Published online Oct 18, 2018. doi: 10.4331/wjbc.v9.i1.1
Peer-review started: July 30, 2018
First decision: August 24, 2018
Revised: September 19, 2018
Accepted: October 11, 2018
Article in press: October 12, 2018
Published online: October 18, 2018
Processing time: 82 Days and 2.6 Hours
Reactive oxygen species (ROS) are produced during normal physiologic processes with the consumption of oxygen. While ROS play signaling roles, when they are produced in excess beyond normal antioxidative capacity this can cause pathogenic damage to cells. The majority of such oxidation occurs in polyunsaturated fatty acids and sulfhydryl group in proteins, resulting in lipid peroxidation and protein misfolding, respectively. The accumulation of misfolded proteins in the endoplasmic reticulum (ER) is enhanced under conditions of oxidative stress and results in ER stress, which, together, leads to the malfunction of cellular homeostasis. Multiple types of defensive machinery are activated in unfolded protein response under ER stress to resolve this unfavorable situation. ER stress triggers the malfunction of protein secretion and is associated with a variety of pathogenic conditions including defective insulin secretion from pancreatic β-cells and accelerated lipid droplet formation in hepatocytes. Herein we use nonalcoholic fatty liver disease (NAFLD) as an illustration of such pathological liver conditions that result from ER stress in association with oxidative stress. Protecting the ER by eliminating excessive ROS via the administration of antioxidants or by enhancing lipid-metabolizing capacity via the activation of peroxisome proliferator-activated receptors represent promising therapeutics for NAFLD.
Core tip: Accumulated experimental data indicate that oxidative stress causes endoplasmic reticulum stress, which together leads to pathogenic damage to cells. The lipid metabolism in the liver is a sensitive target of these types of stress, which appears to be associated with non-alcoholic fatty liver disease.
- Citation: Fujii J, Homma T, Kobayashi S, Seo HG. Mutual interaction between oxidative stress and endoplasmic reticulum stress in the pathogenesis of diseases specifically focusing on non-alcoholic fatty liver disease. World J Biol Chem 2018; 9(1): 1-15
- URL: https://www.wjgnet.com/1949-8454/full/v9/i1/1.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v9.i1.1
Most eukaryotic cells, excluding enucleated cells such as mammalian red blood cells, synthesize proteins based on genetic information from nuclear DNA during their lifespan. Translated secretory and membrane proteins become functional when they undergo appropriate oxidative folding, posttranslational modification, and cellular localization. While the redox status in the cytosol is in a reduced state, the extracellular space is largely in an oxidized state. Premature secretion and exposure to an extracellular environment can cause aberrant conformational changes in proteins due to inappropriate disulfide bond formation. Hence, the appropriate oxidative folding of polypeptides by means of the oxidation of sulfhydryl groups to produce disulfide bridges in proteins within cells is a prerequisite process for producing secretory proteins and the membrane proteins prior to their secretion and translocation to the plasma membrane, respectively (Figure 1). The signaling process for these types of oxidative protein folding reactions was not understood well until the end of the last century[1,2].
The accumulation of misfolded proteins in the endoplasmic reticulum (ER) causes ER stress, which then leads to the malfunction of the ER[3]. Without proper resolution of ER stress, affected cells become dysfunctional and, if not resolved properly, they die. To avoid this unfavorable scenario, multiple defensive machineries, referred to as an unfolded protein response (UPR), are activated under such conditions and play roles in preventing this and permit the cells to recover from this fatal situation[4,5].
Multiple factors, either internally or externally, can cause the accumulation of misfolded proteins in the ER[6,7]. Reactive oxygen species (ROS) are produced during conventional physiological processes accompanied by oxygen consumption and the levels are enhanced under a variety of pathological conditions such as inflammation, high temperature, and a deficit in the antioxidative system, and result in the development of oxidative stress[8]. Both low molecular weight antioxidant compounds and antioxidative enzymes function to control the levels of ROS and reduce their levels to acceptable ranges. However, the antioxidant levels in the ER are relatively low compared to the cytoplasm or other organelles, despite the robust production of hydrogen peroxide via active reduction-oxidation (redox) reactions[9]. Oxidative stress perturbs the usual oxidative protein folding, which results in the ER stress and organ failure. Thus, among a variety of stressful conditions, oxidative stress can occur in any cell and is also responsible for ER stress, and they together lead to the development of a pathogenic state.
Herein we overview recent progress in our understanding of the relationships between oxidative stress and ER stress and attempt to clarify the pathogenic pathways that are involved, by focusing on fatty liver diseases.
Protein conformation is supported by several types of chemical bonds, among which the disulfide bonds formed between cysteine sulfhydryl groups are the primary determinants of overall protein structure for secreted proteins and membrane proteins that face the extracellular space. Endoplasmic reticulum oxidoreductin 1 (ERO1), which contains flavin adenine dinucleotide (FAD) as a redox cofactor, is an evolutionarily conserved protein[10] and, in conjunction with molecular oxygen consumption, catalyzes disulfide formation in nascent proteins via protein disulfide isomerase (PDI) in the ER[11,12]. PDI is a member of the family of chaperone molecules that are specifically responsible for disulfide bond formation in proteins in the ER lumen[13,14]. In addition to PDI, several chaperone molecules are also present in the ER lumen and participate in maintaining ER homeostasis[15]. Mammals have two ERO1 genes, ERO1α and ERO1β, that are transcriptionally regulated by the CCAAT-enhancer-binding protein homologous protein (CHOP)[16]. ERO1 first introduces a disulfide bond in PDI and its family members using molecular oxygen as the oxidant[13,14]. Hydrogen peroxide is produced as a byproduct in this oxidative protein folding process. Because not only secretory proteins but also many membrane proteins that face the extracellular space must undergo oxidative protein folding in the ER, hydrogen peroxide is inevitably produced in the ERO1-mediated sulfoxidation reaction. Thus, the ER is exposed to an oxidative insult to a greater or lesser extent as the result of this type of oxidative protein folding[17].
ERO1 is prerequisite for oxidative protein folding in yeast and the genetic ablation of the gene results in yeast that are hypersensitive to reducing agents such as dithiothreitol[18,19]. However the genetic ablation of genes encoding ERO1α and ERO1β cause only moderate effects in mice, e.g., a decrease in ERO1 activity blunts the cardiomyocyte inotropic response to adrenergic stimulation and sensitized mice to adrenergic blockade[20]. This rather mild phenotype compared to the case of yeast can be explained by the presence of other redundant sulfoxidase enzymes in mammalian cells. Quiescin sulfhydryl oxidase (QSOX), a flavoprotein, catalyzes disulfide formation in PDI family members using an oxygen molecule and produces hydrogen peroxide[21]. The enzymatic properties are somewhat similar to ERO1. However, since QSOX is localized primarily in the Golgi apparatus and secreted fluids it appears to be unique as a multi-domain enzyme[22]. Mammalian cells also possess other sulfoxidases that introduce disulfide bonds to PDI by means of hydrogen peroxide or other oxidizing power, such as peroxiredoxin (PRDX)4 and glutathione peroxidase (GPX)7/GPX8[23], as described below.
The accumulation of inappropriately folded proteins is caused by unfavorable conditions, including the excessive synthesis of proteins, the inhibition of protein glycosylation, and Ca2+ depletion[24]. ER homeostasis is maintained by a variety of factors that include ionic and metabolic conditions. For example, ER is the organelle that stores millimolar ranges of Ca2+ which is maintained by Ca2+-ATPase localized at the ER membrane. Cardiac and skeletal muscle possess specialized forms for this Ca2+ storage, designated as the sarcoplasmic reticulum (SR), that regulates excitation-contraction coupling in these muscular tissues[25]. Ca2+-ATPase in the SR membrane contributes to Ca2+ uptake, so that the inhibition of Ca2+ uptake by thapsigargin decreases the levels of Ca2+ stored in the SR lumen. Similarly thapsigargin inhibits the non-muscular form of Ca2+-ATPase and leads to pathological conditions via ER stress[26]. Some antibiotics, e.g., tunicamycin, inhibit the formation of N-linked glycosylation, leading to the suppression of the translocation of nascent proteins to the Golgi body[27].
Heat stress impairs the proper folding of proteins by affecting the free energy needed for correct protein folding and thereby produces misfolded proteins with a high frequency[28]. As a compensatory mechanism, heat shock proteins (HSPs), a large protein family, are induced at high temperatures and ameliorate protein folding via their ability to function as a chaperone. Repetitive or sustained stimulation of hormonal secretion, such as the case of insulin secretion under hyperglycemic conditions, is a suspected cause for defected insulin production and β-cell dysfunction, which typically leads to type II diabetes[24].
Because the accumulation of misfolded proteins causes dysfunction of the ER and, if not properly resolved, ultimately results in cell death, several systems in the UPR are activated to resolve the stress. Genes involved in ameliorating ER stress are largely induced by several mechanisms via sensor proteins that are localized on the ER membrane. Because this process has been overviewed extensively by experts in this research field[5,29], we briefly mention major machineries and related matters here.
Both secretory proteins and membrane proteins are synthesized on the rough ER where ribosome-mRNA complexes are attached. Eukaryotic initiation factor 2α (eIF2α) plays a key role in the regulation of the translational process from mRNA on the cytoplasmic surface of the rough ER. Upon ER stress, double-stranded RNA-activated eIF2α kinase-like ER kinase (PERK) is activated upon autophosphorylation and phosphorylates eIF2α, which results in the selective translation of activating transcription factor 4 (ATF4)[30]. ATF4 then transcribes activating transcription factor 3 (ATF3), CHOP, and growth arrest and DNA damage-inducible 34 (GADD34). In the meantime phosphorylated eIF2α causes the synthesis of many other protein to be suppressed and restricts further protein influx to the ER lumen via attenuating the guanine nucleotide exchange factor eIF2B[31].
The inositol requiring enzyme 1 (IRE1) was originally reported as a yeast membrane protein that was predicted to be a kinase involved in inositol metabolism[32]. The potential role of IRE1 in the transmission of the unfolded protein signal across the ER or the inner nuclear membrane was then rediscovered later[33]. IRE1 undergoes activation via oligomerization in response to unfolded proteins and autophoshorylation. Active IRE1 then completes the splicing of the X-box-binding-protein 1 (XBP1) precursor mRNA by its RNase activity[34]. The XBP1 protein translated from the mature XBP1 mRNA moves to the nucleus and then transcriptionally activates genes for a protein chaperone, such as the glucose-regulated protein of 78 kDa (GRP78 also referred to as Bip)[35] and PDIs[36].
Activating transcription factor 6 (ATF6) was also originally found in yeast[37], but its localization in the ER membrane was recognized at later period. The localization of ATF6 to the ER membrane in its inactive form and its subsequent transcriptional activation in response to UPR have been reported[38]. ER stress stimulates the translocation of the precursor form of ATF6 to the Golgi membrane where Site-1 (S1P) and the Site-2 proteases (S2P) are located[39,40]. These proteases release the transcriptional activator domain from the precursors, and the resulting active ATF6 moves to the nucleus where it transcriptionally activates the corresponding genes[41]. Astrocytes specifically express a unique ER stress transducer, the old astrocyte specifically induced substance (OASIS), that regulates the signaling of UPR and contributes to the resistance to ER stress[42].
Misfolded proteins that are formed within the ER lumen undergo either refolding to the correct conformation in a chaperone-mediated manner or undergo proteolytic degradation, designated as ER-associated degradation (ERAD)[43]. PDI is the first enzyme that was reported to catalyze protein folding[44]. Nearly 20 PDI family proteins are present in the ER lumen and are involved in correcting disulfide bonds and also the oxidative folding of nascent proteins in the ER in association with ERO1 and other sulfoxidases[45].
Some unfolded proteins are exported out from the ER lumen or the ER membrane, polyubiquitinated and then degraded by proteasomes[46]. ERAD pathways, which are further classified into ERAD-L, -M, and -C, are responsible for misfolded proteins in the ER lumen, within the membrane, or on the cytosolic side of the ER membrane, respectively[43]. RING-finger ligase Hrd1, in association with other components appear to play a major role in ERAD-L and ERAD-M[47]. A retro-translocation channel is formed by Hrd1 in complex with the partner molecule Hrd3 and are involved in the polyubiquitination of target proteins[48]. Thus, among the several candidate channel molecules, the Hrd1-Hrd3 complex appears to play a bona fide role in polyubiquitination/protein translocation in the ERAD. Derin 1 (Der1), which reportedly initiates the export of aberrant proteins from the ER[49], appears to aid luminal substrates to be moved to the Hrd1 channel. The potential involvement of other ubiquitin ligases that span the ER membrane in this manner has also been proposed.
The machinery responsible for ERAD is also involved in cholesterol homeostasis by controlling the protein levels of 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase in the ER membrane[50]. HMG CoA reductase, which is embedded in the ER membrane, limits cholesterol synthesis. The presence of a sufficient amount of cholesterol results in an accelerated degradation of HMG CoA reductase via the activation of polyubiquitination followed by proteasome-catalyzed degradation[51], a process that is independent from ER stress. Hence, the ERAD system appears to be involved in, not only the proteolytic removal of misfolded proteins, but also regulation of the cholesterol metabolic process by means of the same ER-associated machinery. Moreover, because some ER-resident proteins such as Bip[35], PDI[36], and PRDX4[52,53] are reportedly present in the cytosol, the ERAD machinery may be partly responsible for the retro-translocation of these proteins from the ER to the cytosol.
Cell death is induced when these protective mechanisms against the ER stress functions insufficiently and do not rescue ER function[54]. CHOP, also referred to as GADD153, is a developmentally regulated nuclear protein that has a strong sequence similarity to transcription factors C/EBP. CHOP functions as a dominant-negative inhibitor of the activity of C/EBP-like proteins[55]. CHOP induces the production of ERO1α, which then activates the inositol 1,4,5-triphosphate (IP3) receptor and stimulates the release of Ca2+ from the ER via the IP3 receptor channel. The increased levels of cytosolic Ca2+ then trigger apoptosis in cells that are under ER stress[56], and induction of cell death is a well-established function of CHOP[57]. ERO1α was found to be enriched in the mitochondrial associated membrane (MAM), the interface area of the ER with mitochondria[58], and may also regulate mitochondrial Ca2+ flux via controlling redox homeostasis in the ER[59]. The pro-apoptotic roles of CHOP are supported by the finding that the disruption of the gene that encodes CHOP causes delays in the development of ER stress-mediated diabetes and decreases the sensitivity of pancreatic islet cells to nitric oxide toxicity[60,61]. PERK and the IRE1 pathway activate CHOP, which, after translocation to the nucleus, inhibits the expression of anti-apoptotic genes, such a BCL-2, and activates the pro-apoptotic genes, Bim and DR5[62,63]. Nevertheless, CHOP appears to have an additional function, which needs clarification[64].
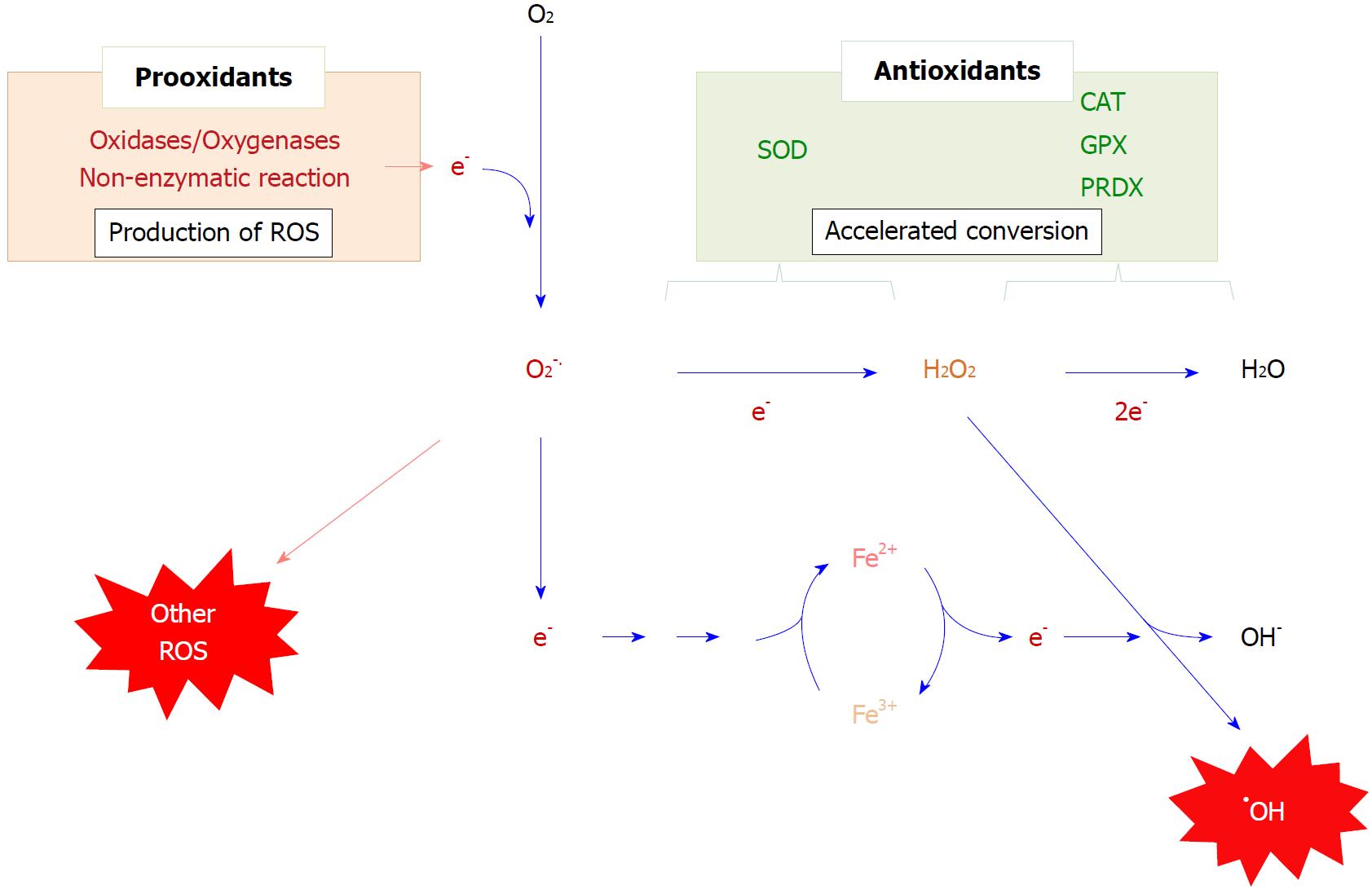
Superoxide radicals are produced primarily via a one-electron donation to molecular oxygen, which is mediated either enzymatic reactions such as oxygenases or non-enzymatic reactions such as glycoxidation[65,66] (Figure 2). While ROS, notably hydrogen peroxide, produced by stimuli function as signal regulators[67], excessively produced ROS cause oxidative stress that can exert a variety of effects over cellular functions. Transferring an unpaired electron together with oxygen to an unsaturated lipid causes lipid peroxidation, which then triggers a radical chain reaction[68]. Superoxide is not reactive as a radical, but it may trigger the production of more reactive free radicals by a reaction referred to as Fenton chemistry, a process that is initiated by the donation of an electron to a transition metal ion, such as Fe3+[69]. The resultant Fe2+ reacts with hydrogen peroxide, leading to the production of hydroxyl radicals, one of the most harmful molecules, which oxidizes lipids that contain unsaturated fatty acids, bases in nucleic acids, and proteins.
Based on the amount of consumed oxygen molecules, mitochondria are generally regarded as the main source of ROS production. MAM appears to be the place where these organelles communicate and provide ROS to the ER from mitochondria[58]. NADPH-oxidases (NOX) are another source of ROS and frequently cause oxidative stress, especially under inflammatory conditions[70]. After mitochondria, the ER is the organelle that consumes the most molecular oxygen because there are several oxygenases that are associated with this organelle. While most NOX members are located in the plasma membrane and produce superoxide as primary ROS, NOX4, a ER-resident enzyme, faces the ER lumen and releases hydrogen peroxide by the two-electron reduction of an oxygen molecule[71]. Cytochrome P450 is largely associated with the ER membrane and releases ROS to the cytoplasmic side[72,73]. However, these genes are selectively expressed in tissues, such as the liver and steroidogenic organs.
In the meantime, oxygen molecule is also consumed inside the ER by ERO1[74,75], which is involved in oxidative protein folding in most cells, as mentioned above. In fact, the ER is the organelle that produces the highest levels of hydrogen peroxide[76]. The levels of hydrogen peroxide increase under conditions where the secretion of proteins is enhanced due to ERO1-mediated oxidative protein folding[77]. Insulin production is maintained at high levels under prolonged hyperglycemic conditions and hydrogen peroxide levels are also kept high, which may cause oxidative stress together with ER stress in pancreatic β-cells and ultimately aberrant insulin secretion[78]. Hydrogen peroxide may also come from other cellular compartments or reactions other than ERO1-catalysed PDI oxidation[79].
To avoid oxidative stress, ROS are either eliminated via interactions with low molecular weight antioxidants, such as glutathione (GSH) and vitamin C, or converted to less-reactive compunds by antioxidative enzymes. Superoxide is exclusively converted to hydrogen peroxide by superoxide dismutase (SOD). In the case of the elimination of hydrogen peroxide, GSH peroxidase (GPX) and peroxiredoxin (PRDX), which are encoded by respective gene families[80,81], largely contribute to the reductive detoxification of hydrogen peroxide. While conventional GPX utilizes GSH as the electron donor, PRDX largely utilizes thioredoxin for this purpose. Extracellular superoxide dismutase (SOD3) and the plasma form of GPX (GPX3) are produced in the ER in the same manner as other secretory proteins. However, while some of them may be retained in the ER lumen, most are excreted. Thus, only few antioxidative systems are present in the ER lumen compared to cytoplasm and mitochondria.
Superoxide is converted to hydrogen peroxide by dismutation, either by a spontaneous reaction between two superoxide molecules or by accelerated production via the SOD-catalyzed reaction[82]. The latter reaction is about 3000 times faster than the spontaneous dismutation and causes superoxide levels to be decreased to less than 0.5% of that of the SOD1-free erythrocytes[83]. However, the physiological relevance of SOD is sometimes a topic of debate because hydrogen peroxide, which is not a radical but is still a reactive compound, is the product of the catalytic conversion of superoxide. While hydrogen peroxide does not itself cause a radical chain reaction, on accepting one electron, hydrogen peroxide is converted to a highly toxic molecule, hydroxyl radical, via the Fenton reaction[69]. The Fenton reaction involves a one-electron reduction of a transition metal ion in the preceding step and can be prevented by either the removal of the free transition metal ion or the elimination of the radical species that serves as the electron donor. Thus, the physiological significance of SOD can be rationally explained by the rapid elimination of superoxide before donating its unpaired electron. The resulting hydrogen peroxide can be reduced immediately to water by peroxidases that abundant in the body.
Among the three SOD isozymes present in mammalian cells, SOD1, which is localized in the cytoplasm and the intermembrane space in mitochondria as well, is expressed ubiquitously in the body and the best characterized from the biochemical and pathological viewpoints[82]. A missense mutation in SOD1 is reportedly a cause of familial amyotrophic lateral sclerosis (ALS)[84]. More than 150 mutations have been identified in the SOD1 gene in familial ALS, and several model mice have been established by the transgenic overexpression of the mutant gene and have been employed in attempts to elucidate the etiology of ALS and to develop possible drugs[85]. On the contrary, the genetic ablation of SOD1 does not induce ALS in mice[86], indicating that ALS develops as a consequence of the gain-of-function of mutant SOD1. Among several proposed mechanisms, an attractive hypothesis has been proposed from viewpoint of ER stress-mediated motor neuron death as follows. ALS-linked mutations of SOD1 cause chronic ER stress through interactions with Der1, a component proposed for ERAD[87], as described above. In addition it is noteworthy that, under zinc-depleted conditions, the wild-type SOD1 interacts with Der1[88,89], suggesting that SOD1 plays a role in the interaction with Der1. Therefore SOD1 may be post-translationally converted into a pathogenic molecule under zinc insufficient conditions and could also cause sporadic ALS. In the meantime, mutant SOD1 interacts with subunits of the coatomer coat protein II (COPII) complex, which is essential for ER-Golgi transport[90]. Thus, mutant SOD1 may aggravate ER stress via the suppression of membrane transport from the ER to the Golgi body.
Because ERO1 and other oxidoreductases that are involved in oxidative protein folding produce hydrogen peroxide, which would be expected to lead to ER stress via oxidative modification, especially sulfhydryls in nascent proteins. While peroxiredoxins exert peroxidase activity using the reducing power derived from reducing agents, such as thioredoxin, PRDX4 possesses a hydrophobic signal peptide[91] and is the only form present in the ER lumen[92,93]. PRDX4 eliminates peroxides in a thioredoxin-dependent manner in vitro analogous to other family members[94], but the thioredoxin-redox system is much less active in the ER compared to the cytosol. Hence, it is not likely that thioredoxin supports the elimination of hydrogen peroxide by PRDX4 in the ER.
PRDX4-catalyzed disulfide formation in PDI proteins by means of hydrogen peroxide was first demonstrated in vitro[95,96]. PRDX4 utilizes hydrogen peroxide to oxidize reactive sulfhydryls in the PDI, rescues an ERO1 deficiency in yeast[95], and catalyzes hydrogen peroxide-mediated oxidative protein folding in the ER[97]. Among the PDI family members, two proteins, P5 and ERp46, are preferential targets of PRDX4[98]. While ERO1 utilizes molecular oxygen as an electron acceptor and produces hydrogen peroxide[95], PRDX4 utilizes electrons from sulfhydryl groups in PDI family proteins and reduces hydrogen peroxide. Thus PRDX4 is beneficial from the standpoint of antioxidation as well[99]. PRDX4 regulates disulfide formation in glycerophosphodiester phosphodiesterase (GDE)2, a membrane protein, and determines its cell surface expression and consequently neurogenesis in mouse embryos as well[100].
The genetic ablation of PRDX4 causes a defect in the development of testis but otherwise shows a moderate phenotypical abnormality in mice[101]. This can be attributed to a functional redundancy of other molecules in the ER. The induction of PRDX4 is observed during the terminal differentiation of B cells to plasma cells after a lipopolysaccharide treatment[102]. In PRDX4-deficient plasma cells, large aggregates of IgM are actually formed in the ER. The ablation of both ERO1 and PRDX4 causes the consumption of ascorbic acid, which donates electrons for the hydroxylation of prolines in collagen, and results in atypical scurvy, which is characterized by an insufficient production of collagen[103]. The pancreas express PRDX4 abundantly[104] probably because protein secretion both as exocrine and endocrine mechanisms is critical in this organ. In fact mice that overexpress PRDX4 show protection against high-dose streptozotocin-induced diabetes in transgenic mice[105,106]. The biosynthesis and secretion of insulin are improved by the overexpression of PRDX4 in INS-1E cells[107]. Moreover, atherosclerosis in apolipoprotein E-knockout mice is attenuated by the excessive expression of PRDX4[108].
Issues regarding the physiological functions of PRDX4 other than ER sulfoxidase remain ambiguous and should be resolved. A portion of the PRDX4 is present in blood plasma[104] and is elevated under some pathological conditions[109]. However, its release from cultivated cells is suppressed by oxidative stress[110], so that the organ source and the mechanism responsible for the release of the PRDX4 are unknown. Moreover, sexually matured testes express a variant form of PRDX4 that is transcribed from the alternative exon 1[111]. The translated variant protein does not possess a secretory signal peptide, which results in this protein being localized in the cytosol. The variant form of PRDX4 exhibits an antioxidative function[112], but its physiological significance in testes is unknown.
Several other proteins that are present in the ER lumen also appear to participate in disulfide formation[113]. GPX7 and GPX8 exhibit sulfoxidase activity in the ER[81,114], and GPX8 is reportedly more selective for the reduction of hydrogen peroxide produced by ERO1. GPX8 appears to form a complex with Ero1α and exerts peroxidase activity that prevents the diffusion of Ero1α-derived hydrogen peroxide into and out of the rough ER[115]. Similar to PRDX4, GPX7 and GPX8 are not expressed in β cells but improves insulin secretion when overexpressed in INS-1E β-cells[116]. Vitamin K epoxide reductase (VKOR) catalyzes the reduction of vitamin K 2,3-epoxide and vitamin K to vitamin K hydroquinone, which is required for the γ-glutamyl carboxylation of coagulation factors[117]. VKOR is an ER protein consisting of four transmembrane helices, employs an electron transfer pathway similar to the bacterial homologue and preferably couples with membrane-bound thioredoxin-like redox partners[118]. The combined depletion of ERO1, PRDX4, and VKOR actually causes cell death, and VKOR alone is capable of supporting cell viability and protein secretion in the absence of ERO1 and PRDX4 activities[119]. Thus, VKOR appears to function sufficiently in oxidative protein folding and compensates for the lack of other sulfoxidases.
Some small molecules other than proteins are also reportedly involved in redox homeostasis in the ER. GSH is the most abundant non-protein thiol in cells and plays pleiotropic roles, such as in antioxidation and in the detoxification of toxicants[120]. The antioxidative functions of GSH are effectively expressed by a donation of electrons to peroxides via GPX[81]. Hence, a GSH insufficiency triggers a redox imbalance and makes cells more vulnerable to an oxidative insult, leading to cell death in severe cases. ER contains 15 mM GSH[121] which reportedly supports the secretion of proteins by its redox capacity. However, GSH depletion, which is caused by expressing an exogenous GSH-metabolizing enzyme in the ER lumen, shows no measurable effect on the induction of UPR, suggesting the existence of an alternative molecule capable of supporting redox reactions in the ER[122].
Ascorbic acid is also present abundantly in the ER and appears to support oxidative protein folding. As mentioned above, a double deficiency of ERO1 and PRDX4 causes atypical scurvy in mice due to ascorbic acid consumption and defects in collagen synthesis[103]. However, the genetic ablation of gulonolactone oxidase (GULO), which is the enzyme that catalyzes the last step of ascorbic synthesis but which is lacking in primates, shows normal collagen status in mouse skin[123]. In mice that lack aldehyde reductase encoded by AKR1A, ascorbic acid synthesis is decreased to approximately 10% of wild-type mice[124] but normal collagen synthesis in the fibrotic kidney after unilateral ureteral obstruction was observed[125]. Thus, these reports suggest the presence of other electron donors, other than ascorbic acid, to support collagen synthesis.
Non-alcoholic fatty liver disease (NAFLD) is a condition where the fat content in the liver exceeds 5%-10% by weight without a history of an excessive ingestion of alcohol[126]. A variety of metabolic dysfunctions including diabetes, dyslipidemia, and the metabolic syndrome are associated with NAFLD, which then degenerates to non-alcoholic steatohepatitis (NASH), a condition associated with inflammation and fibrotic liver damage[127].
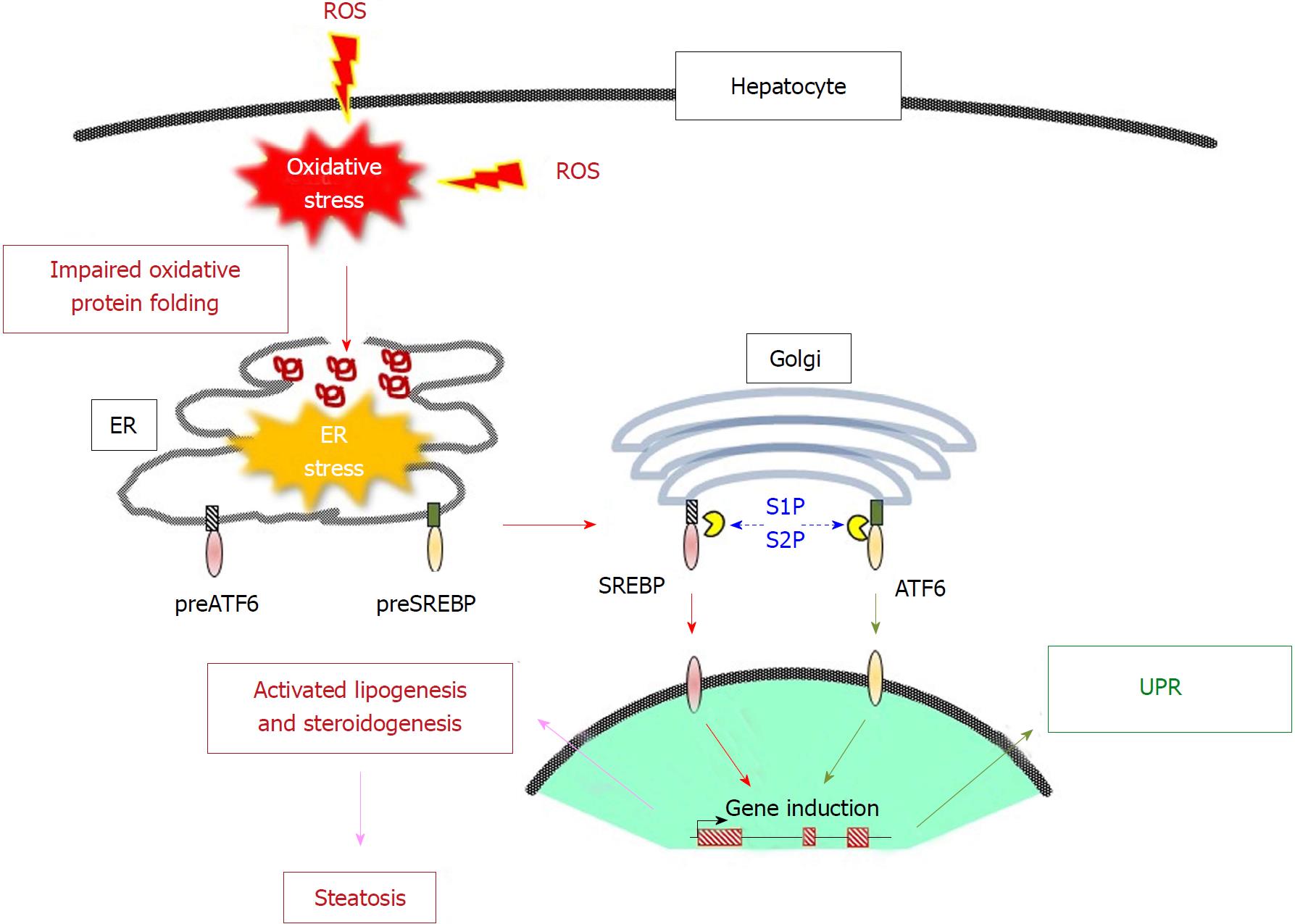
Oxidative stress is commonly regarded as “the second hit” for NASH development[128]. Signaling pathways for the ER stress-induced inflammatory process to NAFLD development has been extensively overviewed[129]. Oxidative stress triggers ER stress by stimulating the formation of misfolded proteins, and they together coordinately trigger NAFLD[130,131]. Below we briefly summarize the processes for lipid droplet accumulation triggered by these types of stress.
Sterol regulatory element-binding proteins (SREBPs) are the main transcriptional regulatory factors that control the synthesis of fatty acids and cholesterol under the control of the sterol status in the ER membrane[51]. Two SREBP genes, SREBP1 and SREBP2, are present in mammals. SREBP1 is alternatively transcribed into two forms SREBP1a and SREBP1c and is mainly involved in fatty acid synthesis. SREBP2 regulates a series of genes that are responsible for cholesterol synthesis. SREBPs are membrane proteins predominantly exposed to the cytoplasmic side and bind to another ER protein, the SREBP cleavage-activating protein (SCAP), which is located in the ER membrane[132]. Intermembrane domains on the SCAP protein bind cholesterol, which enable the formation of the SCAP/SREBP complex. Insig-1, another ER membrane protein, block the lateral movement of SCAP/SREBP into COPII-coated vesicles on ER membranes and prevent them from reaching the Golgi body[51,133]. Upon the dissociation of cholesterol due to an insufficient presence within the ER membrane, the SCAP/SREBP complex is released from Insig-1, translocated to the Golgi apparatus and then cleaved by S1P and S2P there[134]. This proteolytic activation recruits the cytoplasmic domain of the proteins to the nucleus, which then results in the induction of the expression of a series of lipogenic genes[135]. As described above, S1P and S2P are processing enzymes for the ER membrane-bound transcriptional regulatory protein ATF6 and activate it under conditions of ER stress[41]. Because the proteolytic machinery is shared, ER stress actually upregulates SREBP-1c, leading to the accumulation of lipid in hepatic cells[136]. Glycogen synthase kinase (GSK)-3 appears to be involved in signaling downstream of ER stress[137]. In the meantime HMG CoA reductase limits cholesterol synthesis, irrespective of ER stress. The protein levels of HMG CoA reductase are indirectly regulated by cholesterol via an Insig-mediated reaction; i.e., the presence of sufficient amounts of cholesterol consequently drives the polyubiquitination of HMG CoA reductase and proteolytic degradation by proteasomes[50].
The fact that ER stress induces the activation of SREBPs, in turn, suggests that SREBP-mediated lipogenesis also activated under conditions of oxidative stress. In fact, oxidative stress induces SREBP1c activation and lipid accumulation[138]. Thus, oxidative stress and ER stress interdependently stimulates the de novo synthesis and accumulation of triglycerides and cholesterol but, on another front, inhibits the secretion of lipoproteins[139]. Because cell cultures are typically performed under atmospheric oxygen, the spontaneous activation of SREBP1 and the associated expression of genes fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC), and stearoyl-CoA desaturase (SCD)1 are observed in primary hepatocytes and this is greatly enhanced in SOD1-knockout hepatocytes[140]. For the excretion of triglyceride-rich lipoproteins, appropriate oxidative folding of the apoB protein is essential[141,142]. In fact, oxidative stress appears to suppress lipoprotein secretion, which would be most likely caused via the misfolding of apoB and an impaired microsomal transfer protein function[143]. Stimulation of lipogenesis and the inhibition of lipoprotein secretion would cooperatively elevate lipid droplet accumulation, which would consequently result in the development of liver steatosis (Figure 3). This interdependent work of dealing with oxidative stress and ER stress in liver steatosis is further supported by recent observations showing the double knockout of SOD1 and PRDX4 result in aggravated liver damage compared to singly knockout mice[144].
An examination of fatty acid metabolism in NAFLD patients indicates that the inability of the liver to regulate changes in lipogenesis during the transition from the fasted to fed state is the underlying mechanism responsible for this[145]. Fasting induces the formation of lipid droplets, not only in the liver but also other tissues that are dominantly involved in active β-oxidation. While feeding a high fat diet leads to lipid droplet accumulation in the liver[146] and intestinal epithelia[143] more intensely in SOD1-knockout mice than the wild-type mice, fasting induces severe and irreversible damage not only to the liver but to other aerobic organs as well[147,148]. Thus fasting may aggravate oxidative damage in these organs and become a serious pathogenic factor for NAFLD.
These observations raise the next query, i.e., why is lipogenesis elevated as a consequence of UPR in cases of ER stress if lipid accumulation is unfavorable for the liver? We have a clue to this, in that feeding a lard-containing high-calorie diet increases the accumulation of lipid droplets but improves the longevity of the SOD1-deficient mice compared to mice fed a normal diet[149]. This is unexpected because lipid accumulation is generally recognized to be an exacerbating factor for liver function. Based on these observations we hypothesized that lipid droplets that accumulate in response to oxidative stress may have a protective role against the hepatotoxic effects of ROS. Experimental data actually suggest that lipids that transiently accumulate in the liver have a protective function against oxidative injury caused by a liver toxicant thioacetamide in mice and by hydrogen peroxide in cultivated cells[150]. Thus, the accumulation of lipids under ER stress conditions may also be regarded as an adoptive response of hepatocytes to oxidative stress conditions[151], although liver steatosis at a more advanced stage is hazardous and should be treated appropriately.
Multiple processes appear to underlie the pathogenesis of NAFLD, so that a variety of agents and treatment may be applicable for therapeutic purposes. Antioxidants directly eliminate ROS and result in the suppression of oxidative stress and consequently ER stress. Hydrophobic antioxidants, such as vitamin E[68] and coenzyme Q10[152], and their derivatives and hydrophilic antioxidants, such as vitamin C (ascorbic acid) and N-acetylcysteine, a precursor for cysteine and GSH[153], may be promising agents for use in ameliorating the effects of NAFLD. In fact, vitamin E and polyphenol have been reported to be useful in the treatment of NAFLD patients, while the issue of whether vitamin C is beneficial is ambiguous at this moment[154,155].
While antioxidants decrease the levels of pathogenic ROS directly, it would be helpful if cellular capacity to resist malfunctions in lipid metabolism could be additionally enhanced. In this sense, peroxisome proliferator-activated receptors (PPARs) and their binding partner retinoid X-receptors (RXR), appear to have roles in maintaining lipid homeostasis and, hence, represent promising targets for ameliorating the pathogenesis of NAFLD[126]. PPAR family proteins of which PPAR-α, PPAR-β/δ, PPAR-γ are members, can be either activated or inhibited by lipid metabolites and other lipophilic agents in vivo. Regarding the distribution of PPARs in tissues, the highest expression of PPAR-α is observed in brown adipose tissue, liver, kidney, and heart[156]. The expression of PPAR-γ is higher in adipose tissues than other tissues. Compared to these isoforms, the expression of the PPAR-β/δ isoform is rather ubiquitous. Mice in which PPAR-α is knocked out are viable and fertile and show no detectable gross phenotypic defects[157]. Contrary to PPAR-α knockout mice, a PPAR-γ deficiency showed impaired placental vascularization, leading to embryonic death by embryonic day E10.0[158]. Similarly, the genetic ablation of PPAR-β/δ also impairs the placenta and leads to embryonic death at E9.5 - E10.5[159].
The roles of PPAR-β/δ are less established compared to those of PPAR-α and PPAR-γ. In 2003 it was reported that PPAR-δ contributes to fat metabolism[160,161], which then attracted more interest in PPAR-δ than before. For example, PPAR-δ activation enhances fatty acid oxidation and rescues ER stress in pancreatic β-cells[162]. The PPAR-α/δ agonist GFT505 prevents high fat diet-induced liver steatosis and protects the liver from inflammatory reactions in mice[163]. The activation of PPARβ/δ by GW501516 also prevents the inflammation associated with ER stress in skeletal muscle cells and ameliorates insulin resistance in mice through an adenosine monophosphate-activated protein kinase (AMPK)-dependent mechanism[164]. The PPAR-β/δ agonist GW0742 also reportedly attenuates ER stress by improving hepatic energy metabolism in the livers of high fat diet-administered mice[165]. The effectiveness of agonists for PPAR-α/δ has been confirmed in NASH patients as well[166]. Thus, the application of PPAR agonists, notably those for PPAR-β/δ isoforms, appears to be promising therapeutics for the treatment of NAFLD[167]. While the molecular mechanisms for how PPAR-β/δ controls insulin signaling are largely unknown, a stable interaction of PPAR-β/δ with nuclear T-cell protein tyrosine phosphatase 45 (TCPTP45) isoform has been demonstrated as the most upstream component that resolves the downregulation of insulin signaling[168]. Forthcoming experiments directed at elucidating the pathway in more detail would provide a clearer vision on this issue.
As described above, the activation of PPARβ/δ is a largely promising area of therapeutics for fatty live diseases in animal experiments[169]. However, it should be remembered that the most potent and specific activator for PPAR-β/δ, GW501516, also accelerates intestinal adenocarcinoma in Apcmin mice that are predisposed to developing intestinal polyposis[170]. The activation of PPAR-β/δ by GW501516 is also associated with the induction of cancer development, while the PPAR-γ agonist GW7845 causes a moderate delay in tumor formation[171]. At the present stage, the activation of PPAR-β/δ does not always lead to consistent results; i.e., it is either pro-carcinogenic or anti-carcinogenic, depending on carcinogenic model and the animals being used[172,173]. Therefore the development of activating agents that exert a beneficial action on PPAR-β/δ, but do not have other side effects, is awaited for using synthetic activators for therapeutic purposes in the treatment of fatty liver diseases.
Oxidative stress and ER stress are frequently linked and become pathogenic in some diseases such as NAFLD[174]. Antioxidants suppress oxidative stress directly and also subsequent ER stress and, hence, can mitigate the causal factors for NAFLD. In the meantime, the activation of PPARs renders the liver more resistant against stress caused by lipotoxicity. Coordinated treatment with these agents may exert therapeutic effects more efficiently in NAFLD as well as in other diseases that are caused by aberrant lipid metabolism.
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Demonacos C, Moneim AA, Wang K S- Editor: Ji FF L- Editor: A E- Editor: Yin SY
| 1. | Price BD. Signalling across the endoplasmic reticulum membrane: potential mechanisms. Cell Signal. 1992;4:465-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 2. | Little E, Ramakrishnan M, Roy B, Gazit G, Lee AS. The glucose-regulated proteins (GRP78 and GRP94): functions, gene regulation, and applications. Crit Rev Eukaryot Gene Expr. 1994;4:1-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 309] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 3. | Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4586] [Cited by in RCA: 5058] [Article Influence: 281.0] [Reference Citation Analysis (0)] |
| 4. | Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2260] [Cited by in RCA: 2366] [Article Influence: 118.3] [Reference Citation Analysis (0)] |
| 5. | Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146:743-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 309] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 6. | Nagata K. Expression and function of heat shock protein 47: a collagen-specific molecular chaperone in the endoplasmic reticulum. Matrix Biol. 1998;16:379-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 135] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 780] [Cited by in RCA: 809] [Article Influence: 33.7] [Reference Citation Analysis (1)] |
| 8. | Meyer T, Wirtz PH. Mechanisms of Mitochondrial Redox Signaling in Psychosocial Stress-Responsive Systems: New Insights into an Old Story. Antioxid Redox Signal. 2018;28:760-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 9. | Csala M, Margittai E, Bánhegyi G. Redox control of endoplasmic reticulum function. Antioxid Redox Signal. 2010;13:77-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 10. | Araki K, Inaba K. Structure, mechanism, and evolution of Ero1 family enzymes. Antioxid Redox Signal. 2012;16:790-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 343] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 12. | Tu BP, Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 336] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 13. | Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341-346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 763] [Cited by in RCA: 813] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 14. | Inaba K, Masui S, Iida H, Vavassori S, Sitia R, Suzuki M. Crystal structures of human Ero1α reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J. 2010;29:3330-3343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 15. | Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641-3651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 621] [Cited by in RCA: 600] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 16. | Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066-3077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1396] [Cited by in RCA: 1597] [Article Influence: 79.9] [Reference Citation Analysis (0)] |
| 17. | Laurindo FR, Pescatore LA, Fernandes Dde C. Protein disulfide isomerase in redox cell signaling and homeostasis. Free Radic Biol Med. 2012;52:1954-1969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 185] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 18. | Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 392] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 19. | Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 362] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 20. | Chin KT, Kang G, Qu J, Gardner LB, Coetzee WA, Zito E, Fishman GI, Ron D. The sarcoplasmic reticulum luminal thiol oxidase ERO1 regulates cardiomyocyte excitation-coupled calcium release and response to hemodynamic load. FASEB J. 2011;25:2583-2591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Kodali VK, Thorpe C. Oxidative protein folding and the Quiescin-sulfhydryl oxidase family of flavoproteins. Antioxid Redox Signal. 2010;13:1217-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 22. | Alon A, Grossman I, Gat Y, Kodali VK, DiMaio F, Mehlman T, Haran G, Baker D, Thorpe C, Fass D. The dynamic disulphide relay of quiescin sulphydryl oxidase. Nature. 2012;488:414-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 23. | Delaunay-Moisan A, Appenzeller-Herzog C. The antioxidant machinery of the endoplasmic reticulum: Protection and signaling. Free Radic Biol Med. 2015;83:341-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 24. | Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51 Suppl 3:S455-S461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 348] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 25. | MacLennan DH, Abu-Abed M, Kang C. Structure-function relationships in Ca(2+) cycling proteins. J Mol Cell Cardiol. 2002;34:897-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Paschen W. Dependence of vital cell function on endoplasmic reticulum calcium levels: implications for the mechanisms underlying neuronal cell injury in different pathological states. Cell Calcium. 2001;29:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 123] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 27. | Ruddock LW, Molinari M. N-glycan processing in ER quality control. J Cell Sci. 2006;119:4373-4380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 225] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 28. | Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1205] [Cited by in RCA: 1405] [Article Influence: 93.7] [Reference Citation Analysis (0)] |
| 29. | Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3842] [Cited by in RCA: 4539] [Article Influence: 349.2] [Reference Citation Analysis (0)] |
| 30. | Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2430] [Cited by in RCA: 2559] [Article Influence: 98.4] [Reference Citation Analysis (0)] |
| 31. | Zyryanova AF, Weis F, Faille A, Alard AA, Crespillo-Casado A, Sekine Y, Harding HP, Allen F, Parts L, Fromont C. Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science. 2018;359:1533-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 32. | Nikawa J, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1441-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 151] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 970] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 34. | Abdullah A, Ravanan P. The unknown face of IRE1α - Beyond ER stress. Eur J Cell Biol. 2018;97:359-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 35. | Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434:181-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 429] [Article Influence: 30.6] [Reference Citation Analysis (1)] |
| 36. | Soares Moretti AI, Martins Laurindo FR. Protein disulfide isomerases: Redox connections in and out of the endoplasmic reticulum. Arch Biochem Biophys. 2017;617:106-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 93] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 37. | Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957-4966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 141] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787-3799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1484] [Cited by in RCA: 1598] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 39. | Chen X, Shen J, Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045-13052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 371] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 40. | Okada T, Haze K, Nadanaka S, Yoshida H, Seidah NG, Hirano Y, Sato R, Negishi M, Mori K. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J Biol Chem. 2003;278:31024-31032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 176] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 41. | Kohno K. How transmembrane proteins sense endoplasmic reticulum stress. Antioxid Redox Signal. 2007;9:2295-2303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 72] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 42. | Saito A, Hino S, Murakami T, Kondo S, Imaizumi K. A novel ER stress transducer, OASIS, expressed in astrocytes. Antioxid Redox Signal. 2007;9:563-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Wu X, Rapoport TA. Mechanistic insights into ER-associated protein degradation. Curr Opin Cell Biol. 2018;53:22-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 264] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 44. | Goldberger RF, Epstein CJ, Anfinsen CB. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem. 1963;238:628-635. [PubMed] |
| 45. | Okumura M, Kadokura H, Inaba K. Structures and functions of protein disulfide isomerase family members involved in proteostasis in the endoplasmic reticulum. Free Radic Biol Med. 2015;83:314-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 46. | Yu H, Matouschek A. Recognition of Client Proteins by the Proteasome. Annu Rev Biophys. 2017;46:149-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 47. | Baldridge RD, Rapoport TA. Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell. 2016;166:394-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 48. | Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, Baker D, Chambers MG, Su H, Li D. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature. 2017;548:352-355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 154] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 49. | Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16:77-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 50. | Jo Y, Debose-Boyd RA. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev Biochem Mol Biol. 2010;45:185-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 51. | Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1162] [Cited by in RCA: 1293] [Article Influence: 68.1] [Reference Citation Analysis (0)] |
| 52. | Giguère P, Turcotte ME, Hamelin E, Parent A, Brisson J, Laroche G, Labrecque P, Dupuis G, Parent JL. Peroxiredoxin-4 interacts with and regulates the thromboxane A(2) receptor. FEBS Lett. 2007;581:3863-3868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 53. | Palande K, Roovers O, Gits J, Verwijmeren C, Iuchi Y, Fujii J, Neel BG, Karisch R, Tavernier J, Touw IP. Peroxiredoxin-controlled G-CSF signalling at the endoplasmic reticulum-early endosome interface. J Cell Sci. 2011;124:3695-3705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 54. | Lee YS, Lee DH, Choudry HA, Bartlett DL, Lee YJ. Ferroptosis-Induced Endoplasmic Reticulum Stress: Cross-talk between Ferroptosis and Apoptosis. Mol Cancer Res. 2018;16:1073-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 286] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 55. | Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 861] [Cited by in RCA: 937] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 56. | Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR, Tabas I. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol. 2009;186:783-792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 403] [Cited by in RCA: 496] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 57. | Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2001] [Cited by in RCA: 2306] [Article Influence: 109.8] [Reference Citation Analysis (0)] |
| 58. | Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta. 2014;1841:595-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 476] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 59. | Anelli T, Bergamelli L, Margittai E, Rimessi A, Fagioli C, Malgaroli A, Pinton P, Ripamonti M, Rizzuto R, Sitia R. Ero1α regulates Ca(2+) fluxes at the endoplasmic reticulum-mitochondria interface (MAM). Antioxid Redox Signal. 2012;16:1077-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 60. | Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, Wada I, Akira S, Araki E, Mori M. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci USA. 2001;98:10845-10850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 497] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 61. | Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 418] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 62. | McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1430] [Cited by in RCA: 1536] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 63. | Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460-3470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1214] [Cited by in RCA: 1540] [Article Influence: 128.3] [Reference Citation Analysis (1)] |
| 64. | Yang Y, Liu L, Naik I, Braunstein Z, Zhong J, Ren B. Transcription Factor C/EBP Homologous Protein in Health and Diseases. Front Immunol. 2017;8:1612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 154] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 65. | Boyer F, Vidot JB, Dubourg AG, Rondeau P, Essop MF, Bourdon E. Oxidative stress and adipocyte biology: focus on the role of AGEs. Oxid Med Cell Longev. 2015;2015:534873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 66. | He L, He T, Farrar S, Ji L, Liu T, Ma X. Antioxidants Maintain Cellular Redox Homeostasis by Elimination of Reactive Oxygen Species. Cell Physiol Biochem. 2017;44:532-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 1246] [Article Influence: 155.8] [Reference Citation Analysis (0)] |
| 67. | Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1524] [Cited by in RCA: 1704] [Article Influence: 121.7] [Reference Citation Analysis (0)] |
| 68. | Niki E. Lipid peroxidation: physiological levels and dual biological effects. Free Radic Biol Med. 2009;47:469-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 684] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 69. | Toyokuni S, Ito F, Yamashita K, Okazaki Y, Akatsuka S. Iron and thiol redox signaling in cancer: An exquisite balance to escape ferroptosis. Free Radic Biol Med. 2017;108:610-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 70. | Svegliati S, Spadoni T, Moroncini G, Gabrielli A. NADPH oxidase, oxidative stress and fibrosis in systemic sclerosis. Free Radic Biol Med. 2018;125:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 71. | Laurindo FR, Araujo TL, Abrahão TB. Nox NADPH oxidases and the endoplasmic reticulum. Antioxid Redox Signal. 2014;20:2755-2775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 72. | Hrycay EG, Bandiera SM. Monooxygenase, peroxidase and peroxygenase properties and reaction mechanisms of cytochrome P450 enzymes. Adv Exp Med Biol. 2015;851:1-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 73. | Zeeshan HM, Lee GH, Kim HR, Chae HJ. Endoplasmic Reticulum Stress and Associated ROS. Int J Mol Sci. 2016;17:327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 433] [Cited by in RCA: 671] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 74. | Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783:549-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 230] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 75. | Tavender TJ, Bulleid NJ. Molecular mechanisms regulating oxidative activity of the Ero1 family in the endoplasmic reticulum. Antioxid Redox Signal. 2010;13:1177-1187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 76. | Enyedi B, Várnai P, Geiszt M. Redox state of the endoplasmic reticulum is controlled by Ero1L-alpha and intraluminal calcium. Antioxid Redox Signal. 2010;13:721-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 77. | Zito E. ERO1: A protein disulfide oxidase and H2O2 producer. Free Radic Biol Med. 2015;83:299-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 123] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 78. | Oyadomari S, Araki E, Mori M. Endoplasmic reticulum stress-mediated apoptosis in pancreatic beta-cells. Apoptosis. 2002;7:335-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 405] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 79. | Konno T, Pinho Melo E, Lopes C, Mehmeti I, Lenzen S, Ron D, Avezov E. ERO1-independent production of H2O2 within the endoplasmic reticulum fuels Prdx4-mediated oxidative protein folding. J Cell Biol. 2015;211:253-259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 80. | Fujii J, Ikeda Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002;7:123-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 304] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 81. | Brigelius-Flohé R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830:3289-3303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1030] [Cited by in RCA: 1538] [Article Influence: 118.3] [Reference Citation Analysis (0)] |
| 82. | Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2353] [Cited by in RCA: 2190] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 83. | Johnson RM, Goyette G Jr, Ravindranath Y, Ho YS. Hemoglobin autoxidation and regulation of endogenous H2O2 levels in erythrocytes. Free Radic Biol Med. 2005;39:1407-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 141] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 84. | Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4739] [Cited by in RCA: 4844] [Article Influence: 151.4] [Reference Citation Analysis (0)] |
| 85. | Kaur SJ, McKeown SR, Rashid S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene. 2016;577:109-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 238] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 86. | Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, Wilcox HM, Flood DG, Beal MF, Brown RH Jr, Scott RW, Snider WD. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 920] [Cited by in RCA: 879] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 87. | Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, Fukutomi H, Noguchi T, Matsuzawa A, Takeda K, Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451-1464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 407] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 88. | Fujisawa T, Homma K, Yamaguchi N, Kadowaki H, Tsuburaya N, Naguro I, Matsuzawa A, Takeda K, Takahashi Y, Goto J. A novel monoclonal antibody reveals a conformational alteration shared by amyotrophic lateral sclerosis-linked SOD1 mutants. Ann Neurol. 2012;72:739-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 89. | Homma K, Fujisawa T, Tsuburaya N, Yamaguchi N, Kadowaki H, Takeda K, Nishitoh H, Matsuzawa A, Naguro I, Ichijo H. SOD1 as a molecular switch for initiating the homeostatic ER stress response under zinc deficiency. Mol Cell. 2013;52:75-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 90. | Atkin JD, Farg MA, Soo KY, Walker AK, Halloran M, Turner BJ, Nagley P, Horne MK. Mutant SOD1 inhibits ER-Golgi transport in amyotrophic lateral sclerosis. J Neurochem. 2014;129:190-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 91. | Matsumoto A, Okado A, Fujii T, Fujii J, Egashira M, Niikawa N, Taniguchi N. Cloning of the peroxiredoxin gene family in rats and characterization of the fourth member. FEBS Lett. 1999;443:246-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 115] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 92. | Rhee SG, Woo HA, Kil IS, Bae SH. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol Chem. 2012;287:4403-4410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 440] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 93. | Fujii J, Ikeda Y, Kurahashi T, Homma T. Physiological and pathological views of peroxiredoxin 4. Free Radic Biol Med. 2015;83:373-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 94. | Ikeda Y, Nakano M, Ihara H, Ito R, Taniguchi N, Fujii J. Different consequences of reactions with hydrogen peroxide and t-butyl hydroperoxide in the hyperoxidative inactivation of rat peroxiredoxin-4. J Biochem. 2011;149:443-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 95. | Zito E, Melo EP, Yang Y, Wahlander Å, Neubert TA, Ron D. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol Cell. 2010;40:787-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 261] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 96. | Tavender TJ, Springate JJ, Bulleid NJ. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 2010;29:4185-4197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 97. | Zito E. PRDX4, an endoplasmic reticulum-localized peroxiredoxin at the crossroads between enzymatic oxidative protein folding and nonenzymatic protein oxidation. Antioxid Redox Signal. 2013;18:1666-1674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 98. | Sato Y, Kojima R, Okumura M, Hagiwara M, Masui S, Maegawa K, Saiki M, Horibe T, Suzuki M, Inaba K. Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci Rep. 2013;3:2456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 99. | Tavender TJ, Bulleid NJ. Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J Cell Sci. 2010;123:2672-2679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 100. | Yan Y, Wladyka C, Fujii J, Sockanathan S. Prdx4 is a compartment-specific H2O2 sensor that regulates neurogenesis by controlling surface expression of GDE2. Nat Commun. 2015;6:7006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 101. | Iuchi Y, Okada F, Tsunoda S, Kibe N, Shirasawa N, Ikawa M, Okabe M, Ikeda Y, Fujii J. Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem J. 2009;419:149-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 102. | Bertolotti M, Yim SH, Garcia-Manteiga JM, Masciarelli S, Kim YJ, Kang MH, Iuchi Y, Fujii J, Vené R, Rubartelli A. B- to plasma-cell terminal differentiation entails oxidative stress and profound reshaping of the antioxidant responses. Antioxid Redox Signal. 2010;13:1133-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 103. | Zito E, Hansen HG, Yeo GS, Fujii J, Ron D. Endoplasmic reticulum thiol oxidase deficiency leads to ascorbic acid depletion and noncanonical scurvy in mice. Mol Cell. 2012;48:39-51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 104. | Okado-Matsumoto A, Matsumoto A, Fujii J, Taniguchi N. Peroxiredoxin IV is a secretable protein with heparin-binding properties under reduced conditions. J Biochem. 2000;127:493-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 149] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 105. | Ding Y, Yamada S, Wang KY, Shimajiri S, Guo X, Tanimoto A, Murata Y, Kitajima S, Watanabe T, Izumi H. Overexpression of peroxiredoxin 4 protects against high-dose streptozotocin-induced diabetes by suppressing oxidative stress and cytokines in transgenic mice. Antioxid Redox Signal. 2010;13:1477-1490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 106. | Nabeshima A, Yamada S, Guo X, Tanimoto A, Wang KY, Shimajiri S, Kimura S, Tasaki T, Noguchi H, Kitada S. Peroxiredoxin 4 protects against nonalcoholic steatohepatitis and type 2 diabetes in a nongenetic mouse model. Antioxid Redox Signal. 2013;19:1983-1998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 107. | Mehmeti I, Lortz S, Elsner M, Lenzen S. Peroxiredoxin 4 improves insulin biosynthesis and glucose-induced insulin secretion in insulin-secreting INS-1E cells. J Biol Chem. 2014;289:26904-26913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 108. | Guo X, Yamada S, Tanimoto A, Ding Y, Wang KY, Shimajiri S, Murata Y, Kimura S, Tasaki T, Nabeshima A. Overexpression of peroxiredoxin 4 attenuates atherosclerosis in apolipoprotein E knockout mice. Antioxid Redox Signal. 2012;17:1362-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 109. | Schulte J. Peroxiredoxin 4: a multifunctional biomarker worthy of further exploration. BMC Med. 2011;9:137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 110. | Ito R, Takahashi M, Ihara H, Tsukamoto H, Fujii J, Ikeda Y. Measurement of peroxiredoxin-4 serum levels in rat tissue and its use as a potential marker for hepatic disease. Mol Med Rep. 2012;6:379-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 111. | Yim SH, Kim YJ, Oh SY, Fujii J, Zhang Y, Gladyshev VN, Rhee SG. Identification and characterization of alternatively transcribed form of peroxiredoxin IV gene that is specifically expressed in spermatids of postpubertal mouse testis. J Biol Chem. 2011;286:39002-39012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 112. | Tasaki E, Matsumoto S, Tada H, Kurahashi T, Zhang X, Fujii J, Utsumi T, Iuchi Y. Protective role of testis-specific peroxiredoxin 4 against cellular oxidative stress. J Clin Biochem Nutr. 2017;60:156-161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 113. | Bulleid NJ, Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 179] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 114. | Ramming T, Okumura M, Kanemura S, Baday S, Birk J, Moes S, Spiess M, Jenö P, Bernèche S, Inaba K. A PDI-catalyzed thiol-disulfide switch regulates the production of hydrogen peroxide by human Ero1. Free Radic Biol Med. 2015;83:361-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 115. | Ramming T, Hansen HG, Nagata K, Ellgaard L, Appenzeller-Herzog C. GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic Biol Med. 2014;70:106-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 116. | Mehmeti I, Lortz S, Avezov E, Jörns A, Lenzen S. ER-resident antioxidative GPx7 and GPx8 enzyme isoforms protect insulin-secreting INS-1E β-cells against lipotoxicity by improving the ER antioxidative capacity. Free Radic Biol Med. 2017;112:121-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 117. | Tie JK, Stafford DW. Structure and function of vitamin K epoxide reductase. Vitam Horm. 2008;78:103-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 118. | Schulman S, Wang B, Li W, Rapoport TA. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci USA. 2010;107:15027-15032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 119. | Rutkevich LA, Williams DB. Vitamin K epoxide reductase contributes to protein disulfide formation and redox homeostasis within the endoplasmic reticulum. Mol Biol Cell. 2012;23:2017-2027. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 120. | Meister A. Glutathione biosynthesis and its inhibition. Methods Enzymol. 1995;252:26-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 123] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 121. | Montero D, Tachibana C, Rahr Winther J, Appenzeller-Herzog C. Intracellular glutathione pools are heterogeneously concentrated. Redox Biol. 2013;1:508-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 122. | Tsunoda S, Avezov E, Zyryanova A, Konno T, Mendes-Silva L, Pinho Melo E, Harding HP, Ron D. Intact protein folding in the glutathione-depleted endoplasmic reticulum implicates alternative protein thiol reductants. Elife. 2014;3:e03421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 123. | Parsons KK, Maeda N, Yamauchi M, Banes AJ, Koller BH. Ascorbic acid-independent synthesis of collagen in mice. Am J Physiol Endocrinol Metab. 2006;290:E1131-E1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 124. | Takahashi M, Miyata S, Fujii J, Inai Y, Ueyama S, Araki M, Soga T, Fujinawa R, Nishitani C, Ariki S. In vivo role of aldehyde reductase. Biochim Biophys Acta. 2012;1820:1787-1796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 125. | Nishida H, Kurahashi T, Saito Y, Otsuki N, Kwon M, Ohtake H, Yamakawa M, Yamada KI, Miyata S, Tomita Y. Kidney fibrosis is independent of the amount of ascorbic acid in mice with unilateral ureteral obstruction. Free Radic Res. 2014;48:1115-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 126. | Serviddio G, Bellanti F, Vendemiale G. Free radical biology for medicine: learning from nonalcoholic fatty liver disease. Free Radic Biol Med. 2013;65:952-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 127. | Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 1318] [Article Influence: 109.8] [Reference Citation Analysis (0)] |
| 128. | Sumida Y, Niki E, Naito Y, Yoshikawa T. Involvement of free radicals and oxidative stress in NAFLD/NASH. Free Radic Res. 2013;47:869-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 227] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 129. | Lebeaupin C, Vallée D, Hazari Y, Hetz C, Chevet E, Bailly-Maitre B. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. J Hepatol. 2018;69:927-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 659] [Article Influence: 94.1] [Reference Citation Analysis (0)] |
| 130. | Gentile CL, Frye M, Pagliassotti MJ. Endoplasmic reticulum stress and the unfolded protein response in nonalcoholic fatty liver disease. Antioxid Redox Signal. 2011;15:505-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 131. | Ashraf NU, Sheikh TA. Endoplasmic reticulum stress and Oxidative stress in the pathogenesis of Non-alcoholic fatty liver disease. Free Radic Res. 2015;49:1405-1418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 235] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 132. | Brown MS, Radhakrishnan A, Goldstein JL. Retrospective on Cholesterol Homeostasis: The Central Role of Scap. Annu Rev Biochem. 2018;87:783-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 360] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 133. | Sun LP, Li L, Goldstein JL, Brown MS. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J Biol Chem. 2005;280:26483-26490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 182] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 134. | Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355-1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1332] [Cited by in RCA: 1426] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 135. | Goldstein JL, Rawson RB, Brown MS. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch Biochem Biophys. 2002;397:139-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 196] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 136. | Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 561] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 137. | Kim AJ, Shi Y, Austin RC, Werstuck GH. Valproate protects cells from ER stress-induced lipid accumulation and apoptosis by inhibiting glycogen synthase kinase-3. J Cell Sci. 2005;118:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 191] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 138. | Sekiya M, Hiraishi A, Touyama M, Sakamoto K. Oxidative stress induced lipid accumulation via SREBP1c activation in HepG2 cells. Biochem Biophys Res Commun. 2008;375:602-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 140] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 139. | Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal. 2007;9:2277-2293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1149] [Cited by in RCA: 1254] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 140. | Lee J, Homma T, Kurahashi T, Kang ES, Fujii J. Oxidative stress triggers lipid droplet accumulation in primary cultured hepatocytes by activating fatty acid synthesis. Biochem Biophys Res Commun. 2015;464:229-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 141. | Huang XF, Shelness GS. Identification of cysteine pairs within the amino-terminal 5% of apolipoprotein B essential for hepatic lipoprotein assembly and secretion. J Biol Chem. 1997;272:31872-31876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 142. | Tran K, Borén J, Macri J, Wang Y, McLeod R, Avramoglu RK, Adeli K, Yao Z. Functional analysis of disulfide linkages clustered within the amino terminus of human apolipoprotein B. J Biol Chem. 1998;273:7244-7251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 143. | Kurahashi T, Konno T, Otsuki N, Kwon M, Tsunoda S, Ito J, Fujii J. A malfunction in triglyceride transfer from the intracellular lipid pool to apoB in enterocytes of SOD1-deficient mice. FEBS Lett. 2012;586:4289-4295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 144. | Homma T, Kurahashi T, Lee J, Nabeshima A, Yamada S, Fujii J. Double Knockout of Peroxiredoxin 4 (Prdx4) and Superoxide Dismutase 1 (Sod1) in Mice Results in Severe Liver Failure. Oxid Med Cell Longev. 2018;2018:2812904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 145. | Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115:1343-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2112] [Cited by in RCA: 2600] [Article Influence: 130.0] [Reference Citation Analysis (0)] |
| 146. | Uchiyama S, Shimizu T, Shirasawa T. CuZn-SOD deficiency causes ApoB degradation and induces hepatic lipid accumulation by impaired lipoprotein secretion in mice. J Biol Chem. 2006;281:31713-31719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 147. | Kurahashi T, Hamashima S, Shirato T, Lee J, Homma T, Kang ES, Fujii J. An SOD1 deficiency enhances lipid droplet accumulation in the fasted mouse liver by aborting lipophagy. Biochem Biophys Res Commun. 2015;467:866-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 148. | Lee J, Homma T, Kobayashi S, Ishii N, Fujii J. Unveiling systemic organ disorders associated with impaired lipid catabolism in fasted SOD1-deficient mice. Arch Biochem Biophys. 2018;654:163-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 149. | Ito J, Ishii N, Akihara R, Lee J, Kurahashi T, Homma T, Kawasaki R, Fujii J. A high-fat diet temporarily renders Sod1-deficient mice resistant to an oxidative insult. J Nutr Biochem. 2017;40:44-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 150. | Lee J, Homma T, Fujii J. Mice in the early stage of liver steatosis caused by a high fat diet are resistant to thioacetamide-induced hepatotoxicity and oxidative stress. Toxicol Lett. 2017;277:92-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 151. | Welte MA, Gould AP. Lipid droplet functions beyond energy storage. Biochim Biophys Acta. 2017;1862:1260-1272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 374] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 152. | Yamamoto Y. Coenzyme Q10 redox balance and a free radical scavenger drug. Arch Biochem Biophys. 2016;595:132-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 153. | Homma T, Fujii J. Application of Glutathione as Anti-Oxidative and Anti-Aging Drugs. Curr Drug Metab. 2015;16:560-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (1)] |
| 154. | Botham KM, Napolitano M, Bravo E. The Emerging Role of Disturbed CoQ Metabolism in Nonalcoholic Fatty Liver Disease Development and Progression. Nutrients. 2015;7:9834-9846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 155. | Del Ben M, Polimeni L, Baratta F, Pastori D, Angelico F. The role of nutraceuticals for the treatment of non-alcoholic fatty liver disease. Br J Clin Pharmacol. 2017;83:88-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 156. | Silva AKS, Peixoto CA. Role of peroxisome proliferator-activated receptors in non-alcoholic fatty liver disease inflammation. Cell Mol Life Sci. 2018;75:2951-2961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 157. | Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012-3022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1309] [Cited by in RCA: 1287] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 158. | Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1490] [Cited by in RCA: 1511] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 159. | Nadra K, Anghel SI, Joye E, Tan NS, Basu-Modak S, Trono D, Wahli W, Desvergne B. Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor beta/delta. Mol Cell Biol. 2006;26:3266-3281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 160] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 160. | Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci USA. 2003;100:15924-15929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 669] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 161. | Wang YX, Lee CH, Tiep S, Yu RT, Ham J, Kang H, Evans RM. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113:159-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1029] [Cited by in RCA: 1018] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 162. | Cao M, Tong Y, Lv Q, Chen X, Long Y, Jiang L, Wan J, Zhang Y, Zhang F, Tong N. PPARδ Activation Rescues Pancreatic β-Cell Line INS-1E from Palmitate-Induced Endoplasmic Reticulum Stress through Enhanced Fatty Acid Oxidation. PPAR Res. 2012;2012:680684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 163. | Staels B, Rubenstrunk A, Noel B, Rigou G, Delataille P, Millatt LJ, Baron M, Lucas A, Tailleux A, Hum DW. Hepatoprotective effects of the dual peroxisome proliferator-activated receptor alpha/delta agonist, GFT505, in rodent models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology. 2013;58:1941-1952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 344] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 164. | Salvadó L, Barroso E, Gómez-Foix AM, Palomer X, Michalik L, Wahli W, Vázquez-Carrera M. PPARβ/δ prevents endoplasmic reticulum stress-associated inflammation and insulin resistance in skeletal muscle cells through an AMPK-dependent mechanism. Diabetologia. 2014;57:2126-2135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 165. | Silva-Veiga FM, Rachid TL, de Oliveira L, Graus-Nunes F, Mandarim-de-Lacerda CA, Souza-Mello V. GW0742 (PPAR-beta agonist) attenuates hepatic endoplasmic reticulum stress by improving hepatic energy metabolism in high-fat diet fed mice. Mol Cell Endocrinol. 2018;474:227-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 166. | Ratziu V, Harrison SA, Francque S, Bedossa P, Lehert P, Serfaty L, Romero-Gomez M, Boursier J, Abdelmalek M, Caldwell S. Elafibranor, an Agonist of the Peroxisome Proliferator-Activated Receptor-α and -δ, Induces Resolution of Nonalcoholic Steatohepatitis Without Fibrosis Worsening. Gastroenterology. 2016;150:1147-1159.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 845] [Cited by in RCA: 820] [Article Influence: 91.1] [Reference Citation Analysis (0)] |
| 167. | Chen J, Montagner A, Tan NS, Wahli W. Insights into the Role of PPARβ/δ in NAFLD. Int J Mol Sci. 2018;19:pii: E1893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 168. | Yoo T, Ham SA, Lee WJ, Hwang SI, Park JA, Hwang JS, Hur J, Shin HC, Han SG, Lee CH. Ligand-Dependent Interaction of PPARδ With T-Cell Protein Tyrosine Phosphatase 45 Enhances Insulin Signaling. Diabetes. 2018;67:360-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 169. | Palomer X, Barroso E, Pizarro-Delgado J, Peña L, Botteri G, Zarei M, Aguilar D, Montori-Grau M, Vázquez-Carrera M. PPARβ/δ: A Key Therapeutic Target in Metabolic Disorders. Int J Mol Sci. 2018;19:pii: E913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 170. | Gupta RA, Wang D, Katkuri S, Wang H, Dey SK, DuBois RN. Activation of nuclear hormone receptor peroxisome proliferator-activated receptor-delta accelerates intestinal adenoma growth. Nat Med. 2004;10:245-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 220] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 171. | Yin Y, Russell RG, Dettin LE, Bai R, Wei ZL, Kozikowski AP, Kopelovich L, Glazer RI. Peroxisome proliferator-activated receptor delta and gamma agonists differentially alter tumor differentiation and progression during mammary carcinogenesis. Cancer Res. 2005;65:3950-3957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 172. | Peters JM, Foreman JE, Gonzalez FJ. Dissecting the role of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in colon, breast, and lung carcinogenesis. Cancer Metastasis Rev. 2011;30:619-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 173. | Peters JM, Gonzalez FJ, Müller R. Establishing the Role of PPARβ/δ in Carcinogenesis. Trends Endocrinol Metab. 2015;26:595-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 174. | Cao SS, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal. 2014;21:396-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 768] [Cited by in RCA: 1008] [Article Influence: 91.6] [Reference Citation Analysis (0)] |