Published online Jun 15, 2010. doi: 10.4291/wjgp.v1.i2.23
Revised: May 25, 2010
Accepted: June 1, 2010
Published online: June 15, 2010
Cholangiocarcinoma (CCA) is a notoriously lethal epithelial cancer originating from the biliary system. As radical resection offers a poor success rate and limited effective adjuvant modalities exist in its advanced stage, the disease leads to a fairly poor prognosis. As the incidence of CCA is increasing, although the mortality rate remains stable, and few other definite etiologies have yet to be established, renewing our knowledge of its fundamental carcinogenesis is advisable. The latest advances in molecular carcinogenesis have highlighted the roles of epigenetic perturbations and cancer-related inflammation in CCA. This review focuses on the reciprocal effects between aberrant DNA methylation and inflammatory microenvironment in CCA.
- Citation: Huang L, Frampton G, Liang LJ, DeMorrow S. Aberrant DNA methylation profile in cholangiocarcinoma. World J Gastrointest Pathophysiol 2010; 1(2): 23-29
- URL: https://www.wjgnet.com/2150-5330/full/v1/i2/23.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v1.i2.23
Cholangiocarcinoma (CCA) is the second most common epithelial cancer originating from the biliary system, accounting for 10% to 20% of primary liver cancer[1]. As the presentation of symptoms is delayed, an R0 resection (both gross and microscope negative margin) can be achieved in less than 80% of those 10% early-stage patients in whom radical surgical intervention can be applied[2,3]. Additionally, the limited adjuvant modalities available for advanced patients have failed to show substantial benefit[3]. The aforementioned factors offer CCA a fairly poor prognosis, as overall 5-year survival rates in the resectable cases are less than 40% in both extrahepatic cholangiocarcinoma (ECC) and intrahepatic cholangiocarcinoma (ICC), and the median survival time is less than 12 mo in unresectable or metastatic cases[2,3]. Incidences of CCA, especially ICC, are increasing worldwide, although mortality has remained stable during the last four decades, with the exception of the decline in gallbladder carcinoma[1,4]. To date, though some well-documented risk factors have been identified, the majority of CCA etiology has remained unknown[1,5,6]. Emerging advances in CCA molecular research have highlighted the roles of epigenetic perturbations and cancer-related inflammation[1,6-11]. The aberrant DNA methylation of CCA, which regarded as one of the best-characterized, mitotically heritable and reversible epigenetic modulations, has been seen to affect multiple steps of cholangiocarcinogenesis[7-12].
Epigenetic controls of gene expression orchestrate changes of chromatin architecture tissue-specifically and dynamically without affecting gene sequences, encompassing some basic mechanisms like post-translational modification of histones, displacement of nucleosomes and DNA methylations. Broadly, it may also include RNAi and non-coding RNAs. Within the above epigenetic modulations, DNA methylation is best characterized and delineated as a post-replicative addition of a methyl (-CH3) group to the cytosine-5-carbon position, which is catalyzed by at least three DNA methyltransferases (DNMTs): DNMT1, DNMT3a and DNMT3b. Methylation occurs mainly at CpG dinucleotides, CpNpG and rarely at non-CpG dinucleotides like CpA, CpT and CpC[13-15] of the CpG islands or the CpG island shores[16] located in the promoter or encoding regions of genes[17]. Methylated cytosine will allow binding with methyl-CpG binding domain (MBD) proteins (MeCP1 or MeCP2) to remodel the chromatin architecture, a process that has been recognized as essential and versatile for epigenetic modification[18]. To date, global genome hypomethylation and local tumor suppressor genes hypermethylation have been noticed in tumorigenesis[19,20]. Furthermore, abundant bench and bedside evidence supports the putative association between inflammation and cancer[12], which potentially leads to dysregulated DNA methylation in CCA[7-11]. Though clinicopathological and epidemiological differences exist between ICC and ECC, emerging evidence has revealed that both of them are closely related to chronic inflammation[1,2,5-10]. This review aims to summarize current reported aberrant DNA methylation profiles of CCA and outline the involving role of cancer-related inflammation.
Briefly, there are two pathways bridging inflammation to CCA: the extrinsic pathway (risk factors or related environmental exposures of CCA) and the intrinsic pathway (congenital or acquired genetic alterations, e.g. activation of oncogenes, inactivation of tumor suppressors, senescence-related perturbations, etc). The milieu of chronic inflammation may environmentally select the adaptive transformed cholangiocytes, thereby initiating cholangiocarcinogenesis. For instance, inactivation of 9p21 gene cluster (p16INK4a/p14ARF/p15Ink4b) has been unraveled in liver fluke-related CCA[21] and primary sclerosing cholangitis-associated CCA[22]. Also, a high frequency of microsatellite instability (MSI) and inactivation of hMLH1 has been observed in thorotrast-related CCA[23]. Although a recent study of thyroid carcinoma has uncovered that an early genetic event is necessary and sufficient for initiating a cancerous development by promoting an inflammatory microenvironment[24], similar instances of an intrinsic pathway have yet to be addressed in CCA. Instead, a mounting body of evidence of genetic alterations in CCA has indicated these pathways indirectly, such as mutations or deletions of K-ras, p53, p16INK4a, p15Ink4b, p14ARF or Smad4; loss of heterozygosity (LOH) of adenomatous polyposis coli gene (APC) or allelic losses on 3p13-p21 and 8q22[25-30].
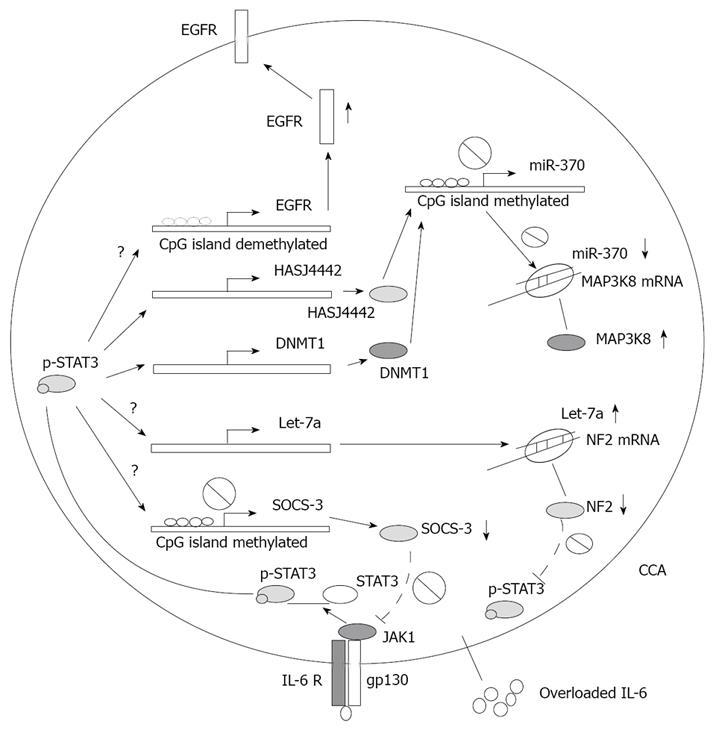
Some key intrinsic factors can mediate inflammation-related gene regulation in CCA, including transcription factors [signal transducer and activator of transcription 3 (STAT3), etc], cytokines (IL-6, TNF-α, etc), growth factors, nitric oxide, reactive nitrogen oxide species (RNOS) and bile acids[1]. Among the aforementioned mediators, IL-6 plays a crucial role in many cancers, especially in epithelial cancers[31]. Upregulation of IL-6 in carcinogenesis is triggered by an autocrine[32,33] or paracrine loop[34,35], or even by an intrinsic somatic mutation of epidermal growth factor receptor (EGFR)[36]. It is also well documented that in chronic cholangiopathies or biliary infection, the level of IL-6 is increased in bile. Briefly, in CCA, the negative feedback of the IL-6 pathway is deficient and replaced by an unlimited autocrine loop owing to aberrant epigenetic silence of suppressors of cytokine signaling 3 (SOCS-3), which is mediated by the IL-6/STAT3 pathway to maintain hypermethylation of the gene promoter[33]. Aberrant activated IL-6 in CCA leads to carcinogenesis promotion, through mechanisms such as up-regulating anti-apoptotic myeloid cell leukemia-1 (Mcl-1) mediated by phosphorylated STAT3[33,37]; up-regulating EGFR expression through decreasing its promoter methylation level mediated by undefined mechanisms[38]; and activating telomerase[39,40]. In breast cancer, overloaded IL-6 also enhances expression of stem cell survival regulator Notch-3 and activates the hypoxia-resistant gene carbonic anhydrase IX (CA-IX) through the Notch-3/Jagged-1 pathway[41]. However, effect of IL-6 such as these on cancer stem/progenitor cells in CCA has yet to be clearly defined. Moreover, IL-6 had been shown to affect microRNA profiles of CCA. Overloaded IL-6 also increased let-7a expression via an undetermined mechanism, resulting in suppression of neurofibromatosis 2 (NF2) and a subsequent increase of phosphorylated STAT3[42]. Recently, Meng and colleagues have revealed that overload of IL-6 in CCA can up-regulate expressions of two DNA methyltransferases, DNMT1 and HASJ4442, leading to hypermethylation of the CpG island where the miR-370 encoding gene was embedded. Consequently, oncogenic mitogen-activated protein kinase kinase kinase 8 (MAP3K8) suppressing effect of miR-370 was abrogated in CCA[43]. (The role of IL-6 signaling in CCA inflammation-related epigenetic regulation is summarized in Figure 1).
Infiltrated leukocytes also fuel the inflammation-related epigenetic perturbations in CCA. The mechanisms of leukocyte recruitment and homing in cholangiopathies, like primary biliary cirrhosis (PBC), primary sclerosing cholangitis (PSC) and chronic viral hepatitis C, have long been investigated. Briefly, chemokines CCL21, CCL28, CX3CL1, CXCL9 and CXCL10 mediate the recruitment of leukocytes into the portal tract and subsequently CX3CL1, CXCL12, CXCL16 and CCL28 retain these inflammatory cells in the bile duct to serve their under-determined roles[44,45]. Little research has been conducted regarding the immunosuppressive milieu of CCA-related inflammation. Recently, the roles of PD-L1 (also termed B7-H1 or CD274, an inhibitor of activated effector T cells)[46] and Foxp3+ regulatory T cells[47] in ICC have been explored. Meanwhile, Opisthorchis viverrini infection has been shown to upregulate inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) expression in rodent macrophages through the Toll-like receptor 2/NF-κB pathway[48]. However, epigenetic modulations on immunity against CCA have yet to be unraveled. Actually, in the Foxp3+ regulatory T cells of the mouse colitis model, the immunosuppressive effect related to Foxp3 was mediated by its co-repressors, Eos and C-terminal binding protein-1 (CtBP1), by promoting hypermethylation of IL-2 gene promoter, trimethylation of histone 3 lysine 4 and acetylation of histone 3 and histone 4[49]. Furthermore, both of the antigen-presentation molecule major histocompatibility complex class 2 (MHC-II) and its co-activator, class II transactivator (CIITA) can be epigenetically modulated in cancer[50]. Briefly, in renal carcinoma and pancreatic carcinoma, dysregulated IL-10 and Decoy receptor 3 (DcR3) can suppress CIITA expression respectively in immune cells through epigenetic modifications on CIITA gene promoter[51-52].
Besides leukocytes, much research has also highlighted the pivotal roles of other stroma constituents in the progression of chronic cholangiopathies, encompassing fibroblasts, hepatic stellate cells (HSC), extracellular matrix (ECM), etc[53-57]. Recently stromal effect on parenchyma epithelial-to-mesenchymal transition (EMT) has also been noticed in cholangiopathies[54,58-60]. However, in CCA, less evidence involving the contribution of remodeled stroma to the epigenetic perturbations than in other cancers could be obtained. Briefly, cancer-associated fibroblasts (CAFs) have been proven to exert their epigenetic modulations on neighboring immortalized human breast epithelial cells MCF10A by direct cell-cell contact, simultaneously activating Akt1 and suppressing Akt1 repressor, inositol polyphosphate-4-phosphatase type II (INPP4B), subsequently leading to de novo promoter hypermethylation of the tumor suppressor gene Cystatin M (CST6)[61]. Furthermore, mechanical forces of the remodeled ECM or tissue architecture may also exert potential modulated effects on epigenetic perturbations of cholangiocarcinogenesis. In this context, some details about dynamic biochemical pathways and mechanotransduction, termed “mechanoreciprocity”, have been explained in other cancers, like breast cancer[61-64]. Effects of ECM on the cellular epigenome are increasingly being deciphered[62-64]. Briefly, the ECM exert their cis- and trans-regulations on transcription via specific membrane receptors such as integrin, and various intracellular molecules like focal adhension kinase (FAK), RhoGTPases, ATP-dependent chromatin remodeling complexes (SWI/SNF, ISWI, CHD, INO80 and SWR1), which couple extracellular signals to the cytoskeleton and chromatin and mainly mediate by targeting the ECM-responsive elements (EREs) in the genome as well as transcription factors[62-65]. Moreover, it is increasingly accepted that ECM-mediated mechanotransduction can “prepare” chromatin structures to receive specific biochemical signals and can control common sets of genes in cells possessing similar morphology. Subsequently, defined biochemical signaling networks permit further tissue-specific transcription in differentiation-specific genes[62-64]. One typical example of this intricate reciprocity is nuclear lamina-associated transcriptional silencing[62]. The nuclear lamina is made of intermediate filament proteins like lamins A/C and B, which can bind smaller nesprins that belong to the inner nuclear membrane proteins. Smaller nesprins can shuttle between the outer and inner nuclear membrane, while the biggest nesprins are anchored in the outer nuclear membrane where they can extend into the cytoplasm and bind actin microfilaments and intermediate filaments[62]. Moreover, associations of negative transcription factors with components of the nuclear lamina have also been explained. For instance, the lamin B receptor can bind histone 3 methylated lysine 9 and heterochromatin component HP1[66], while lamin A/C and lamin B can bind MOK2 and Oct-1, respectively[67,68]. Recently, a genome-wide high-resolution mapping of lamin B1-associated DNA domains further implied this putative association between remodeled ECM and epigenetic control, revealing that lamin B1-associated DNA domains contain H3K27me3, the insulator protein CTCF or methylated CpG islands[69].
As the inflammatory microenvironment may vary with diverse cholangiopathies, disease-specific DNA methylation profiles or epigenome can reasonably be expected. Some of the aberrantly methylated genes reported in currently available literature are summarized in Table 1[70-74], according to respective specific risk factors of CCA.
| Methylated gene (proportion rate) | |
| Definite risk factors | |
| Primary sclerosing cholangitis | p16 (25%)[22] |
| Liver fluke infection (opisthorchis viverrini, or less frequently, clonorchis sinensis) | RUNX3 (49.1%)[70]; p14 (40.2%), p15 (48.9%), p16 (28.3%)[21]; hMLH1 (44.6%)[71] |
| Hepatolithiasis | p16 (100%[72], 54.6%[73]); TFF1 (37.5%)[74] |
| Biliary malformation (congenital choledochal cysts, caroli’s disease, etc) | NA |
| Thorotrast | hMLH1 (45.8%), hMSH2 (25.0%)[23] |
| Probable risk factors | |
| Hepatitis C or hepatitis B | NA |
| Cirrhosis | NA |
| Toxins (dioxin, polyvinyl chloride, nitrosamines, etc) | NA |
| Biliary-enteric anastomosis or cholangiojejunostomy | NA |
From the above Table 1, more details about aberrant DNA methylation profiles in CCA remain to be unraveled. To date, although the mechanism of demethylation and the candidate enzymes exhibiting direct demethylase activity and related cofactors are not yet firmly established in mammalian cells, recent trends in research about DNA demethylation have revealed the putative role of the BER/NER pathways and the association with DNMT1[75,76]. Moreover, repair-mediated DNA demethylation of Oct-4 gene promoter by Gadd45a has been observed[77]. Recently, Gadd45a has also been shown to connect neuronal circuit activity with DNA demethylation in mature neurons for extrinsic modulation of adult neurogenesis[78]. Further progresses in this field will help to facilitate our understanding about mechanisms of aberrant DNA methylation in CCA further.
In conclusion, chronic inflammation of cholangiopathies predisposes individuals to CCA. Since alterations of the cellular epigenome usually precede morphologic changes and genetic alterations, identification of related aberrant DNA methylation profiles according to specific inflammation milieu may serve as a reasonable early diagnostic marker and an intervention target for CCA.
Peer reviewers: Bob Roger Olsson, PhD, Associate Professor, Department of Internal Medicine, University of Gothenburg, Vita Stråket 15, Göteborg 41345, Sweden; Marçal Pastor Anglada, PhD, MSc, Professor, Department of Biochemistry and Molecular Biology, Institute of Biomedicine (IBUB) and CIBER EHD, University of Barcelona, Diagonal 645, Barcelona 08028, Spain
| 1. | Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308-321. |
| 2. | Aljiffry M, Abdulelah A, Walsh M, Peltekian K, Alwayn I, Molinari M. Evidence-based approach to cholangiocarcinoma: a systematic review of the current literature. J Am Coll Surg. 2009;208:134-147. |
| 3. | Hezel AF, Zhu AX. Systemic therapy for biliary tract cancers. Oncologist. 2008;13:415-423. |
| 4. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225-249. |
| 5. | Broomé U, Olsson R, Lööf L, Bodemar G, Hultcrantz R, Danielsson A, Prytz H, Sandberg-Gertzén H, Wallerstedt S, Lindberg G. Natural history and prognostic factors in 305 Swedish patients with primary sclerosing cholangitis. Gut. 1996;38:610-615. |
| 6. | Lazaridis KN, Gores GJ. Cholangiocarcinoma. Gastroenterology. 2005;128:1655-1667. |
| 7. | Rashid A. Cellular and molecular biology of biliary tract cancers. Surg Oncol Clin N Am. 2002;11:995-1009. |
| 8. | Lee S, Kim WH, Jung HY, Yang MH, Kang GH. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol. 2002;161:1015-1022. |
| 9. | Tischoff I, Wittekind C, Tannapfel A. Role of epigenetic alterations in cholangiocarcinoma. J Hepatobiliary Pancreat Surg. 2006;13:274-279. |
| 10. | Stutes M, Tran S, DeMorrow S. Genetic and epigenetic changes associated with cholangiocarcinoma: from DNA methylation to microRNAs. World J Gastroenterol. 2007;13:6465-6469. |
| 11. | Sandhu DS, Shire AM, Roberts LR. Epigenetic DNA hy-permethylation in cholangiocarcinoma: potential roles in pathogenesis, diagnosis and identification of treatment targets. Liver Int. 2008;28:12-27. |
| 12. | Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436-444. |
| 13. | Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237-5242. |
| 14. | Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41-48. |
| 15. | Kouidou S, Agidou T, Kyrkou A, Andreou A, Katopodi T, Georgiou E, Krikelis D, Dimitriadou A, Spanos P, Tsilikas C. Non-CpG cytosine methylation of p53 exon 5 in non-small cell lung carcinoma. Lung Cancer. 2005;50:299-307. |
| 16. | Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178-186. |
| 17. | Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378-1385. |
| 18. | Bogdanovi O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549-565. |
| 19. | Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148-1159. |
| 20. | Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286-298. |
| 21. | Chinnasri P, Pairojkul C, Jearanaikoon P, Sripa B, Bhudhisawasdi V, Tantimavanich S, Limpaiboon T. Preferentially different mechanisms of inactivation of 9p21 gene cluster in liver fluke-related cholangiocarcinoma. Hum Pathol. 2009;40:817-826. |
| 22. | Ahrendt SA, Eisenberger CF, Yip L, Rashid A, Chow JT, Pitt HA, Sidransky D. Chromosome 9p21 loss and p16 inactivation in primary sclerosing cholangitis-associated cholangiocarcinoma. J Surg Res. 1999;84:88-93. |
| 23. | Liu D, Momoi H, Li L, Ishikawa Y, Fukumoto M. Microsatellite instability in thorotrast-induced human intrahepatic cholangiocarcinoma. Int J Cancer. 2002;102:366-371. |
| 24. | Borrello MG, Alberti L, Fischer A, Degl’innocenti D, Ferrario C, Gariboldi M, Marchesi F, Allavena P, Greco A, Collini P. Induction of a proinflammatory program in normal human thyrocytes by the RET/PTC1 oncogene. Proc Natl Acad Sci USA. 2005;102:14825-14830. |
| 25. | Kang YK, Kim WH, Lee HW, Lee HK, Kim YI. Mutation of p53 and K-ras, and loss of heterozygosity of APC in intrahepatic cholangiocarcinoma. Lab Invest. 1999;79:477-483. |
| 26. | Sturm PD, Baas IO, Clement MJ, Nakeeb A, Johan G, Offerhaus A, Hruban RH, Pitt HA. Alterations of the p53 tumor-suppressor gene and K-ras oncogene in perihilar cholangiocarcinomas from a high-incidence area. Int J Cancer. 1998;78:695-698. |
| 27. | Yoshida S, Todoroki T, Ichikawa Y, Hanai S, Suzuki H, Hori M, Fukao K, Miwa M, Uchida K. Mutations of p16Ink4/CDKN2 and p15Ink4B/MTS2 genes in biliary tract cancers. Cancer Res. 1995;55:2756-2760. |
| 28. | Hahn SA, Bartsch D, Schroers A, Galehdari H, Becker M, Ramaswamy A, Schwarte-Waldhoff I, Maschek H, Schmiegel W. Mutations of the DPC4/Smad4 gene in biliary tract carcinoma. Cancer Res. 1998;58:1124-1126. |
| 29. | Shiraishi K, Okita K, Kusano N, Harada T, Kondoh S, Okita S, Ryozawa S, Ohmura R, Noguchi T, Iida Y. A comparison of DNA copy number changes detected by comparative genomic hybridization in malignancies of the liver, biliary tract and pancreas. Oncology. 2001;60:151-161. |
| 30. | Kawaki J, Miyazaki M, Ito H, Nakagawa K, Shimizu H, Yoshidome H, Uzawa K, Tanzawa H, Nakajima N. Allelic loss in human intrahepatic cholangiocarcinoma: correlation between chromosome 8p22 and tumor progression. Int J Cancer. 2000;88:228-231. |
| 31. | Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. 2007;117:3660-3363. |
| 32. | Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13:7-9. |
| 33. | Isomoto H, Mott JL, Kobayashi S, Werneburg NW, Bronk SF, Haan S, Gores GJ. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology. 2007;132:384-396. |
| 34. | Song L, Asgharzadeh S, Salo J, Engell K, Wu HW, Sposto R, Ara T, Silverman AM, DeClerck YA, Seeger RC. Valpha24-invariant NKT cells mediate antitumor activity via killing of tumor-associated macrophages. J Clin Invest. 2009;119:1524-1536. |
| 35. | Li YY, Hsieh LL, Tang RP, Liao SK, Yeh KY. Macrophage-derived interleukin-6 up-regulates MUC1, but down-regulates MUC2 expression in the human colon cancer HT-29 cell line. Cell Immunol. 2009;256:19-26. |
| 36. | Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846-3856. |
| 37. | Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054-2065. |
| 38. | Wehbe H, Henson R, Meng F, Mize-Berge J, Patel T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res. 2006;66:10517-10524. |
| 39. | Yamagiwa Y, Meng F, Patel T. Interleukin-6 decreases senescence and increases telomerase activity in malignant human cholangiocytes. Life Sci. 2006;78:2494-2502. |
| 40. | Park J, Tadlock L, Gores GJ, Patel T. Inhibition of interleukin 6-mediated mitogen-activated protein kinase activation attenuates growth of a cholangiocarcinoma cell line. Hepatology. 1999;30:1128-1133. |
| 41. | Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988-4002. |
| 42. | Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256-8264. |
| 43. | Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27:378-386. |
| 44. | Isse K, Harada K, Zen Y, Kamihira T, Shimoda S, Harada M, Nakanuma Y. Fractalkine and CX3CR1 are involved in the recruitment of intraepithelial lymphocytes of intrahepatic bile ducts. Hepatology. 2005;41:506-516. |
| 45. | Borchers AT, Shimoda S, Bowlus C, Keen CL, Gershwin ME. Lymphocyte recruitment and homing to the liver in primary biliary cirrhosis and primary sclerosing cholangitis. Semin Immunopathol. 2009;31:309-322. |
| 46. | Ye Y, Zhou L, Xie X, Jiang G, Xie H, Zheng S. Interaction of B7-H1 on intrahepatic cholangiocarcinoma cells with PD-1 on tumor-infiltrating T cells as a mechanism of immune evasion. J Surg Oncol. 2009;100:500-504. |
| 47. | Kobayashi N, Hiraoka N, Yamagami W, Ojima H, Kanai Y, Kosuge T, Nakajima A, Hirohashi S. FOXP3+ regulatory T cells affect the development and progression of hepatocarcinogenesis. Clin Cancer Res. 2007;13:902-911. |
| 48. | Pinlaor S, Tada-Oikawa S, Hiraku Y, Pinlaor P, Ma N, Sithithaworn P, Kawanishi S. Opisthorchis viverrini antigen induces the expression of Toll-like receptor 2 in macrophage RAW cell line. Int J Parasitol. 2005;35:591-596. |
| 49. | Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142-1146. |
| 50. | Wright KL, Ting JP. Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol. 2006;27:405-412. |
| 51. | Choi YE, Yu HN, Yoon CH, Bae YS. Tumor-mediated down-regulation of MHC class II in DC development is attributable to the epigenetic control of the CIITA type I promoter. Eur J Immunol. 2009;39:858-868. |
| 52. | Chang YC, Chen TC, Lee CT, Yang CY, Wang HW, Wang CC, Hsieh SL. Epigenetic control of MHC class II expression in tumor-associated macrophages by decoy receptor 3. Blood. 2008;111:5054-5063. |
| 53. | Ohira S, Sasaki M, Harada K, Sato Y, Zen Y, Isse K, Kozaka K, Ishikawa A, Oda K, Nimura Y. Possible regulation of migration of intrahepatic cholangiocarcinoma cells by interaction of CXCR4 expressed in carcinoma cells with tumor necrosis factor-alpha and stromal-derived factor-1 released in stroma. Am J Pathol. 2006;168:1155-1168. |
| 54. | Omenetti A, Porrello A, Jung Y, Yang L, Popov Y, Choi SS, Witek RP, Alpini G, Venter J, Vandongen HM. Hedgehog signaling regulates epithelial-mesenchymal transition during biliary fibrosis in rodents and humans. J Clin Invest. 2008;118:3331-3342. |
| 55. | Roderfeld M, Rath T, Voswinckel R, Dierkes C, Dietrich H, Zahner D, Graf J, Roeb E. Bone marrow transplantation demonstrates medullar origin of CD34+ fibrocytes and ameliorates hepatic fibrosis in Abcb4-/- mice. Hepatology. 2010;51:267-276. |
| 56. | Farazi PA, Zeisberg M, Glickman J, Zhang Y, Kalluri R, DePinho RA. Chronic bile duct injury associated with fibrotic matrix microenvironment provokes cholangiocarcinoma in p53-deficient mice. Cancer Res. 2006;66:6622-6627. |
| 57. | Yasoshima M, Sato Y, Furubo S, Kizawa K, Sanzen T, Ozaki S, Harada K, Nakanuma Y. Matrix proteins of basement membrane of intrahepatic bile ducts are degraded in congenital hepatic fibrosis and Caroli’s disease. J Pathol. 2009;217:442-451. |
| 58. | Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology. 2009;50:2007-2013. |
| 59. | Lee MJ, Yu GR, Yoo HJ, Kim JH, Yoon BI, Choi YK, Kim DG. ANXA8 down-regulation by EGF-FOXO4 signaling is involved in cell scattering and tumor metastasis of cholangiocarcinoma. Gastroenterology. 2009;137:1138-1150, 1150.e1-e9. |
| 60. | Harada K, Sato Y, Ikeda H, Isse K, Ozaki S, Enomae M, Ohama K, Katayanagi K, Kurumaya H, Matsui A. Epithelial-mesenchymal transition induced by biliary innate immunity contributes to the sclerosing cholangiopathy of biliary atresia. J Pathol. 2009;217:654-664. |
| 61. | Lin HJ, Zuo T, Lin CH, Kuo CT, Liyanarachchi S, Sun S, Shen R, Deatherage DE, Potter D, Asamoto L. Breast cancer-associated fibroblasts confer AKT1-mediated epigenetic silencing of Cystatin M in epithelial cells. Cancer Res. 2008;68:10257-10266. |
| 62. | Lelièvre SA. Contributions of extracellular matrix signaling and tissue architecture to nuclear mechanisms and spatial organization of gene expression control. Biochim Biophys Acta. 2009;1790:925-935. |
| 63. | Xu R, Boudreau A, Bissell MJ. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer Metastasis Rev. 2009;28:167-176. |
| 64. | Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9:108-122. |
| 66. | Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983-14989. |
| 67. | Dreuillet C, Tillit J, Kress M, Ernoult-Lange M. In vivo and in vitro interaction between human transcription factor MOK2 and nuclear lamin A/C. Nucleic Acids Res. 2002;30:4634-4642. |
| 68. | Imai S, Nishibayashi S, Takao K, Tomifuji M, Fujino T, Hasegawa M, Takano T. Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression. Mol Biol Cell. 1997;8:2407-2419. |
| 69. | Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948-951. |
| 70. | Dachrut S, Banthaisong S, Sripa M, Paeyao A, Ho C, Lee SA, Kosinski C, Patil MA, Zhang J, Chen X. DNA copy-number loss on 1p36.1 harboring RUNX3 with promoter hypermethylation and associated loss of RUNX3 expression in liver fluke-associated intrahepatic cholangiocarcinoma. Asian Pac J Cancer Prev. 2009;10:575-582. |
| 71. | Limpaiboon T, Khaenam P, Chinnasri P, Soonklang M, Jearanaikoon P, Sripa B, Pairojkul C, Bhudhisawasdi V. Promoter hypermethylation is a major event of hMLH1 gene inactivation in liver fluke related cholangiocarcinoma. Cancer Lett. 2005;217:213-219. |
| 72. | Sasaki M, Yamaguchi J, Itatsu K, Ikeda H, Nakanuma Y. Over-expression of polycomb group protein EZH2 relates to decreased expression of p16 INK4a in cholangiocarcinogenesis in hepatolithiasis. J Pathol. 2008;215:175-183. |
| 73. | Ishikawa A, Sasaki M, Sato Y, Ohira S, Chen MF, Huang SF, Oda K, Nimura Y, Nakanuma Y. Frequent p16ink4a inactivation is an early and frequent event of intraductal papillary neoplasm of the liver arising in hepatolithiasis. Hum Pathol. 2004;35:1505-1514. |
| 74. | Sasaki M, Tsuneyama K, Nakanuma Y. Aberrant expression of trefoil factor family 1 in biliary epithelium in hepatolithiasis and cholangiocarcinoma. Lab Invest. 2003;83:1403-1413. |
| 75. | Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45-50. |
| 76. | Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112-115. |