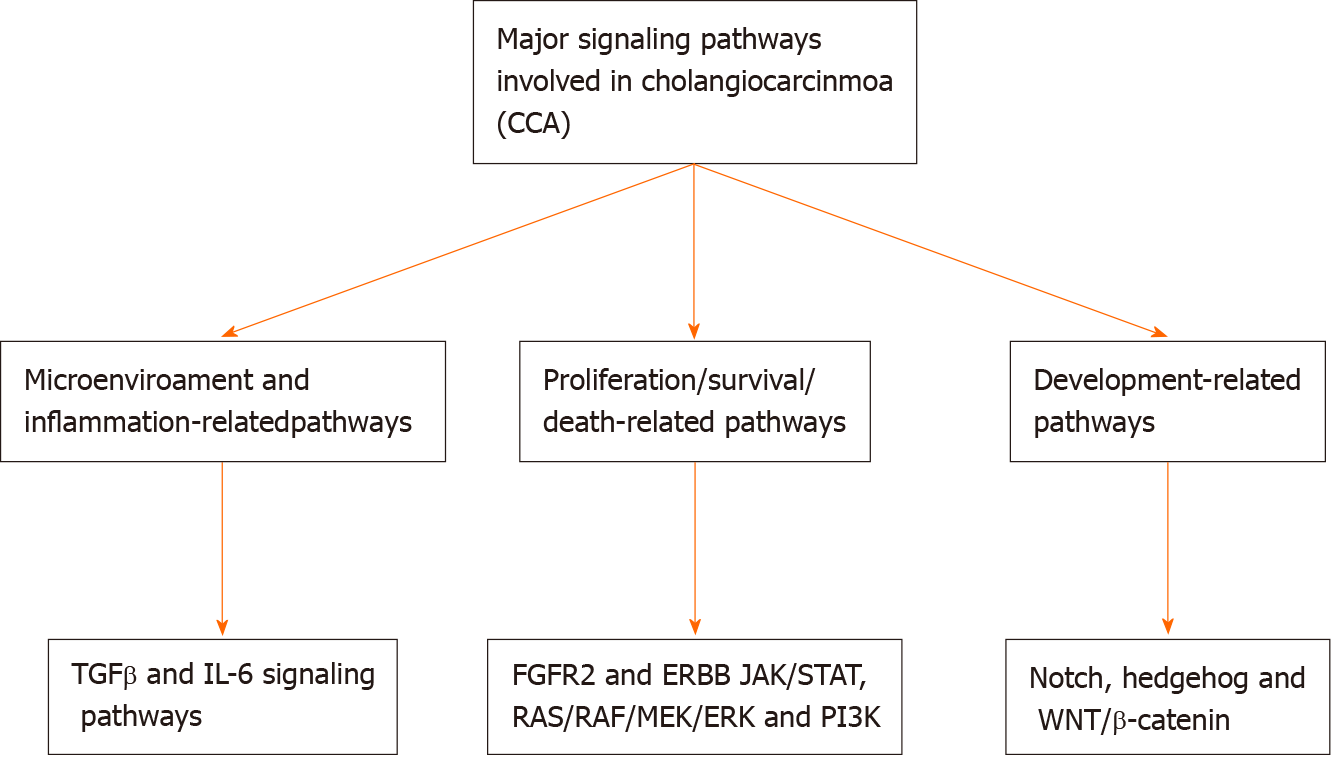
Cholangiocarcinogenesis is linked not only with genetic and epigenetic alterations but also with major changes in the microenvironment of the tumor. These modifications contribute to the triggering of different signaling pathways that are able to drive the initiation and progression of tumors[25]. Chronic inflammation contributes to increased exposure of cholangiocytes to Wnt inflammatory mediators, interleukin-6, cyclo-oxygenase-2, and tumor necrosis factor-alpha, leading to progressive mutations in some critical cancer-related genes including tumor suppressors, proto-oncogenes, and DNA mismatch-repair[26]. Increased apoptosis, decreased pH, and activation of extracellular signal-regulated kinase 1/2, Akt, and nuclear factor-kappa B signaling pathways following the accumulation of bile acids from cholestasis lead to promotion of survival, cell proliferation, and migration. Vascular endothelial growth factor, transforming growth factor-β, hepatocyte growth factor, and other microRNAs (miRNAs) are other mediators that are upregulated in CCA. Tumor development, angiogenesis, and migration are triggered by increased expression of the glucose transporter protein type 1, the cell surface receptor c-Met, and the sodium iodide symporter. The composition of the extracellular matrix and macrophage/fibroblast recruitment result in stromal shifts that establish a microenvironment to promotes cell survival, invasion, and metastasis[25,27-29]. The major signaling pathways involved in CCA are illustrated in Figure 1.
Few studies have described chromosomal abnormalities in CCA, and, due to the limited number of samples and large genetic variation between the population groups examined, the findings have been difficult to interpret. Data have revealed gains at 1q, 7p, 8q, 17q, and/or 20q and losses at 1p, 3p, 4q, 6q, 8p, 9pq, 13q, 14q, 17p, 18q, and/or 21q[24,30]. Curiously, genetic heterogeneity may be correlated with CCA in cells other than cholangiocytes. Natural killer cells and T-lymphocytes, for instance, express the natural killer group 2D receptor that plays an important role in cytotoxicity and tumor surveillance regulated by cells. One study indicated that the risk of experiencing CCA ranged significantly in patients with PSC, according to the patient's natural killer group 2D alleles; some were protective and others more than doubled the risk[31]. As potential risk factors for CCA, host genetic factors, alone or combined with environmental factors, have been investigated. For polymorphic variants that may be correlated with greater vulnerability to CCA, genes coding for xenobiotic detoxification, multidrug resistance, enzymes responsible for carcinogen metabolism, DNA repair, folate metabolism, and inflammation have been investigated. However, due to the inclusion of gallbladder and ampullary cancers in their evaluation in some of these reports and the lack of replication in separate cohorts, no conclusive conclusions can be taken. Multiple gene polymorphisms have been correlated with greater and reduced danger of experiencing CCA in many hospital-based, case-control studies. Due to the different populations of the sample and the lack of replication of the study in separate cohorts, it is hard to draw definite conclusions about these results. Table 2 summarizes genetic mutations and polymorphisms associated with CCA.
| Gene (Full name) | Protein (Full name) | Normal function(s) | Ref. |
| ATP8B1 (ATPase Phospholipid Transporting 8B1) | FIC1 (Familial Intrahepatic Cholestasis type 1) | Transmembrane phospholipid transfer | Wadsworth et al[88], 2011 |
| ABCB11 (ATP Binding Cassette Subfamily B Member 11) | BSEP (Bile Salt Exporter Pump) | Transport of cholate conjugates from hepatocytes to bile | Wadsworth et al[88], 2011 |
| ABCC2 (ATP Binding Cassette Subfamily C Member 2) | MRP2 (Multidrug resistance-associated protein 2) | Transport of endogenous and xenobiotic compounds from hepatocytes to bile | Hoblinger et al[89], 2009 |
| ABCB4 (ATP Binding Cassette Subfamily B Member 4) | MDR3 (MHC class I polypeptide-related sequence A) | Transport of lipids from hepatocytes to bile | Khabou et al[90], 2019 |
| COX-2 (Cyclooxygenase 2) | COX-2 (Cyclooxygenase 2) | Inflammatory cytokine | Kim et al[91], 2002 |
| CYP1A2 (Cytochrome P450 1A2) | CYP1A2 (Cytochrome P450 1A2) | Xenobiotic metabolism | Prawan et al[92], 2005 |
| KLRK1 (Killer Cell Lectin Like Receptor K1) | NKG2D (NKG2-D type II integral membrane protein) | Tumor surveillance | Melum et al[93], 2008 |
| MTHFR (Methylenetetrahydrofolate Reductase) | MTHFR (5,10-Methylenetetrahydrofolate reductase) | DNA methylation | Ko et al[94], 2006 |
| NAT2 (N-Acetyltransferase 2) | ARY2(Arylamine N-acetyltransferase 2) | Drug and carcinogen metabolism | Prawan et al[92], 2005 |
| PTGS2 (Prostaglandin-endoperoxide synthase 2) | PTGS2 (Prostaglandin G/H synthase 2) | The key enzyme in prostaglandin biosynthesis, and acts both as a dioxygenase and as a peroxidase | Sakoda et al[95], 2006 |
| XRCC1 (X-ray repair cross complementing 1) | XRCC1 (DNA repair protein XRCC1) | Involved in DNA single-strand break repair by mediating the assembly of DNA break repair protein complexes | Huang et al[96], 2008 |
| GSTO1(Glutathione S-transferase omega-1) | GST01 (Glutathione S-transferase omega-1) | Detoxification of endogenous and xenobiotic compounds | Marahatta et al[97], 2006 |
| MICA (MICA PERB11.1) | MICA (MHC class I polypeptide-related sequence A) | Stress-induced self-antigen and Ligand for the KLRK1/NKG2D receptor | Melum et al[93], 2008 |
| NR1H4(Nuclear Receptor Subfamily 1 Group H Member 4) | BAR (FXR) (Bile acid receptor (Farnesoid X receptor) | Negative feedback inhibitor of bile acid synthesis | Wadsworth et al[88], 2011 |
| TYMS (Thymidylate Synthetase) | TYMS (Thymidylate synthase) | DNA repair | Razumilava et al[61], 2014 |
| XRCC1 (X-Ray Repair Complementing Defective Repair in Chinese Hamster Cells 1) | XRCC1 (DNA repair protein XRCC1) | DNA repair | Gong et al[98], 2015 |
| APC (Adenomatous polyposis coli) | APC (Adenomatous polyposis coli) | Tumor suppressor | Kang et al[99], 1999 |
| ARID1A (AT-Rich Interaction Domain 1A) | ARID1a (AT-rich interactive domain-containing protein 1A) | Transcription factor | Razumilava et al[61], 2014 |
| BAP1 (BRCA1 Associated Protein 1) | BAP1 (Ubiquitin carboxyl-terminal hydrolase BAP1) | Regulates cell growth | Yoshino et al[100], 2020 |
| BCL-2 (B cell Lymphoma-2) | Bcl-2 (B-cell lymphoma 2) | Regulates apoptosis | Fingas et al[101], 2010 |
| BRAF (B Rapidly Accelerated Fibrosarcoma) | B-Raf (B-Rapidly Accelerated Fibrosarcoma) | Proto-oncogene | Sia et al[24], 2013 |
| BRCA1 (Breast Cancer 1) | BRCA1 (Breast cancer type 1 susceptibility protein) | Tumor suppressor and DNA repair | Paradiso et al[102], 2020 |
| BRCA2 (Breast Cancer 2) | BRCA2 (Breast cancer type 2 susceptibility protein) | DNA repair | |
| CCND1(Cyclin D1) | CCND1 (G1/S-specific cyclin-D1) | Regulates cell growth | Yoshino et al[100], 2020 |
| CDH1(Cadherin 1) | E-cadherin (Epithelial cadherin) | Tumor suppressor, cell adhesion | Ross et al[103], 2014 |
| CDK6 (Cyclin-Dependent Kinase 6) | CDK6 (Cyclin-Dependent Kinase 6) | Controls cell cycle and differentiation | |
| CTNNB1 (Catenin Beta 1) | Β-catenin | Proto-oncogene | O'Dell et al[104], 2012 |
| EGFR (ERBB1) (Epidermal Growth Factor Receptor) | EGFR (ErbB-1) (Epidermal Growth Factor Receptor) | Proto-oncogene | |
| ERBB2 (HER2) (Avian Erythroblastosis oncogene B2) | ErbB-2 (HER2) (Receptor tyrosine-protein kinase erbB-2) | Proto-oncogene | |
| FBXW7 (F-Box and WD Repeat Domain Containing 7) | FBXW7 (F-box/WD repeat-containing protein 7) | Component of proteasomal protein degradation pathway | Ross et al[103], 2014 |
| FGF19 (Fibroblast Growth Factor 19) | FGF19 (Fibroblast Growth Factor 19) | Regulation of bile salt synthesis | |
| FGFR2 (Fibroblast Growth Factor Receptor 2) | FGFR2 (Fibroblast Growth Factor Receptor 2) | Cell surface receptor regulating cell proliferation, differentiation, migration and apoptosis | |
| IDH1 (Isocitrate dehydrogenase 1) | Isocitrate de-hydrogenase 1 (Isocitrate dehydrogenase (cytoplasmic)) | Glucose metabolism, indirectly mitigates oxidative stress | Nabeshima et al[105], 2020 |
| IDH2 (Isocitrate dehydrogenase 2) | Isocitrate de-hydrogenase 2 (Isocitrate dehydrogenase (mitochondrial)) | Glucose metabolism, indirectly mitigates oxidative stress | |
| Keap1 (Kelch-like ECH-associated protein 1) | KEAP1 (Kelch-like ECH-associated protein 1) | Prevents Nrf2-driven transcription | Ma et al[106], 2020 |
| KRAS (Kirsten Rat Sarcoma) | K-Ras (Kirsten Rat Sarcoma) | Proto-oncogene | Tannapfel et al[107], 2000 |
| MDM2 (Mouse Double Minute 2) | Mdm2 (E3 ubiquitin-protein ligase Mdm2) | Proto-oncogene, p53 inhibitor | Ross et al[103], 2014 |
| MYC (Avian myelocytomatosis virus oncogene cellular homolog) | Myc (Myc proto-oncogene protein) | Proto-oncogene | Zhou et al[108], 2019 |
| NF1 (Neurofibromin 1) | NF1 (Neurofibromin) | Stimulates Ras activity | Ross et al[103], 2014 |
| PBRM1 (Polybromo 1) | PBRM1 (Protein polybromo-1) | Negative regulator of cell proliferation | Luchini et al[109], 2017 |
| PIK3CA (Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha) | PIK3CA (Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform) | Generates PIP3 that activates signalling cascades for cell growth, survival and motility | Xu et al[110], 2011 |
| PTEN (Phosphatase and Tensin Homolog) | PTEN (Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN) | Tumor suppressor | Zhu et al[111], 2014 |
| RAD51AP1 (RAD51 Associated Protein 1) | RAD51AP1 (RAD51 Associated Protein 1) | DNA damage repair | Liu et al[112], 2021 |
| RASSF1A (Ras association domain family 1 isoform A) | RASSF1A (Ras association domain-containing protein 1 isoform A) | Tumor suppressor | Chen et al[113], 2005 |
| SMAD4 (Small Mothers Against Decapentaplegic 4) | SMAD4 (Small Mothers Against Decapentaplegic 4) | Tumor suppressor, transcription factor | Yoshino et al[100], 2020 |
| SOCS3 (Suppressor of Cytokine Signaling 3) | SOCS3 (Suppressor of Cytokine Signaling 3) | Signal transduction inhibitor | Andersen et al[114], 2012 |
| TP53 (Tumor Protein 53) | p53 (Protein 53) | Tumor suppressor | O'Dell et al[104], 2012 |
By the advent of array-based and deep sequencing techniques, technological advances have taken epigenetics into the omics-age, emphasizing the role of the epigenome in the human carcinogenesis process, including DNA CpG methylation, histone modifications, and non-coding RNA organisms. Only few systematic CCA epigenomic reports have been conducted, and data on abnormal CpG promoter methylation have mainly focused on individual genes in the CCA regulation[32]. In various important cancer-associated genes in CCA, abnormal epigenetic modulation such as promoter hypermethylation, was reported[32,33]. Studies examining these modifications to existing prognostic and predictive gene signatures have not yet been investigated in CCA to predict the therapeutic benefits of agents targeting the cancer epigenome. In CCA, the well-studied epigenetic process is DNA methylation. The promoter regions of tumor suppressor genes are highly methylated (promoter hypermethylation) in CCA tumorigenesis, which contributes to gene silencing. The promoter hypermethylation of genes involved in the repair of DNA, cell cycle, apoptosis, metabolism of carcinogen/drugs, and cell adhesion has been documented in CCA[33,34]. Some of the most frequent epigenetic events reported in CCA by methylation is summarized in Table 3.
| Gene (location) | Function | Epigenetic modification/effect | Outcome | Ref. |
| p16INK4A or CDKN2A (9p21) | Tumor suppressor gene Regulates cell proliferation and oncogenesis | Promoter region hypermethylation of the p16INK4A results in gene inactivation. Common event in PSC-associated CCA | More frequent in ECC cases. More commonly observed in tumors with vascular invasion. Poor clinical outcome | Ueki et al[115], 2004 |
| p14ARF(9p21) | Encoded by the β transcript of CDKN2A (p16/CDKN2A) | Methylation of p14ARF MF = 38 and 25% (32.35); 40.2% liver fluke CCA (37) | Increased tumorigenesis in CCA | Kim et al[116], 2007 |
| p15INK4b or p15 (9p21) | Effecter of TGF-β-mediated cell cycle arrest | Promoter hypermethylation of p15 gene | Increased tumorigenesis in CCA | Yang et al[117], 2005 |
| p73 gene (1p36.3) | Tumor suppressor gene and related to the p53 gene | Promoter region hypermethylation increased tumorigenesis | Increased tumorigenesis in CCA | |
| TMS1/ASC (16p11.2) | Tumor suppressor gene | Aberrant methylation of the TMS1/ASC cause inactivation of gene | Associated with CCA | Liu et al[118], 2006 |
| FHIT (3p14.2) | Tumor suppressor gene | Promoter hypermethylation of the FHIT gene results in epigenetic silencing of the FHIT promoter region | Development of intrahepatic CCAs | Foja et al[119], 2005 |
| RASSF1A (3p21.3) | Tumor suppressor gene induces cell cycle arrest by inhibiting the accumulation of cyclin D1 | Hypermethylation of its CpG island promoter region results in inactivation | Promoter methylation is more common in ECC than | Wong et al[120], 2002 |
| hMLH1 (3p21.3) | DNA mismatch repair gene | Promoter methylation/hypermethylation of the hMLH1 gene | Methylation frequencies vary in sporadic CCA, biliary papillary, neoplasms, and liver fluke-related CCA. Associated with poorly differentiated subtype of CCA with vascular invasion | Yang et al[117], 2005 |
| APC (5q21–q22) | Tumor suppressor gene Controls cell division, cell-cell interactions and cell migration and invasion, and conservation of chromosomal number during cell division | APC gene hypermethylation | Worse clinical outcome in CCA | Yang et al[117], 2005 |
| RAR-β (or HAP, RRB2 and NR1B2) (3p24) | Mediates cellular signaling in embryonic morphogenesis, cell growth and differentiation by regulating gene expression | Gene silencing by promoter region hypermethylation Results in increased tumorigenesis | Increased tumorigenesis in CCA | |
| Epithelial (E) cadherin gene (16q22.1) | Tumor suppressor gene | Hypermethylation of the promoter region of E gene Results in loss of function and contribute to progression of cancer by increasing proliferation, invasion and metastasis | Development of intrahepatic CCA | Lee et al[121], 2002 |
| DAPK (9q34.1) | Tumor suppressor gene Positive mediator of interferon-γ (IFN-γ)-induced programmed cell death | DAPK gene hypermethylation | Associated with poorly differentiated CCAs and with a poor prognosis | Tozawa et al[122], 2004 |
| CHFR gene (12q24.33) | Tumor suppressor gene Delays the entry into the metaphase | Gene silencing by promoter hypermethylation | Increased tumorigenesis in CCA | |
| RUNX3 gene (Ip36) | Tumor suppressor gene Regulate proliferation of the biliary tract epithelium | Methylation of RUNX3 results in gene silencing | Associated with poorer survival | |
| GSTP gene (1q43) | Regulate drug and xenobiotic. metabolism | Promoter region hypermethylation | Hypermethylation more frequent in ICCA than in ECC | Lee et al[121], 2002 |
| MGMT gene (10q26) | Responsible for repairing alkylation. DNA damage inhibits estrogen receptor-mediated cell proliferation | Methylation of discrete regions of the MGMT CpG island, results in heterochromatinization of the MGMT transcription start site and silencing of the gene | Increased frequency of GC to AT transitions in oncogenes and tumor suppressor genes and a poor prognosis | Koga et al[123], 2005 |
| BLU gene (3p21.3) | Tumor suppressor gene | Gene methylation | Increased tumorigenesis in CCA | Tischoff et al[124], 2005 |
| SEMA3B (3p21.3) | Tumor suppressor gene by inducing apoptosis. Plays a critical role in the guidance of growth cones during neuronal development | Methylation of SEMA3B gene | Increased tumorigenesis in CCA | |
| TIMP3 gene (22q12.3) | Plays a role in the induction of apoptosis | CpG island methylation of TIMP3 gene | Associated with worse survival | Lee et al[121], 2002 |
| RIZ1 | Tumor suppressor gene | Methylation of RIZ1 Results in chromatin compaction and gene silencing MF = 38% liver fluke CCA (47) | Increased proliferation and migration of CCA cell line | Khaenam et al[125], 2010 |
| OPCML | Tumor suppressor gene | Hypermethylation of OPCML | Increased tumorigenesis in CCA | Sriraksa et al[126], 2011 |
| GSTP1 | Tumor suppressor gene | Methylation of GSTP1 | Increased tumorigenesis in CCA | Yang et al[117], 2005 |
| COX-2/PTGS2 (1q25.2–q25.3) | Acts both as a dioxygenase and as a peroxidase | Methylation of COX-2 gene | Increased tumorigenesis in CCA | Lee et al[121], 2002 |
| THBS1 gene (15q15) | Mediates cell-to-cell and cell-to-matrix interactions and play roles in platelet aggregation, angiogenesis and tumorigenesis | Hypermethylation in the promoter region of THBS1 gene | Increased tumorigenesis in CCA | Tischoff et al[124], 2005 |
| SOCS3 | responsible for sustained IL-6/STAT-3 signaling and enhanced Mcl-1 expression in cholangiocarcinoma | Hypermethylation in the promoter region of SOCS3 gene | Increased tumorigenesis in CCA | Zhang et al[127], 2012 |
MiRNAs are a type of small non-coding RNA that is involved in the post-transcriptional regulation of gene expression. The upregulation/downregulation in multiple miRNAs have been reported in CCA, wherein dysregulated miRNAs led to mitosis, increased cell survival, and metastasis[35]. However, whether the alteration in miRNA expression in CCA is part of the process of carcinogenesis or the consequence of established CCA remains to be fully understood[36]. Long non-coding RNAs (lncRNAs) widely transcribed in the genome are evolving as key cancer regulators and play crucial roles in almost every facet of cell biology, including tumorigenesis. Via their association with DNA, proteins, and RNA, lncRNAs control cells' malignant transformation. The molecular mechanisms of lncRNA involved in CCA tumorigenesis may therefore be promising targets for therapeutic intervention and diagnostic applications in the battle against cancer[37,38]. The majority of upregulated genes are involved in carcinogenesis, diseases of the hepatic system, and transduction of signals. The miRNAs and lncRNAs related to the promotion of the pathogenesis of CCA are indicated in Tables 4 and 5.
| miRNAs | Target gene | Correlation with CCA tumorigenesis | Upregulated/downregulated | Ref. |
| miR-26a | GSK-3b | Tumor growth | Upregulated | Zhang et al[127], 2012 |
| miR-24 | MEN1(11q13) | Tumor suppressor gene | Upregulated | Ehrlich et al[128], 2017 |
| miR-29b | MCL-1 | Tumor suppressor gene | Downregulated | Stutes et al[129], 2007 |
| let-7a | NF2 | Tumor suppressor gene | Upregulated | |
| miR-148a | DNMT-1 | Regulate methyltransferase | Downregulated | Braconi et al[130], 2010 |
| miR-124 | SMYD3 | Migration and invasion of CCA cells | Downregulated | Zeng et al[131], 2012 |
| miR-21 | PTEN | Tumor suppressor gene | Upregulated | Meng et al[132], 2006 |
| miR-152 | DNMT-1 | Regulate methyltransferase | Downregulated | Braconi et al[130], 2010 |
| miR-200b | PTPN12 | Tumor suppressor gene | Upregulated | Meng et al[132], 2006 |
| miR-429 | CDH-6 | Tumor suppressor gene | Upregulated | Goeppert et al[133], 2016 |
| miR-122, miR-145, miR-200c, miR-221, and miR-222 | Multiple | Associated with tumorigenesis of ICCA | Downregulated | Karakatsanis et al[134], 2013 |
| miR-21, miR-31, and miR-223 | Multiple | No association with clinic-pathological parameters of CCA | Upregulated | |
| miR-370 | MAP3K8 | Tumor suppressor gene | Downregulated | Stutes et al[129], 2007 |
| miR-141 | CLOCK | Tumor suppressor gene | Upregulated | Meng et al[132], 2006 |
| miR-214 | Twist | Oncogene | Downregulated | Li et al[135], 2012 |
| LncRNA | Possible mechanism | Clinical relevance | Ref. |
| AFAP1-AS1 | (1) Decreasing the expression of c-Myc, Cyclin D1, MMP-2 and MMP-9; and (2) Decreasing the AFAP1 expression and promoting cell stress filament integrity | Unfavorable prognostic biomarker; potential therapeutic target | Lu et al[136], 2017 |
| CCAT2 | - | Unfavorable prognostic biomarker; potential therapeutic target | Xu et al[137], 2018 |
| HULC | Activating CXCR4 by sponging to miR-372/miR-373 as ceRNA | Potential therapeutic target | Wang et al[138], 2016 |
| ASAP1-IT1 | Interacting with hedgehog signaling pathway | Unfavorable prognostic biomarker; potential therapeutic target | Guo et al[139], 2018 |
| CPS1IT1 | Coexpressed with host gene CPS1 | Unfavorable prognostic biomarker; potential therapeutic target | Lu et al[136], 2017 |
| EPIC1 | Directly interacting with Mys | - | Li et al[140], 2018 |
| H19 | Activating IL-6 by sponging to let-7a/let-7b as ceRNA | Unfavorable prognostic biomarker; potential therapeutic target | Xu et al[141], 2017 |
| CCAT1 | Sponging to miR-152 as ceRNA | Independent prognostic factor; potential therapeutic target | Jiang et al[142], 2017 |
| LINC01296 | Modulating MYCN transcription by sponge miR-5095 as ceRNA | Potential therapeutic target | Jiang et al[142], 2017 |
| PCAT1 | Enhancing Wnt/β-catenin signaling through miR-122 repression and WNT1 expression | Potential therapeutic target | Zhang et al[143], 2017 |
| SNHG1 | Modulating cancer-related gene like CDKN1A by co-operating with chromatin-modifying enzymes as EZH2 | Unfavorable prognostic biomarker; potential therapeutic target | Yu et al[144], 2018 |
| MALAT1 | (1) Activating PI3K/Akt pathway; and (2) miR-204-dependent CXCR4 regulation as ceRNA | Unfavorable prognostic biomarker; potential therapeutic target | Tan et al[145], 2017 |
| PVT1 | Binding to epigenetic modification complexes, adjusting the expression of ANGPTL4 | Potential therapeutic target | Yang et al[146], 2018 |
| UCA1 | (1) Facilitating apoptosis via Bcl-2/caspase-3 pathway; (2) Activating AKT/GSK-3β/CCND1 axis; and (3) Upregulating MMP-9 | Unfavorable prognostic biomarker; potential therapeutic target | Xu et al[147], 2017 |
| SPRY4-IT1 | Recruiting EZH2, LSD1 or DNMT1 via sponging to miR-101-3p | Unfavorable prognostic biomarker; potential therapeutic target | Xu et al[148], 2018 |
| T-UCRs | Downstream of Wnt pathway and sponging to miR-193b | Unfavorable prognostic biomarker; potential therapeutic target | Carotenuto et al[149], 2017 |
Several publications have shown that PSC has an annual incidence rate of 0.77 per 100000 persons. PSC is more prevalent in adults between 25-years-old and 45-years-old; the median age of diagnosis of PSC is 41 years. Patients with PSC have a considerably higher risk of CCA, with an estimated incidence rate ranging from approximately 0.5% to 1.5% annually and lifetime incidence of 20%[4,39,40]. The estimated prevalence of CCA in patients with PSC ranges from 6.5% to 13.3%[4,41,42]. A recent cohort study on 7121 patients from 37 countries showed the prevalence of CCA in patients with PSC to be 8.3%[43]. In high prevalence regions, such as Scandinavian countries, PSC is the most common indication for liver transplantation[44]. Death attributed to PSC is increased nearly four-fold as compared to the general population, in part because of end-stage liver disease; however, more than 40% of deaths in PSC patients have been attributed to cancer development[4].
In Western countries, PSC is the most common known predisposing factor for CCA. The risk of CCA development per year among patients with PSC is 0.5% to 1.5%, with estimated lifetime prevalence of 5%–10%[45]. Several potential risk factors for CCA in PSC patients have been evaluated; smoking and alcohol consumption are increasingly recognized as risk factors for CCA[46].
Epidemiologic data studies regarding CCA mortality risk indicate that age-adjusted death rate for iCCA is increasing while trend mortality from pCCA and dCCA is expected to decrease worldwide[47]. Although the recorded rise in the incidence of CCA during the past 30 years has been observed as an increase in iCCA, it might be due to potential misclassification of perihilar tumors as iCCAs[48]. The age-adjusted incidence rate according to the United States database for iCCA enhanced from 0.59 per 100000 population in 1990 to 0.91 in 2001. Subsequently, the age-adjusted incidence rate decreased to 0.6 per 100000 population by 2007. Contrarily, the incidence rate among pCCA plus dCCA patients remained approximately 0.8 per 100000 population until 2001 then steadily increased to 0.97 until 2007. Perihilar CCA was identified as iCCAs before 2001 and subsequently was recognized as pCCA after releasing the 3rd edition of Classification of Tumors. This amendment plausibly affected the aforementioned alterations in specific incidence rates of both CCA subtypes[49].
iCCA is a primary carcinoma of the liver with rare entity, accounting for about 3% of global gastrointestinal cancers[50]. iCCA comprises 8%–10% of all CCA and has a distinguished disease course, incidence, and prevalence of disease from hilar and eCCA[51]. In addition, in spite of the fact that iCCA has been historically mistaken for other HCC[52], previous studies have shown that ICC accounts for 10%–20% of primary liver malignancies[53]. iCCA is uncommon in individuals under 40 years of age; it occurs primarily at an old age with the peak incidence in the 5th and 7th decade of life[54]. In the United States it is estimated a slight male predominance in iCCA cases (1.5 fold) over women[54].
Despite the low frequency of iCCA vs HCC, the incidence of iCCA appears to be increasing worldwide[55]. This increased risk of incidence rate is independent of tumor size and staging, and it is implausibly secondary to earlier diagnosis[55]. In the United States the incidence of iCCA during the past 30-year period enhanced 165% to 0.95 cases/100000 population[55]. A similar rise in iCCA incidence rate has also been reported in the United Kingdom, Japan, and Crete[56].
Globally, there is a certain disparity incidence of iCCA, with markedly lower rates of iCCA reported in Western nations when compared to East Asian countries[50]. This demographic variation is explained mainly by the prevalence of risk factors for iCCA in these East Asian countries[57].
In addition, Hispanic-Americans (1.22 per 100000 population) were considered to be significantly susceptible to high incidence of iCCA compared to other ethnic groups; for instance, African-Americans have a low rate of incidence (0.3 per 100000 population). The researchers have shown that this disparity may reflect genetic diversity, cultural differences, and socio-economic status in iCCA susceptibility[58].
Several risk factors implicated in iCCA pathogenesis have demographical prevalence. A previous report indicated that approximately 40% of iCCA patients will have no detectable risk factor, suggesting the need to be explored for further research in this regard[59].
It is believed that PSC is a predisposing factor for the development of iCCA. Both biliary inflammation and subsequent chronic proliferative activation of hepatic stem cells potentially predispose to iCCA formation[60]. It has been reported that PSC patients possess a lifetime incidence of CCA from approximately 5%–10%, while 50% of cases are recognized during 2 years of the course of PSC[61]. Additionally, researchers showed a predisposing risk of iCCA (odds ratio: 2.2; 95% confidence interval: 1.2–3.9) in ulcerative colitis patients[59]. The iCCA arose in PSC patients earlier, despite most individuals diagnosed between the ages of 30 and 50.
Recent results indicated that cancer risk is higher among patients with primary biliary stones and chronic biliary tract inflammation. Furthermore, incidence risk of iCCA has been found to be approximately 7% in hepaticolithiasis patients[61]. Another Asian study demonstrated that hepaticolithiasis in CCA patients followed by surgical resection is nearly 70% in Taiwan[60].
Furthermore, congenital anomalies of biliary tree, like Caroli’s disease and fibrocystic hepatic disorder, reveal approximately 15% lifetime risk factors of iCCA following the 2nd decade of life[60]. Caroli’s disease is a rare inherited disorder characterized by cystic widening of ducts in the liver, usually in a bilobar pattern. iCCA risk has been shown to be rising among subjects with bile stasis, cholangitis, and chronic inflammation[62].
CCA represents approximately 3% of all gastrointestinal cancers. The total incidence rate of CCA appears to have increased dramatically over the past 30 years[49]. The 5-year overall survival rates after diagnosis remained at 10% during this span of time[46].
CCA is a highly fatal malignancy tumor due to late clinical presentation[57]. While it is generally believed that standard of care is resection, most patients who present with metastatic disease are deemed unresectable[63]. The liver transplantation outcomes alone for unresectable conditions have been disappointing[64]. A previous study examined the effectiveness of a novel modality combining neoadjuvant chemoradiotherapy followed by liver transplantation. Survival outcomes from a combination of neoadjuvant chemoradiotherapy and liver transplantation for CCA are considerably excellent in comparison with resection[65]. Thus, even if transplantation may be a useful cure for unresectable iCCA, survival output remains poor. Orthotopic liver transplantation utilization is increasing within the United States and appears promising because it may obviate complications to achieve surgical margins into the liver. Unfortunately, efforts during the past decades were poor. In addition, according to the registry between 1968 and 1997, researchers have reported a 28% 5-year survival rate with a 51% risk of tumor recurrence rate after liver transplantation[64]. Further
There are many benefits for liver transplantation vs conventional resection to acquire complete elimination of tumor. There is some difficulty in evaluating hepatic duct tumor involvement before resection, and this is the most frequent reason for failure towards the achievement of an R0 resection. This problem is considerably obviated by liver transplantation. Liver transplantation promotes extirpation of all adjacent tissue and resection of the caudate. Liver transplantation facilitates arterial and portal venous inflow preservation to the remaining liver. Liver transplantation provides wide local excision and higher patient survival than what could be achieved with conventional resection[8].
Researchers conclude that neoadjuvant supportive treatment therapy in combination with liver transplantation presently appears to have fared far better than resection for selected patients with regional lymph node negative hCCA. Surgical staging information is essential; 23% of patients had localized lymph node metastases and concomitant extrahepatic disorder, which increased subsequent risk for transplantation. In a quarter of patients with underlying PSC, pancreatoduodenectomy may be required to obtain complete removal of the patient’s tumor with biliary tract involvement at the time of transplantation. Liver transplantation in combination with neoadjuvant treatment should be considered as an alternative option to surgical resection for patients with hCCA[8].
Liver transplantation as an important therapeutic option for iCCA is still debated. It has been reported that iCCA recurs within 5 years of liver transplantation among 70% of patients[68]. Locoregional interventions, such as radiofrequency ablation and transarterial chemoembolization, have garnered attention as a therapeutic alternative for localized, unresectable iCCA patients[69]. The standard treatment for patients with advanced-stage iCCA is the most common combinations, which includes systemic chemotherapy regimen of gemcitabine and cisplatin. According to a recent clinical study, liver transplantation could be a treatment choice for patients with early detected unresectable iCCA (i.e. ≤ 2 cm), with better survival results compared with those of HCC[70].
iCCA remains a contraindication for liver transplantation in most clinical centers around the world because of very poor prognosis, with a 2-year overall survival rate of approximately 30%[71]. The lack of standardization due to different patients’ selection and the absence of neoadjuvant treatments are expected to change outcomes[72]. The best survival was achieved in hCCA thanks to careful patient selection for neoadjuvant radiotherapy. Results from cohort studies after 2014 confirmed promising results after liver transplantation for iCCA[70]. The significance of proper patient selection criteria was first evaluated in a global multicentric report among iCCA patients who underwent liver transplantation[70]. The only curative treatments available for pCCA are surgical resection and neoadjuvant chemoradiation therapy after liver transplantation. Owing to the existence of parenchymal liver disease, PSC patients in most cases need liver transplantation as the preferred choice when compared to surgical resection[1]. Besides this, recent studies confirm that in PSC patients, intense immunosuppression ensuing liver transplantation increased risk of disease recurrence[73]. Liver transplantation for pCCA patients following neoadjuvant chemoradiation treatment establishes a proper long-term survival rate in a group of selected candidates with unresectable early stage pCCA and patients with PSC–related pCCA. Commitment to appropriate selection criteria, heavy neoadjuvant intervention, operative staging before liver transplantation, and specified technical procedures throughout the transplant process are required for success[74].
The research evidence shows that neoadjuvant therapy for liver transplantation is an effective treatment for unresectable early stage pCCA and pCCA occurring in the setting of PSC[75]. Recently, programmed cell death protein 1 inhibitors are noticed as a promising therapeutic option for CCA. Chimeric antigen receptor T cells, oncolytic viruses cancer vaccines and bispecific antibodies, show a remarkable ability to achieve satisfactory results.
Furthermore, the combinations of immunotherapy with other immunotherapeutics such as conventional therapies display some efficacy, and various studies have provided new insights into their administration in antitumor therapy[76]. The main barrier to successful liver transplantation and effective treatment is the availability of donor organs[75]. According to data from the Mayo Clinic and several other centers, from the start of therapy, a promising survival rate between 5-10 years was reached[77]. Post-transplant survival is approaching 50% at 5 years for both pCCA–related PSC and de novo pCCA, and these findings rationalize the use of both deceased and living donors.
After onset of this therapy in 1993, significant increases were observed in the time elapsed between the end of neoadjuvant treatment and liver transplantation. This interval can differ widely between patients by blood type compatibility, transplant center address, and availability of living donor organs. It has been shown that longer time elapsed between neoadjuvant therapy and liver transplantation results in reducing local recurrence[78]. Selection of patients with prolonged intervals and better oncologic biography, who are less susceptible to advancing the disease following neoadjuvant treatment, are less prone to develop recurrence post-transplantation. However, patients with radiation-induced fibrosis and longer intervals can significantly complicate the staging and transplant operations. Living donor liver transplant (LDLT) may solve these problems by removing the need to waitlist for a deceased donor and help physicians for optimal timing of liver transplantation. Recent findings based on clinical study demonstrated that LDLT and deceased donor liver transplant (DDLT) outcomes for pCCA-associated PSC are similar. In addition, LDLT for de novo pCCA shows a recurrence tendency and slightly worse patient survival outcomes vs DDLT. Despite these minor differences, researchers have been looking into possible mechanisms of disease progression following neoadjuvant treatment for a period to choose those patients who are at risk due to disease progression in order to prevent post-transplant disease recurrences[79]. A previous report indicated that liver transplantation is more effective and achieved better survival and less recurrence than surgical resection, and that the indications for liver transplantation and neoadjuvant treatment should advocate for resectable pCCA patients. According to these favorable findings, physicians have advocated for this viewpoint for patients with pCCA-associated PSC and transplanted many such patients at many transplant centers.
The role of neoadjuvant chemoradiation therapy and liver transplantation remains a consideration though, especially in de novo pCCA patients. Earlier studies were equivocal and unable to detect whether a subset of patients with de novo pCCA may benefit from liver transplantation vs surgical resection[80]. In 2015, American Hepato-Pancreato-Biliary Association recommended that surgical resection can be standard curative treatment for patients with resectable de novo pCCA[81]. Recent reports have suggested that liver transplantation vs surgical resection for hCCA patients who may need a liver transplant had better prognosis than those found after resection[82]. Analysis of results obtained from multicenter study between 2000 to 2015 showed that patients with pCCA not associated with PSC continued to show superiority of transplant compared to resection with promising post-transplant survival outcomes at 3 and 5 years (54% vs 44%, P = 0.03; 54% vs 29%, P = 0.03)[82]. Additionally, researchers pointed out 5-year estimated overall survival of 41% for patients enrolled onto clinical trials of neoadjuvant treatment/transplant procedure vs 27% among those patients who underwent surgical resection[83]. This discrepancy (14%) is too minor to approve the use of a donor liver for resectable non-PSC related pCCA[83]. In France a multi-center randomized clinical trial evaluating neoadjuvant chemoradiation and liver transplantation in comparison with resection will further elucidate pivotal details on these equivocal results.
In brief, over the past 2 decades liver transplantation has been currently considered the proven treatment of unresectable early stage pCCA and pCCA associated with PSC. Outstanding findings can be attained by stringent adherence to patient selection criteria and clinical management, application of high-dose neoadjuvant radiation therapy, and clinical staging before liver transplantation. Liver transplantation in combination with neoadjuvant treatment can obtain outcomes similar to surgical resection for unresectable early stage pCCA patients, and this is the treatment of choice administered for patients with pCCA arising in the setting of PSC[84]. Approximately 5% of all cases affected by pCCA require liver transplantation under the Mayo eligibility criteria. If the liver does not work properly, without transplantation, a median survival time is approximately 1 year. pCCA is reported as the most common malignancy and aggressive type of the biliary duct and arises from biliary lining the liver hilum[85]. The Mayo Clinic and other international centers are recently selecting the optimal subgroup to treat patients with locally advanced pCCA by neoadjuvant chemoradiation in combination with liver transplantation[65,86]. Outcome of patients treated according to this guideline, a 5-year survival rate of 53%, marginally improves the survival rate of patients after surgery for resectable type of disease[86,87].
It is most important to understand oncological suitability, donor liver organ availability, as well as ability to obtain appropriate long-term results in patients with CCA with or without PSC. In pCCA not associated with PSC, liver transplantation seems to provide promising survival. In resectable types of pCCA patients, neoadjuvant chemoradiotherapy and liver transplantation by strict selection criteria may improve the survival rate of patients compared to unresectable early stage pCCA patients. Owing to the shortage of available organs, it still remains unknown whether liver transplantation and neoadjuvant chemoradiotherapy should be increasingly considered for other classifications of CCA. Imbalance between organ supply and demand further conducts a need for stringent indications and contraindications in recognizing liver transplantation proper status. It is also essential for doctors to stay up to date with the general indications for liver transplantation and to consider when it is suitable or unsuitable to refer patients for transplant evaluation.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Iran
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dawood R S-Editor: Fan JR L-Editor: Filipodia P-Editor: Wu RR
| 1. | Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 966] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 2. | Saha SK, Zhu AX, Fuchs CS, Brooks GA. Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.: Intrahepatic Disease on the Rise. Oncologist. 2016;21:594-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 561] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 3. | Boberg KM, Lind GE. Primary sclerosing cholangitis and malignancy. Best Pract Res Clin Gastroenterol. 2011;25:753-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 4. | Boonstra K, Weersma RK, van Erpecum KJ, Rauws EA, Spanier BW, Poen AC, van Nieuwkerk KM, Drenth JP, Witteman BJ, Tuynman HA, Naber AH, Kingma PJ, van Buuren HR, van Hoek B, Vleggaar FP, van Geloven N, Beuers U, Ponsioen CY; EpiPSCPBC Study Group. Population-based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis. Hepatology. 2013;58:2045-2055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 504] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 5. | Boberg KM, Bergquist A, Mitchell S, Pares A, Rosina F, Broomé U, Chapman R, Fausa O, Egeland T, Rocca G, Schrumpf E. Cholangiocarcinoma in primary sclerosing cholangitis: risk factors and clinical presentation. Scand J Gastroenterol. 2002;37:1205-1211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 208] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 6. | Chapman MH, Webster GJ, Bannoo S, Johnson GJ, Wittmann J, Pereira SP. Cholangiocarcinoma and dominant strictures in patients with primary sclerosing cholangitis: a 25-year single-centre experience. Eur J Gastroenterol Hepatol. 2012;24:1051-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 152] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 7. | Welzel TM, McGlynn KA, Hsing AW, O'Brien TR, Pfeiffer RM. Impact of classification of hilar cholangiocarcinomas (Klatskin tumors) on the incidence of intra- and extrahepatic cholangiocarcinoma in the United States. J Natl Cancer Inst. 2006;98:873-875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 267] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 8. | Rea DJ, Heimbach JK, Rosen CB, Haddock MG, Alberts SR, Kremers WK, Gores GJ, Nagorney DM. Liver transplantation with neoadjuvant chemoradiation is more effective than resection for hilar cholangiocarcinoma. Ann Surg. 2005;242:451-8; discussion 458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 428] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 9. | Jarnagin WR, Fong Y, DeMatteo RP, Gonen M, Burke EC, Bodniewicz BS J, Youssef BA M, Klimstra D, Blumgart LH. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg. 2001;234:507-17; discussion 517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 964] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 10. | Barr Fritcher EG, Voss JS, Brankley SM, Campion MB, Jenkins SM, Keeney ME, Henry MR, Kerr SM, Chaiteerakij R, Pestova EV, Clayton AC, Zhang J, Roberts LR, Gores GJ, Halling KC, Kipp BR. An Optimized Set of Fluorescence In Situ Hybridization Probes for Detection of Pancreatobiliary Tract Cancer in Cytology Brush Samples. Gastroenterology. 2015;149:1813-1824.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 11. | Goldaracena N, Gorgen A, Sapisochin G. Current status of liver transplantation for cholangiocarcinoma. Liver Transpl. 2018;24:294-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 12. | Nakanuma Y, Kakuda Y. Pathologic classification of cholangiocarcinoma: New concepts. Best Pract Res Clin Gastroenterol. 2015;29:277-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 13. | Krasinskas AM. Cholangiocarcinoma. Surg Pathol Clin. 2018;11:403-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 14. | Louis C, Papoutsoglou P, Coulouarn C. Molecular classification of cholangiocarcinoma. Curr Opin Gastroenterol. 2020;36:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Nakanuma Y, Sato Y, Harada K, Sasaki M, Xu J, Ikeda H. Pathological classification of intrahepatic cholangiocarcinoma based on a new concept. World J Hepatol. 2010;2:419-427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 233] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 16. | Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 969] [Article Influence: 107.7] [Reference Citation Analysis (0)] |
| 17. | Kendall T, Verheij J, Gaudio E, Evert M, Guido M, Goeppert B, Carpino G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019;39 Suppl 1:7-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 236] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 18. | Cardinale V, Wang Y, Carpino G, Reid LM, Gaudio E, Alvaro D. Mucin-producing cholangiocarcinoma might derive from biliary tree stem/progenitor cells located in peribiliary glands. Hepatology. 2012;55:2041-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, Hiraoka N, Ojima H, Shimada K, Okusaka T, Kosuge T, Miyagawa S, Shibata T. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 651] [Cited by in RCA: 941] [Article Influence: 94.1] [Reference Citation Analysis (0)] |
| 20. | Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov. 2017;7:943-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 455] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 21. | Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, Kwak EL, Clark JW, Ryan DP, Deshpande V, Dias-Santagata D, Ellisen LW, Zhu AX, Iafrate AJ. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 598] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 22. | Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, Calvisi DF, Perugorria MJ, Fabris L, Boulter L, Macias RIR, Gaudio E, Alvaro D, Gradilone SA, Strazzabosco M, Marzioni M, Coulouarn C, Fouassier L, Raggi C, Invernizzi P, Mertens JC, Moncsek A, Rizvi S, Heimbach J, Koerkamp BG, Bruix J, Forner A, Bridgewater J, Valle JW, Gores GJ. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1555] [Cited by in RCA: 1539] [Article Influence: 307.8] [Reference Citation Analysis (0)] |
| 23. | Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, Choo SP, Myint SS, Thanan R, Nagarajan S, Lim WK, Ng CCY, Boot A, Liu M, Ong CK, Rajasegaran V, Lie S, Lim AST, Lim TH, Tan J, Loh JL, McPherson JR, Khuntikeo N, Bhudhisawasdi V, Yongvanit P, Wongkham S, Totoki Y, Nakamura H, Arai Y, Yamasaki S, Chow PK, Chung AYF, Ooi LLPJ, Lim KH, Dima S, Duda DG, Popescu I, Broet P, Hsieh SY, Yu MC, Scarpa A, Lai J, Luo DX, Carvalho AL, Vettore AL, Rhee H, Park YN, Alexandrov LB, Gordân R, Rozen SG, Shibata T, Pairojkul C, Teh BT, Tan P. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017;7:1116-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 677] [Article Influence: 84.6] [Reference Citation Analysis (0)] |
| 24. | Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, Cornella H, Klotzle B, Fan JB, Cotsoglou C, Thung SN, Fuster J, Waxman S, Garcia-Valdecasas JC, Bruix J, Schwartz ME, Beroukhim R, Mazzaferro V, Llovet JM. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 433] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 25. | Fouassier L, Marzioni M, Afonso MB, Dooley S, Gaston K, Giannelli G, Rodrigues CMP, Lozano E, Mancarella S, Segatto O, Vaquero J, Marin JJG, Coulouarn C. Signalling networks in cholangiocarcinoma: Molecular pathogenesis, targeted therapies and drug resistance. Liver Int. 2019;39 Suppl 1:43-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 26. | Zabron A, Edwards RJ, Khan SA. The challenge of cholangiocarcinoma: dissecting the molecular mechanisms of an insidious cancer. Dis Model Mech. 2013;6:281-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 27. | Rizvi S, Borad MJ, Patel T, Gores GJ. Cholangiocarcinoma: molecular pathways and therapeutic opportunities. Semin Liver Dis. 2014;34:456-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 28. | Kiguchi K. Molecular aspects of cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2014;21:371-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 29. | Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, Covington KR, Wu CC, Shinbrot E, Stransky N, Hegde A, Yang JD, Reznik E, Sadeghi S, Pedamallu CS, Ojesina AI, Hess JM, Auman JT, Rhie SK, Bowlby R, Borad MJ; Cancer Genome Atlas Network, Zhu AX, Stuart JM, Sander C, Akbani R, Cherniack AD, Deshpande V, Mounajjed T, Foo WC, Torbenson MS, Kleiner DE, Laird PW, Wheeler DA, McRee AJ, Bathe OF, Andersen JB, Bardeesy N, Roberts LR, Kwong LN. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18:2780-2794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 380] [Article Influence: 47.5] [Reference Citation Analysis (1)] |
| 30. | Sia D, Tovar V, Moeini A, Llovet JM. Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene. 2013;32:4861-4870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 183] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 31. | Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5322] [Cited by in RCA: 7533] [Article Influence: 837.0] [Reference Citation Analysis (0)] |
| 32. | Isomoto H. Epigenetic alterations associated with cholangiocarcinoma (review). Oncol Rep. 2009;22:227-232. [PubMed] |
| 33. | Sandhu DS, Shire AM, Roberts LR. Epigenetic DNA hypermethylation in cholangiocarcinoma: potential roles in pathogenesis, diagnosis and identification of treatment targets. Liver Int. 2008;28:12-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Limpaiboon T. Epigenetic aberrations in cholangiocarcinoma: potential biomarkers and promising target for novel therapeutic strategies. Asian Pac J Cancer Prev. 2012;13 Suppl:41-45. [PubMed] |
| 35. | O'Rourke CJ, Munoz-Garrido P, Aguayo EL, Andersen JB. Epigenome dysregulation in cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1423-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 36. | Wangyang Z, Daolin J, Yi X, Zhenglong L, Lining H, Yunfu C, Xingming J. NcRNAs and Cholangiocarcinoma. J Cancer. 2018;9:100-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2202] [Cited by in RCA: 2421] [Article Influence: 269.0] [Reference Citation Analysis (0)] |
| 38. | Yang W, Li Y, Song X, Xu J, Xie J. Genome-wide analysis of long noncoding RNA and mRNA co-expression profile in intrahepatic cholangiocarcinoma tissue by RNA sequencing. Oncotarget. 2017;8:26591-26599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 39. | Takakura WR, Tabibian JH, Bowlus CL. The evolution of natural history of primary sclerosing cholangitis. Curr Opin Gastroenterol. 2017;33:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 40. | Fevery J, Verslype C, Lai G, Aerts R, Van Steenbergen W. Incidence, diagnosis, and therapy of cholangiocarcinoma in patients with primary sclerosing cholangitis. Dig Dis Sci. 2007;52:3123-3135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 97] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 41. | Bergquist A, Ekbom A, Olsson R, Kornfeldt D, Lööf L, Danielsson A, Hultcrantz R, Lindgren S, Prytz H, Sandberg-Gertzén H, Almer S, Granath F, Broomé U. Hepatic and extrahepatic malignancies in primary sclerosing cholangitis. J Hepatol. 2002;36:321-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 469] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 42. | Bakhshi Z, Hilscher MB, Gores GJ, Harmsen WS, Viehman JK, LaRusso NF, Gossard AA, Lazaridis KN, Lindor KD, Eaton JE. An update on primary sclerosing cholangitis epidemiology, outcomes and quantification of alkaline phosphatase variability in a population-based cohort. J Gastroenterol. 2020;55:523-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 43. | Weismüller TJ, Trivedi PJ, Bergquist A, Imam M, Lenzen H, Ponsioen CY, Holm K, Gotthardt D, Färkkilä MA, Marschall HU, Thorburn D, Weersma RK, Fevery J, Mueller T, Chazouillères O, Schulze K, Lazaridis KN, Almer S, Pereira SP, Levy C, Mason A, Naess S, Bowlus CL, Floreani A, Halilbasic E, Yimam KK, Milkiewicz P, Beuers U, Huynh DK, Pares A, Manser CN, Dalekos GN, Eksteen B, Invernizzi P, Berg CP, Kirchner GI, Sarrazin C, Zimmer V, Fabris L, Braun F, Marzioni M, Juran BD, Said K, Rupp C, Jokelainen K, Benito de Valle M, Saffioti F, Cheung A, Trauner M, Schramm C, Chapman RW, Karlsen TH, Schrumpf E, Strassburg CP, Manns MP, Lindor KD, Hirschfield GM, Hansen BE, Boberg KM; International PSC Study Group. Patient Age, Sex, and Inflammatory Bowel Disease Phenotype Associate With Course of Primary Sclerosing Cholangitis. Gastroenterology. 2017;152:1975-1984.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 364] [Article Influence: 45.5] [Reference Citation Analysis (1)] |
| 44. | Fosby B, Melum E, Bjøro K, Bennet W, Rasmussen A, Andersen IM, Castedal M, Olausson M, Wibeck C, Gotlieb M, Gjertsen H, Toivonen L, Foss S, Makisalo H, Nordin A, Sanengen T, Bergquist A, Larsson ME, Soderdahl G, Nowak G, Boberg KM, Isoniemi H, Keiding S, Foss A, Line PD, Friman S, Schrumpf E, Ericzon BG, Höckerstedt K, Karlsen TH. Liver transplantation in the Nordic countries - An intention to treat and post-transplant analysis from The Nordic Liver Transplant Registry 1982-2013. Scand J Gastroenterol. 2015;50:797-808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 45. | Razumilava N, Gores GJ. Classification, diagnosis, and management of cholangiocarcinoma. Clin Gastroenterol Hepatol. 2013;11:13-21.e1; quiz e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 216] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 46. | Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology. 2011;54:173-184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 722] [Cited by in RCA: 689] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 47. | Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology. 2001;33:1353-1357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 773] [Cited by in RCA: 797] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 48. | Khan SA, Emadossadaty S, Ladep NG, Thomas HC, Elliott P, Taylor-Robinson SD, Toledano MB. Rising trends in cholangiocarcinoma: is the ICD classification system misleading us? J Hepatol. 2012;56:848-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 220] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 49. | Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD, Thillainayagam AV, Thomas HC, Wasan H; British Society of Gastroenterology. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut. 2012;61:1657-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 593] [Article Influence: 45.6] [Reference Citation Analysis (1)] |
| 50. | Maithel SK, Gamblin TC, Kamel I, Corona-Villalobos CP, Thomas M, Pawlik TM. Multidisciplinary approaches to intrahepatic cholangiocarcinoma. Cancer. 2013;119:3929-3942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 51. | Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Proceedings of the Seminars in liver disease; 2004. New York: Thieme Medical Publishers, 2004: 115-125. |
| 52. | Palmer WC, Patel T. Are common factors involved in the pathogenesis of primary liver cancers? J Hepatol. 2012;57:69-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 397] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 53. | Poultsides GA, Zhu AX, Choti MA, Pawlik TM. Intrahepatic cholangiocarcinoma. Surg Clin North Am. 2010;90:817-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 54. | Dodson RM, Weiss MJ, Cosgrove D, Herman JM, Kamel I, Anders R, Geschwind JF, Pawlik TM. Intrahepatic cholangiocarcinoma: management options and emerging therapies. J Am Coll Surg. 2013;217:736-750.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 55. | Shaib YH, Davila JA, McGlynn K, El-Serag HB. Rising incidence of intrahepatic cholangiocarcinoma in the United States: a true increase? J Hepatol. 2004;40:472-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 543] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 56. | Mouzas IA, Dimoulios P, Vlachonikolis IG, Skordilis P, Zoras O, Kouroumalis E. Increasing incidence of cholangiocarcinoma in Crete 1992-2000. Anticancer Res. 2002;22:3637-3641. [PubMed] |
| 57. | Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24:115-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 839] [Cited by in RCA: 848] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 58. | McLean L, Patel T. Racial and ethnic variations in the epidemiology of intrahepatic cholangiocarcinoma in the United States. Liver Int. 2006;26:1047-1053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 59. | Shaib YH, El-Serag HB, Davila JA, Morgan R, McGlynn KA. Risk factors of intrahepatic cholangiocarcinoma in the United States: a case-control study. Gastroenterology. 2005;128:620-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 385] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 60. | Portolani N, Baiocchi GL, Coniglio A, Piardi T, Grazioli L, Benetti A, Ferrari Bravo A, Giulini SM. Intrahepatic cholangiocarcinoma and combined hepatocellular-cholangiocarcinoma: a Western experience. Ann Surg Oncol. 2008;15:1880-1890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 61. | Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1072] [Cited by in RCA: 1378] [Article Influence: 125.3] [Reference Citation Analysis (1)] |
| 62. | Shimonishi T, Sasaki M, Nakanuma Y. Precancerous lesions of intrahepatic cholangiocarcinoma. J Hepatobiliary Pancreat Surg. 2000;7:542-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 80] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 63. | Marchan EM, Landry JC. Neoadjuvant chemoradiation followed by orthotopic liver transplantation in cholangiocarcinomas: the emory experience. J Gastrointest Oncol. 2016;7:248-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 64. | Meyer CG, Penn I, James L. Liver transplantation for cholangiocarcinoma: results in 207 patients. Transplantation. 2000;69:1633-1637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 348] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 65. | Darwish Murad S, Kim WR, Harnois DM, Douglas DD, Burton J, Kulik LM, Botha JF, Mezrich JD, Chapman WC, Schwartz JJ, Hong JC, Emond JC, Jeon H, Rosen CB, Gores GJ, Heimbach JK. Efficacy of neoadjuvant chemoradiation, followed by liver transplantation, for perihilar cholangiocarcinoma at 12 US centers. Gastroenterology. 2012;143:88-98.e3; quiz e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 395] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 66. | Robles R, Figueras J, Turrión VS, Margarit C, Moya A, Varo E, Calleja J, Valdivieso A, Valdecasas JC, López P, Gómez M, de Vicente E, Loinaz C, Santoyo J, Fleitas M, Bernardos A, Lladó L, Ramírez P, Bueno FS, Jaurrieta E, Parrilla P. Spanish experience in liver transplantation for hilar and peripheral cholangiocarcinoma. Ann Surg. 2004;239:265-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 217] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 67. | Goldstein RM, Stone M, Tillery GW, Senzer N, Levy M, Husberg BS, Gonwa T, Klintmalm G. Is liver transplantation indicated for cholangiocarcinoma? Am J Surg. 1993;166:768-71; discussion 771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 120] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 68. | Sapisochin G, Fidelman N, Roberts JP, Yao FY. Mixed hepatocellular cholangiocarcinoma and intrahepatic cholangiocarcinoma in patients undergoing transplantation for hepatocellular carcinoma. Liver Transpl. 2011;17:934-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 69. | Kuhlmann JB, Blum HE. Locoregional therapy for cholangiocarcinoma. Curr Opin Gastroenterol. 2013;29:324-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 70. | Mazzaferro V, Gorgen A, Roayaie S, Droz Dit Busset M, Sapisochin G. Liver resection and transplantation for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72:364-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 240] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 71. | Becker NS, Rodriguez JA, Barshes NR, O'Mahony CA, Goss JA, Aloia TA. Outcomes analysis for 280 patients with cholangiocarcinoma treated with liver transplantation over an 18-year period. J Gastrointest Surg. 2008;12:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 72. | Mazzaferro V, Regalia E, Doci R, Andreola S, Pulvirenti A, Bozzetti F, Montalto F, Ammatuna M, Morabito A, Gennari L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5110] [Cited by in RCA: 5308] [Article Influence: 183.0] [Reference Citation Analysis (0)] |
| 73. | Fosby B, Karlsen TH, Melum E. Recurrence and rejection in liver transplantation for primary sclerosing cholangitis. World J Gastroenterol. 2012;18:1-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 103] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 74. | Tan EK, Taner T, Heimbach JK, Gores GJ, Rosen CB. Liver Transplantation for Peri-hilar Cholangiocarcinoma. J Gastrointest Surg. 2020;24:2679-2685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 75. | Schaefer B, Zoller H, Schneeberger S. Con: Liver transplantation for expanded criteria malignant diseases. Liver Transpl. 2018;24:104-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 76. | Guo X, Shen W. Latest evidence on immunotherapy for cholangiocarcinoma. Oncol Lett. 2020;20:381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 77. | Heimbach JK, Gores GJ, Haddock MG, Alberts SR, Pedersen R, Kremers W, Nyberg SL, Ishitani MB, Rosen CB. Predictors of disease recurrence following neoadjuvant chemoradiotherapy and liver transplantation for unresectable perihilar cholangiocarcinoma. Transplantation. 2006;82:1703-1707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 146] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 78. | Duignan S, Maguire D, Ravichand CS, Geoghegan J, Hoti E, Fennelly D, Armstrong J, Rock K, Mohan H, Traynor O. Neoadjuvant chemoradiotherapy followed by liver transplantation for unresectable cholangiocarcinoma: a single-centre national experience. HPB (Oxford). 2014;16:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 79. | Croome KP, Rosen CB, Heimbach JK, Nagorney DM. Is Liver Transplantation Appropriate for Patients with Potentially Resectable De Novo Hilar Cholangiocarcinoma? J Am Coll Surg. 2015;221:130-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 80. | Chen XP, Lau WY, Huang ZY, Zhang ZW, Chen YF, Zhang WG, Qiu FZ. Extent of liver resection for hilar cholangiocarcinoma. Br J Surg. 2009;96:1167-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 81. | Mansour JC, Aloia TA, Crane CH, Heimbach JK, Nagino M, Vauthey JN. Hilar cholangiocarcinoma: expert consensus statement. HPB (Oxford). 2015;17:691-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 82. | Ethun CG, Lopez-Aguiar AG, Anderson DJ, Adams AB, Fields RC, Doyle MB, Chapman WC, Krasnick BA, Weber SM, Mezrich JD, Salem A, Pawlik TM, Poultsides G, Tran TB, Idrees K, Isom CA, Martin RCG, Scoggins CR, Shen P, Mogal HD, Schmidt C, Beal E, Hatzaras I, Shenoy R, Cardona K, Maithel SK. Transplantation Versus Resection for Hilar Cholangiocarcinoma: An Argument for Shifting Treatment Paradigms for Resectable Disease. Ann Surg. 2018;267:797-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 83. | Rosen CB. Transplantation Versus Resection for Hilar Cholangiocarcinoma: An Argument for Shifting Paradigms for Resectable Disease in Annals of Surgery 2018. Ann Surg. 2018;267:808-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 84. | Tan EK, Rosen CB, Heimbach JK, Gores GJ, Zamora-Valdes D, Taner T. Living Donor Liver Transplantation for Perihilar Cholangiocarcinoma: Outcomes and Complications. J Am Coll Surg. 2020;231:98-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 85. | Edge SB, Byrd DR, Carducci MA, Compton CC, Fritz A, Greene F. AJCC cancer staging manual. New York: Springer, 2010. |
| 86. | Heimbach JK, Gores GJ, Haddock MG, Alberts SR, Nyberg SL, Ishitani MB, Rosen CB. Liver transplantation for unresectable perihilar cholangiocarcinoma. Semin Liver Dis. 2004;24:201-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 191] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 87. | Vugts JJA, Gaspersz MP, Roos E, Franken LC, Olthof PB, Coelen RJS, van Vugt JLA, Labeur TA, Brouwer L, Besselink MGH, IJzermans JNM, Darwish Murad S, van Gulik TM, de Jonge J, Polak WG, Busch ORC, Erdmann JL, Groot Koerkamp B, Buettner S. Eligibility for Liver Transplantation in Patients with Perihilar Cholangiocarcinoma. Ann Surg Oncol. 2021;28:1483-1492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 88. | Wadsworth CA, Dixon PH, Wong JH, Chapman MH, McKay SC, Sharif A, Spalding DR, Pereira SP, Thomas HC, Taylor-Robinson SD, Whittaker J, Williamson C, Khan SA. Genetic factors in the pathogenesis of cholangiocarcinoma. Dig Dis. 2011;29:93-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 89. | Hoblinger A, Grunhage F, Sauerbruch T, Lammert F. Association of the c.3972C>T variant of the multidrug resistance-associated protein 2 Gene (MRP2/ABCC2) with susceptibility to bile duct cancer. Digestion. 2009;80:36-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 90. | Khabou B, Trigui A, Boudawara TS, Keskes L, Kamoun H, Barbu V, Fakhfakh F. A homozygous ABCB4 mutation causing an LPAC syndrome evolves into cholangiocarcinoma. Clin Chim Acta. 2019;495:598-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 91. | Kim CJ, Cox C, Dupont E, Reintgen DS. Accurate staging of women with breast cancer. J Surg Oncol. 2002;79:2-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 92. | Prawan A, Kukongviriyapan V, Tassaneeyakul W, Pairojkul C, Bhudhisawasdi V. Association between genetic polymorphisms of CYP1A2, arylamine N-acetyltransferase 1 and 2 and susceptibility to cholangiocarcinoma. Eur J Cancer Prev. 2005;14:245-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 93. | Melum E, Karlsen TH, Schrumpf E, Bergquist A, Thorsby E, Boberg KM, Lie BA. Cholangiocarcinoma in primary sclerosing cholangitis is associated with NKG2D polymorphisms. Hepatology. 2008;47:90-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 94] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 94. | Ko KH, Kim NK, Yim DJ, Hong SP, Park PW, Rim KS, Kim S, Hwang SG. Polymorphisms of 5,10-methylenetetrahydrofolate reductase (MTHFR C677T) and thymidylate synthase enhancer region (TSER) as a risk factor of cholangiocarcinoma in a Korean population. Anticancer Res. 2006;26:4229-4233. [PubMed] |
| 95. | Sakoda LC, Gao YT, Chen BE, Chen J, Rosenberg PS, Rashid A, Deng J, Shen MC, Wang BS, Han TQ, Zhang BH, Cohen-Webb H, Yeager M, Welch R, Chanock S, Fraumeni JF Jr, Hsing AW. Prostaglandin-endoperoxide synthase 2 (PTGS2) gene polymorphisms and risk of biliary tract cancer and gallstones: a population-based study in Shanghai, China. Carcinogenesis. 2006;27:1251-1256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 96. | Huang WY, Gao YT, Rashid A, Sakoda LC, Deng J, Shen MC, Wang BS, Han TQ, Zhang BH, Chen BE, Rosenberg PS, Chanock SJ, Hsing AW. Selected base excision repair gene polymorphisms and susceptibility to biliary tract cancer and biliary stones: a population-based case-control study in China. Carcinogenesis. 2008;29:100-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 97. | Marahatta SB, Punyarit P, Bhudisawasdi V, Paupairoj A, Wongkham S, Petmitr S. Polymorphism of glutathione S-transferase omega gene and risk of cancer. Cancer Lett. 2006;236:276-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 98. | Gong Y, Qi M, Chen J, Fang R, Mai C, Chen T, Tang H, Tang Y. XRCC1 Arg194Trp and Arg399Gln polymorphisms and risk of extrahepatic cholangiocarcinoma: a hospital-based case-control study in China. Int J Clin Exp Med. 2015;8:19339-19345. [PubMed] |
| 99. | Kang YK, Kim WH, Lee HW, Lee HK, Kim YI. Mutation of p53 and K-ras, and loss of heterozygosity of APC in intrahepatic cholangiocarcinoma. Lab Invest. 1999;79:477-483. [PubMed] |
| 100. | Yoshino J, Akiyama Y, Shimada S, Ogura T, Ogawa K, Ono H, Mitsunori Y, Ban D, Kudo A, Yamaoka S, Tanabe M, Tanaka S. Loss of ARID1A induces a stemness gene ALDH1A1 expression with histone acetylation in the malignant subtype of cholangiocarcinoma. Carcinogenesis. 2020;41:734-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 101. | Fingas CD, Katsounas A, Kahraman A, Siffert W, Jochum C, Gerken G, Nückel H, Canbay A. Prognostic assessment of three single-nucleotide polymorphisms (GNB3 825C>T, BCL2-938C>A, MCL1-386C>G) in extrahepatic cholangiocarcinoma. Cancer Invest. 2010;28:472-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 102. | Paradiso AV, Patruno M, Digennaro M, Tommasi S, Pilato B, Argentiero A, Brunetti O, Silvestris N. Somatic BRCA Mutation in a Cholangiocarcinoma Patient for HBOC Syndrome Detection. Front Oncol. 2020;10:1292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 103. | Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, Lee HJ, Sheehan CE, Otto GA, Palmer G, Yelensky R, Lipson D, Morosini D, Hawryluk M, Catenacci DV, Miller VA, Churi C, Ali S, Stephens PJ. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19:235-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 366] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 104. | O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, Rossi RM, Zhu AX, Land H, Bardeesy N, Hezel AF. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 171] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 105. | Nabeshima T, Hamada S, Taguchi K, Tanaka Y, Matsumoto R, Yamamoto M, Masamune A. Keap1 deletion accelerates mutant K-ras/p53-driven cholangiocarcinoma. Am J Physiol Gastrointest Liver Physiol. 2020;318:G419-G427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 106. | Ma B, Meng H, Tian Y, Wang Y, Song T, Zhang T, Wu Q, Cui Y, Li H, Zhang W, Li Q. Distinct clinical and prognostic implication of IDH1/2 mutation and other most frequent mutations in large duct and small duct subtypes of intrahepatic cholangiocarcinoma. BMC Cancer. 2020;20:318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 107. | Tannapfel A, Benicke M, Katalinic A, Uhlmann D, Köckerling F, Hauss J, Wittekind C. Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut. 2000;47:721-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 149] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 108. | Zhou M, Zhu Y, Hou R, Mou X, Tan J. Identification of candidate genes for the diagnosis and treatment of cholangiocarcinoma using a bioinformatics approach. Oncol Lett. 2019;18:5459-5467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 109. | Luchini C, Robertson SA, Hong SM, Felsenstein M, Anders RA, Pea A, Nottegar A, Veronese N, He J, Weiss MJ, Capelli P, Scarpa A, Argani P, Kapur P, Wood LD. PBRM1 loss is a late event during the development of cholangiocarcinoma. Histopathology. 2017;71:375-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 110. | Xu RF, Sun JP, Zhang SR, Zhu GS, Li LB, Liao YL, Xie JM, Liao WJ. KRAS and PIK3CA but not BRAF genes are frequently mutated in Chinese cholangiocarcinoma patients. Biomed Pharmacother. 2011;65:22-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 111. | Zhu AX, Borger DR, Kim Y, Cosgrove D, Ejaz A, Alexandrescu S, Groeschl RT, Deshpande V, Lindberg JM, Ferrone C, Sempoux C, Yau T, Poon R, Popescu I, Bauer TW, Gamblin TC, Gigot JF, Anders RA, Pawlik TM. Genomic profiling of intrahepatic cholangiocarcinoma: refining prognosis and identifying therapeutic targets. Ann Surg Oncol. 2014;21:3827-3834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 112. | Liu CC, Veeraraghavan J, Tan Y, Kim JA, Wang X, Loo SK, Lee S, Hu Y, Wang XS. A Novel Neoplastic Fusion Transcript, RAD51AP1-DYRK4, Confers Sensitivity to the MEK Inhibitor Trametinib in Aggressive Breast Cancers. Clin Cancer Res. 2021;27:785-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 113. | Chen YJ, Tang QB, Zou SQ. Inactivation of RASSF1A, the tumor suppressor gene at 3p21.3 in extrahepatic cholangiocarcinoma. World J Gastroenterol. 2005;11:1333-1338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 114. | Andersen JB, Thorgeirsson SS. Genetic profiling of intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol. 2012;28:266-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 115. | Ueki T, Hsing AW, Gao YT, Wang BS, Shen MC, Cheng J, Deng J, Fraumeni JF Jr, Rashid A. Alterations of p16 and prognosis in biliary tract cancers from a population-based study in China. Clin Cancer Res. 2004;10:1717-1725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 116. | Kim BH, Cho NY, Choi M, Lee S, Jang JJ, Kang GH. Methylation profiles of multiple CpG island loci in extrahepatic cholangiocarcinoma versus those of intrahepatic cholangiocarcinomas. Arch Pathol Lab Med. 2007;131:923-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 117. | Yang B, House MG, Guo M, Herman JG, Clark DP. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol. 2005;18:412-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 118. | Liu XF, Zhu SG, Zhang H, Xu Z, Su HL, Li SJ, Zhou XT. The methylation status of the TMS1/ASC gene in cholangiocarcinoma and its clinical significance. Hepatobiliary Pancreat Dis Int. 2006;5:449-453. [PubMed] |
| 119. | Foja S, Goldberg M, Schagdarsurengin U, Dammann R, Tannapfel A, Ballhausen WG. Promoter methylation and loss of coding exons of the fragile histidine triad (FHIT) gene in intrahepatic cholangiocarcinomas. Liver Int. 2005;25:1202-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 120. | Wong N, Li L, Tsang K, Lai PB, To KF, Johnson PJ. Frequent loss of chromosome 3p and hypermethylation of RASSF1A in cholangiocarcinoma. J Hepatol. 2002;37:633-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 121. | Lee S, Kim WH, Jung HY, Yang MH, Kang GH. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol. 2002;161:1015-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 112] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 122. | Tozawa T, Tamura G, Honda T, Nawata S, Kimura W, Makino N, Kawata S, Sugai T, Suto T, Motoyama T. Promoter hypermethylation of DAP-kinase is associated with poor survival in primary biliary tract carcinoma patients. Cancer Sci. 2004;95:736-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 123. | Koga Y, Kitajima Y, Miyoshi A, Sato K, Kitahara K, Soejima H, Miyazaki K. Tumor progression through epigenetic gene silencing of O(6)-methylguanine-DNA methyltransferase in human biliary tract cancers. Ann Surg Oncol. 2005;12:354-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 124. | Tischoff I, Markwarth A, Witzigmann H, Uhlmann D, Hauss J, Mirmohammadsadegh A, Wittekind C, Hengge UR, Tannapfel A. Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. Int J Cancer. 2005;115:684-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 125. | Khaenam P, Jearanaikoon P, Pairojkul C, Bhudhisawasdi V, Limpaiboon T. Genetic and epigenetic alterations of RIZ1 and the correlation to clinicopathological parameters in liver fluke-related cholangiocarcinoma. Exp Ther Med. 2010;1:385-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 126. | Sriraksa R, Zeller C, El-Bahrawy MA, Dai W, Daduang J, Jearanaikoon P, Chau-In S, Brown R, Limpaiboon T. CpG-island methylation study of liver fluke-related cholangiocarcinoma. Br J Cancer. 2011;104:1313-1318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 127. | Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating β-catenin. Gastroenterology. 2012;143:246-56.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 149] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 128. | Ehrlich L, Hall C, Venter J, Dostal D, Bernuzzi F, Invernizzi P, Meng F, Trzeciakowski JP, Zhou T, Standeford H, Alpini G, Lairmore TC, Glaser S. miR-24 Inhibition Increases Menin Expression and Decreases Cholangiocarcinoma Proliferation. Am J Pathol. 2017;187:570-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 129. | Stutes M, Tran S, DeMorrow S. Genetic and epigenetic changes associated with cholangiocarcinoma: from DNA methylation to microRNAs. World J Gastroenterol. 2007;13:6465-6469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 130. | Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881-890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 131. | Zeng B, Li Z, Chen R, Guo N, Zhou J, Zhou Q, Lin Q, Cheng D, Liao Q, Zheng L, Gong Y. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012;586:3271-3278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 132. | Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113-2129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 782] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 133. | Goeppert B, Ernst C, Baer C, Roessler S, Renner M, Mehrabi A, Hafezi M, Pathil A, Warth A, Stenzinger A, Weichert W, Bähr M, Will R, Schirmacher P, Plass C, Weichenhan D. Cadherin-6 is a putative tumor suppressor and target of epigenetically dysregulated miR-429 in cholangiocarcinoma. Epigenetics. 2016;11:780-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 134. | Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 277] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 135. | Li B, Han Q, Zhu Y, Yu Y, Wang J, Jiang X. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. FEBS J. 2012;279:2393-2398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 136. | Lu X, Zhou C, Li R, Deng Y, Zhao L, Zhai W. Long Noncoding RNA AFAP1-AS1 Promoted Tumor Growth and Invasion in Cholangiocarcinoma. Cell Physiol Biochem. 2017;42:222-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 137. | Xu Y, Yao Y, Qin W, Zhong X, Jiang X, Cui Y. Long non-coding RNA CCAT2 promotes cholangiocarcinoma cells migration and invasion by induction of epithelial-to-mesenchymal transition. Biomed Pharmacother. 2018;99:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 138. | Wang WT, Ye H, Wei PP, Han BW, He B, Chen ZH, Chen YQ. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 194] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 139. | Guo L, Zhou Y, Chen Y, Sun H, Wang Y, Qu Y. LncRNA ASAP1-IT1 positively modulates the development of cholangiocarcinoma via hedgehog signaling pathway. Biomed Pharmacother. 2018;103:167-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 140. | Li Y, Cai Q, Li W, Feng F, Yang L. Long non-coding RNA EPIC1 promotes cholangiocarcinoma cell growth. Biochem Biophys Res Commun. 2018;504:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 141. | Xu Y, Wang Z, Jiang X, Cui Y. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed Pharmacother. 2017;92:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 142. | Jiang XM, Li ZL, Li JL, Zheng WY, Li XH, Cui YF, Sun DJ. LncRNA CCAT1 as the unfavorable prognostic biomarker for cholangiocarcinoma. Eur Rev Med Pharmacol Sci. 2017;21:1242-1247. [PubMed] |
| 143. | Zhang F, Wan M, Xu Y, Li Z, Leng K, Kang P, Cui Y, Jiang X. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/β-catenin-signaling pathway. Biomed Pharmacother. 2017;94:55-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 144. | Yu Y, Zhang M, Wang N, Li Q, Yang J, Yan S, He X, Ji G, Miao L. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell Death Dis. 2018;9:746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 145. | Tan X, Huang Z, Li X. Long Non-Coding RNA MALAT1 Interacts With miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. J Cell Biochem. 2017;118:3643-3653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 146. | Yang Q, Yu Y, Sun Z, Pan Y. Long non-coding RNA PVT1 promotes cell proliferation and invasion through regulating miR-133a in ovarian cancer. Biomed Pharmacother. 2018;106:61-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 147. | Xu Y, Yao Y, Leng K, Li Z, Qin W, Zhong X, Kang P, Wan M, Jiang X, Cui Y. Long non-coding RNA UCA1 indicates an unfavorable prognosis and promotes tumorigenesis via regulating AKT/GSK-3β signaling pathway in cholangiocarcinoma. Oncotarget. 2017;8:96203-96214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 148. | Xu Y, Yao Y, Jiang X, Zhong X, Wang Z, Li C, Kang P, Leng K, Ji D, Li Z, Huang L, Qin W, Cui Y. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 149. | Carotenuto P, Fassan M, Pandolfo R, Lampis A, Vicentini C, Cascione L, Paulus-Hock V, Boulter L, Guest R, Quagliata L, Hahne JC, Ridgway R, Jamieson T, Athineos D, Veronese A, Visone R, Murgia C, Ferrari G, Guzzardo V, Evans TRJ, MacLeod M, Feng GJ, Dale T, Negrini M, Forbes SJ, Terracciano L, Scarpa A, Patel T, Valeri N, Workman P, Sansom O, Braconi C. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2017;66:1268-1277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |