Published online Sep 27, 2020. doi: 10.4331/wjbc.v11.i2.62
Peer-review started: May 13, 2020
First decision: June 15, 2020
Revised: June 30, 2020
Accepted: September 10, 2020
Article in press: September 10, 2020
Published online: September 27, 2020
Processing time: 134 Days and 6.9 Hours
Alzheimer disease (AD) is the primary form of dementia that occurs spontaneously in older adults. Interestingly, the epigenetic profile of the cells forming the central nervous system changes during aging and may contribute to the progression of some neurodegenerative diseases such as AD. In this review, we present general insights into relevant epigenetic mechanisms and their relationship with aging and AD. The data suggest that some epigenetic changes during aging could be utilized as biomarkers and target molecules for the prevention and control of AD.
Core Tip: The deregulation of non-coding ribonucleic acids and epigenetic modifications have been described in Alzheimer disease (AD). These changes have been observed in different brain regions related to learning and memory, processes that are affected in AD. The epigenetic basics in the progression of AD were integrated into this review.
- Citation: Tecalco-Cruz AC, Ramírez-Jarquín JO, Alvarez-Sánchez ME, Zepeda-Cervantes J. Epigenetic basis of Alzheimer disease. World J Biol Chem 2020; 11(2): 62-75
- URL: https://www.wjgnet.com/1949-8454/full/v11/i2/62.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v11.i2.62
Alzheimer disease (AD) is the most common form of neurodegenerative dementia. AD is characterized by memory loss and rapidly progresses to symptoms such as personality changes and language problems, leading to a loss of the ability to perform routine activities and eventual death of the individual. Diagnosed AD progresses over approximately 8-10 years, but the first events of this disease can occur up to 20 years beforehand. Of all AD cases, more than 95% occur sporadically in adults aged 65 years or older, defined as late-onset AD. Multiple factors, such as environmental, biological, and genetic susceptibility, are associated with the development of AD. Less than 1% of AD cases are related to genetic mutations; these cases generally occur in younger adults (approximately 45 years old). Furthermore, some polymorphisms have been implicated in the development of AD; for instance, the apolipoprotein E 4 (APOE4) variant is associated with an increase of risk to develop AD, while APOE2 variant seems to decrease this risk[1-4].
The development and progression of AD are linked to the dysfunction and death of neurons, which generally appear to originate in the hippocampus, frontal and temporal lobes, and the limbic system, extending to neocortex regions as the disease progresses. In turn, these events are linked to the detection of two neuropathological structures: β-amyloid plaques and neurofibrillary tangles (NFTs) (Figure 1).
The primary component of β-amyloid plaques is the β-amyloid peptide, which is generated and secreted via proteolysis of the β-amyloid precursor protein (APP) by enzyme complexes known as β-secretases (BACE) and γ-secretases, which contain presenilin 1 (PS1, encoded by PSEN1) and PS2 (encoded by PSEN2) as subunits[5]. β-amyloid peptides have lengths ranging from 38 to 43 amino acids and are generated in neurons and released in the extracellular space[6]. Although their function is unclear, they may play a role in synaptic plasticity. Under normal conditions, β-amyloid peptide is eliminated via several mechanisms such as: (1) Ubiquitin-proteasome system; (2) Autophagy-lysosome system; (3) Proteases; (4) Microglial or astrocytic phagocytosis; and (5) Blood circulatory clearance[1,7]. Additionally, it has been suggested that APOE expression improves β-amyloid clearance[8]. However, these mechanisms may deteriorate upon aging and may thus contribute to the accumulation of β-amyloid peptides, forming neurotoxic plaques. These amyloidogenic plaques are surrounded by glial cells and are associated with dystrophic neurites (random neuron prolongations caused by the accumulation of abnormal filaments); therefore, these plaques lead to fibrillar degeneration in nerve cells. The degree of AD is apparently correlated with the proportion of β-amyloid plaques leading to neurodegeneration[9-11].
NFTs are formed by paired helical filaments comprising dense accumulations of insoluble polymers, with the hyperphosphorylated tau protein as the primary component. The tau protein is a 50-64 kDa thermostable protein associated with tubulin. Tau promotes the assembly of microtubules in the neuron cytoskeleton, and the tau-phosphorylation regulates this function. NFTs are formed in the perinuclear region of hippocampal neurons, and according to analyses of postmortem samples from individuals with AD, the quantity of NFTs correlates with the severity of AD. It has been proposed that β-amyloid plaques can also promote intracellular tau aggregation. Additionally, the release of tau damages other cells. Thus, AD is characterized by the presence of extracellular plaques containing insoluble β-amyloid filament accumulations, NFTs formed by hyperphosphorylated tau, and neuroinflammation. These elements are critical markers of AD and contribute to the neurotoxicity of this disease[9,12].
Currently, AD can be diagnosed in living patients by positron emission tomography (PET) and cerebrospinal fluid (CSF) techniques. In PET, a radionuclide as florbetapir
Chromatin consists of deoxyribonucleic acid (DNA) mainly associated with histone proteins; an octamer of histones surrounded by 147 base pairs of DNA forms the nucleosome, which is the basic unit of chromatin. Epigenetics refers to chromatin structure changes that affect gene expression. The conformation of chromatin is highly dynamic, oscillating from an open, lax, or relaxed state to a compact, non-relaxed, and closed state and vice versa. A compact chromatin structure inhibits transcription, while the relaxed form of chromatin promotes this process. Conformational changes in chromatin are orchestrated by the action of co-regulatory proteins, which are divided into co-repressors and co-activators. Co-regulators are proteins that do not bind to DNA directly, but through their interaction with transcription factors. Importantly, co-regulators may have an enzymatic activity to modify chromatin and/or recruit other co-regulatory proteins with the catalytic ability to produce these changes in chromatin structure[13,14].
It has been demonstrated that epigenetic regulatory events involve histone acetyltransferase and histone deacetyltransferase (HDAC) enzymes. For example, some co-activator complexes exhibit histone acetyltransferase activity, in which an acetyl group is added to the lysines at the N-terminus of histones to neutralize their positive charge, thus weakening their strong interaction with DNA and relaxing the chromatin to promote gene expression. In contrast, co-repressor complexes with HDAC activity remove the acetyl group and promote chromatin compaction to inhibit gene expression[13]. In addition to acetylation/deacetylation, histones also undergo other modifications by adding or removing other functional groups (e.g., methylation or phosphorylation) or small proteins (e.g., ubiquitination or sumoylation), which affects the chromatin conformation. It has been suggested that the combination of modifications generates a histone code for chromatin restructuring, which regulates gene expression[13,15].
Another epigenetic modification occurs in the cytosine residues of the CpG dinucleotide of DNA through the addition of a methyl group by DNA methyltransferases. This modification affects the binding of transcription factors to consensus sites, and recruits methylated DNA-binding proteins that bind to co-repressors and HDACs, compacting the chromatin and inhibiting gene expression[16]. Active DNA demethylation is mediated by the methylcytosine dioxygenase enzyme known as ten-eleven translocation, which oxidizes 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC), followed by DNA repair to generate an unmodified cytosine. Therefore, DNA methylation is detected by 5-mC marks and is associated with transcriptional repression, while DNA with 5-hmC marks is related to co-activator recruitment and transcriptional activity[17,18]. Moreover, ATP-dependent chromatin remodelers displace DNA and reposition it within nucleosomes, creating areas of DNA with access to transcriptional machinery. It is worth noting that epigenetic modifications are linked, as there are proteins that modify chromatin and others that can read these modifications, generating a dynamic system coordinated by several marks and regulatory complexes that modulate gene expression[14].
Other molecules involved in epigenetics include non-coding ribonucleic acids (RNAs), which can be small [less than 200 nucleotides (nt) or long (more than 200 nt)[19,20]. MicroRNAs (miRNAs) are small non-coding RNAs with a 19-25 nt hairpin structure that can imperfectly match (partial complementarity with 6-8 nt) to the sequence of messenger RNA (mRNA), resulting in inhibition of its translation or mRNA degradation[21]. Genes located in active chromatin regions appear to be more commonly regulated by miRNAs[22]. In contrast, long non-coding RNAs (lncRNA) regulate the chromatin structure by interacting with proteins, RNA, and DNA[23-25]. Therefore, lncRNAs can act as hooks for proteins or miRNAs, competitive inhibitors of molecular interactions, scaffolding to bring proteins closer for interaction, guides for binding of protein complexes, and gene transcription activators by favoring promoter–enhancer interactions[23-25].
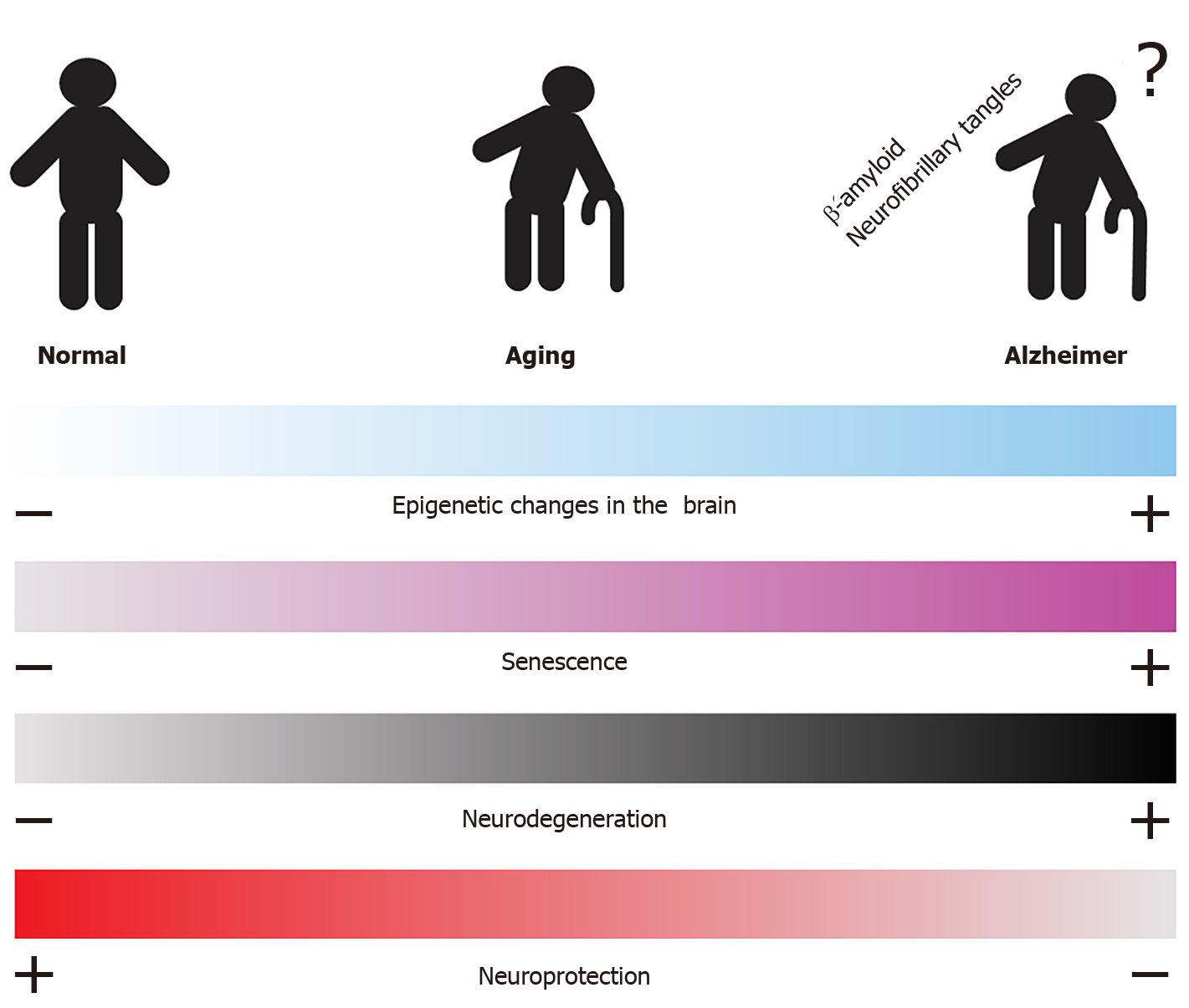
Neurodegenerative diseases are caused by the degeneration, dysfunction, and irreversible death of neurons in specific regions of the central nervous system. In this system, glial cells that include astrocytes, microglia, and oligodendrocytes, are important for the support and proper functioning of neural connections, and these are also affected in neurodegenerative diseases[26]. AD (which affects memory), Parkinson’s disease, and amyotrophic lateral sclerosis (which affects motor activities) are some examples of neurodegenerative diseases[27]. These diseases frequently manifest in older adults; thus, the first risk factor for their development is aging, which has been associated with processes of cellular senescence. Senescence is a viable and metabolically active state of cells, but it is also a non-proliferative state accompanied by pro-inflammatory secretory activity. Senescence is a mechanism for controlling damaged cells, but the accumulation of senescent cells during aging competes with normal cells, blocking the capacity for tissue regeneration and releasing factors that stimulate chronic inflammation and contribute to neuronal degeneration[27-29].
During senescence, epigenetic changes are detected, such as the formation of senescence-associated heterochromatic foci, i.e., compacted chromatin linked to a reduced expression of histone-encoding genes. However, senescent cells also show a lower number of repressive heterochromatin marks such as DNA methylation and histone methylation (H3K9me3, H3K27me3, and H4K20me3), suggesting an increased expression of many other genes[30,31]. Taken together, epigenetic modifications lead to drastic changes in the pattern of gene deactivation and activation in cellular senescence, which, when enriched in cells of the nervous system during aging, may lead to the development of neurodegeneration[30,31].
During aging, epigenetic changes related to acetylation processes can be detected; for example, the H4K16ac mark has been associated with a state of chromatin compaction, stress response, gene expression, and DNA repair. The H4K16ac mark is also enriched during normal aging in the temporal lobes of the human brain, as well as in senescent mammalian cells and aging models in yeast. Interestingly, the H4K16ac mark is reduced in AD. The correlation of H4K16ac levels with normal aging suggests that this modification may protect against neurodegenerative diseases and that changes in its levels may predispose individuals to the development of such diseases, primarily AD[32].
Interestingly, silencing transcription factor RE1 (REST), also known as the neuron restrictive silencer factor, is a specific transcription factor that binds to the RE-1/NRSE response elements and recruits a set of co-repressors to silence the transcription of neural genes. In this respect, REST is associated with HDAC1/2 enzymes, histones methyltransferase such as G9a (methyltransferase of H3K9), as well as CoREST, LSD1, MeCP2, and C-terminal binding protein, which are related to transcriptional repression. REST is expressed at low levels in differentiated neurons[33]; therefore, REST represses genes that promote cell death and AD, induces stress response genes, and protects neurons against oxidative stress and β-amyloid toxicity. REST induction is a result of normal aging in human cortical and hippocampal neurons, and the involved signaling pathways include the Wnt signaling. However, the expression of REST is reduced in AD. In AD, REST is not detected in the cell nucleus but appears in autophagosomes along with misfolded pathological proteins[34]. Thus, the presence of REST during aging is correlated with preservation and longevity, suggesting that REST may generate neuroprotection in the aging of the brain while its decrease may lead to neurodegeneration[34].
The importance of epigenetic mechanisms in AD has been demonstrated through studies using cell cultures, transgenic animal models for AD, and induction of AD by the injection of β-amyloid 1-42 in rats, as well as antemortem and postmortem studies of samples from AD patients. For example, antemortem and postmortem studies have been performed on monozygotic twins, in which one twin was diagnosed with AD, while the other twin did not present any type of dementia. The twin with AD was a chemical engineer who, due to his work, maintained constant contact with pesticides. This twin developed his first symptom of the disease at 60 years old, characterized by a progressive memory loss, with death at 76 years. His twin brother was also a chemical engineer, with the same education, but in a different work environment; this twin died at the age of 79 years of prostate cancer, but without cognitive damage. The presence or absence of AD was confirmed after death[35]. These studies highlight the importance of epigenetics in AD progression in individuals, even among those with identical genetics.
Although models of transgenic mice are a key component of AD studies, none of these models have been able to produce all characteristics of AD. There is still a need for an “ideal” model that develops all clinical and pathological features, ranging from cognitive and behavioral deficits to molecular aspects, including b-amyloid plaques, tau tangles, synaptic and neuronal loss, and neurodegeneration. Different transgenic mouse models have been generated with several modifications to promote the production and accumulation of β-amyloid and tau protein (Table 1).
| Strain | Promoter used | Proteins expressed | Pathogeny | Ref. |
| 3xTG–AD | Thy1 and mPS1 | Mutant of APP (hAPP695, Swedish mutation), PS1 (PSEN1, M146V) and tau (hTau-4R0N, P301L) | Mice containing these mutations develop β-amyloid plaques and NFTs resembling the brain with AD | [36,60,96,97] |
| CK-p25 | tetO (tet operator) | These mice overexpress the truncated form of p35, p25 | p25 activates CDK5 (cyclin-dependent kinase 5), implicated in AD. CK-p25 mice develop neuronal loss, β-amyloid accumulation and loss of synaptic terminations in the hippocampus and cortex as well as memory deficits | [57,98,99] |
| APPPS 1-21 /HDAC6–/– crossbred | Thy1 | Mutated APP (KM670/671NL) and the mutated presenilin 1 (L166P) | Mice develop β-amyloid plaques leading to cerebral amyloidosis, dystrophic synaptic boutons, hyper-phosphorylated tau, inflammatory responses and the impairment of cognitive function | [64,100,101] |
| TgCRND8 | Hamster PrP | hAPP695 Swe/Ind | The brain of mice contains plaques formed by depositions of β-amyloid, leading to inflammation and cognitive impairments. There is also neuronal loss, accumulation of NFTs, and neuritic changes similar to those observed in AD | [93,102,103] |
| Tg19959 | Hamster PrP | hAPP695 with two familial mutations (Swedish and Indiana mutations: K670N/M671L and V717F, respectively). (FVB X 129S6F1 background) | Mice overexpress β-amyloid 1-42 peptide and Bace1 forming plaques | [5,93,102,104] |
| Tg2576-APPswe crossbred | Hamster PrP and Mouse PrP | The Swedish mutation (hAPP695) and m/hAPP6953 (extra and intracellular regions of mouse β-amyloid, a human β-amyloid sequence and the Swedish mutations of β-amyloid, K594N/M595L) | These mice develop β-amyloid plaques deposition and memory deficits | [94,105,106] |
| APP/PSI | Thy | Mutated APP (KM670/671 NL) and mutated presenilin 1 (L166P) | Mice show dystrophic synaptic, hyperphosphorylation of tau, gliosis, and neuronal loss in the dentate gyrus as well as impairment in reversal learning | [95,101] |
The methylation and demethylation processes of DNA are altered in AD. In one study, the levels and distribution of 5-mC and 5-hmC were evaluated in several regions of the brain during the aging of wild-type and triple transgenic (3xTG–AD). The researchers observed a global reduction in 5-mC and an increase in 5-hmC in the brain of aged 3xTG–AD mice in comparison with the wild-type[36]. These data suggest an abnormal establishment of permissive chromatin, which may lead to an increase in several markers linked to AD[36] (Figure 1).
Decreases in the DNA methylation of hippocampal and cerebral cortex cells have generally been observed in AD. For instance, in cortical neurons from postmortem AD brains, the 5-mC levels were lower than those of healthy controls. Similarly, low 5-mC levels have been reported in the hippocampus, cerebral cortex, and cerebellum of AD patients[37-39]. Furthermore, some methylation maintenance factors, such as DNA methyltransferase 1 and methylated DNA-binding 2, were found to be decreased in the AD hippocampus in contrast to healthy controls[39]. Interestingly, in the above-mentioned study on monozygotic twins, 5-mC was found in the neurons, microglia, and astrocytes of the healthy twin, but not in the brain of the AD twin, demonstrating a reduction in DNA methylation in several cell types in the AD brain[35].
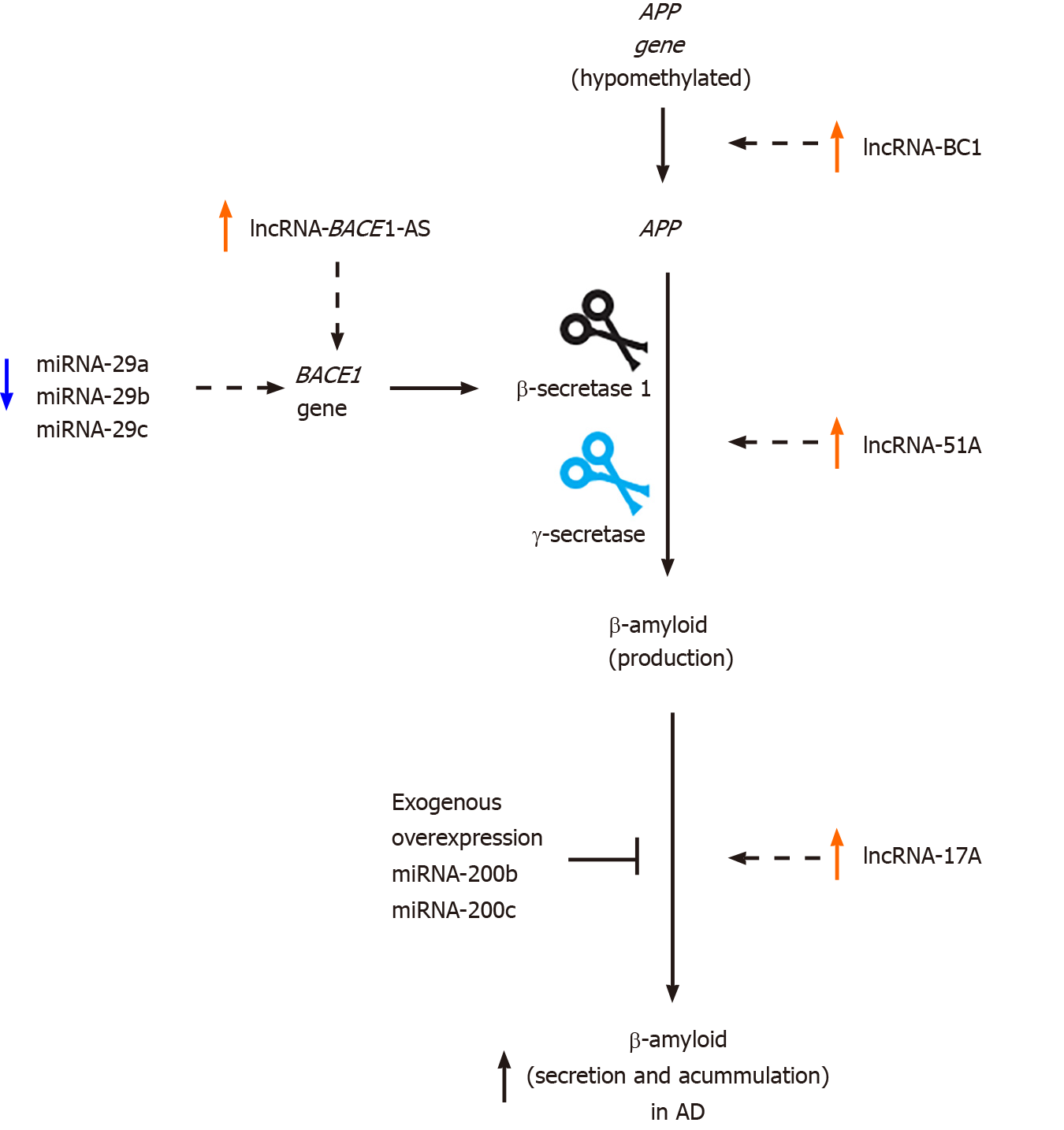
In contrast, the expression of several genes increases upon reduced DNA methylation (hypomethylation), although many of these reports have not been fully validated. Some examples include the APP gene, which encodes the APP. APP gene is silenced by methylation of its promoter region; however, during aging, this gene is demethylated, promoting its expression and consequently, the accumulation of β-amyloid in the brain[40-43]. Nevertheless, some studies suggest no changes in the DNA methylation status of the APP gene for a healthy brain in comparison with AD[44,45]. Moreover, normal brain samples were compared with postmortem AD brain samples, and it was reported that the promoter of the BRCA1 gene, which encodes a DNA repair protein, is hypomethylated in the AD brain. Under these conditions, this gene has a high expression level, whereas the BRCA1 protein appears to be sequestered by tau aggregates. Thus, alterations in the expression and functions of BRCA1 may be involved in the deterioration of AD[46]. In addition, it has been reported that the hypomethylation of intron 1 of the triggering receptor expressed on myeloid cells 2 gene, which is principally expressed in microglia, may induce inflammation pathways associated with AD development. Importantly, triggering receptor expressed on myeloid cells 2 expression is augmented in the hippocampus and leukocytes of AD patients, suggesting its potential as a biomarker for this disease[47-49].
However, studies have also reported augmented DNA methylation in regulatory regions for some genes involved in AD. For instance, methylation of an alternative promoter for the rare coding variant in the phospholipase D3 gene is increased in the AD hippocampus[50], affecting the function of rare coding variant in the phospholipase D3 protein in the processing of APP. Furthermore, increased methylation of the promotor for the brain-derived neurotrophic factor (BDNF) gene, which encodes a key protein implicated in the maintenance of adult cortical neurons and cognoscitive functions, has also been reported in the brain and peripheral blood of AD patients[51-53]. In contrast, it has been reported that methylation of the phosphatidylinositol binding clathrin assembly protein gene in blood cells from AD patients is most likely related to disrupted cognitive functions, as the phosphatidylinositol binding clathrin assembly protein is involved in modulating the production, transport, and abundance of β-amyloid peptide[54]. Therefore, the DNA methylation status associated with the expression of several specific genes is altered in AD, occurring in the hippocampus, cerebral cortex, and some in peripheral blood cells, demonstrating their potential as putative biomarkers for this disease.
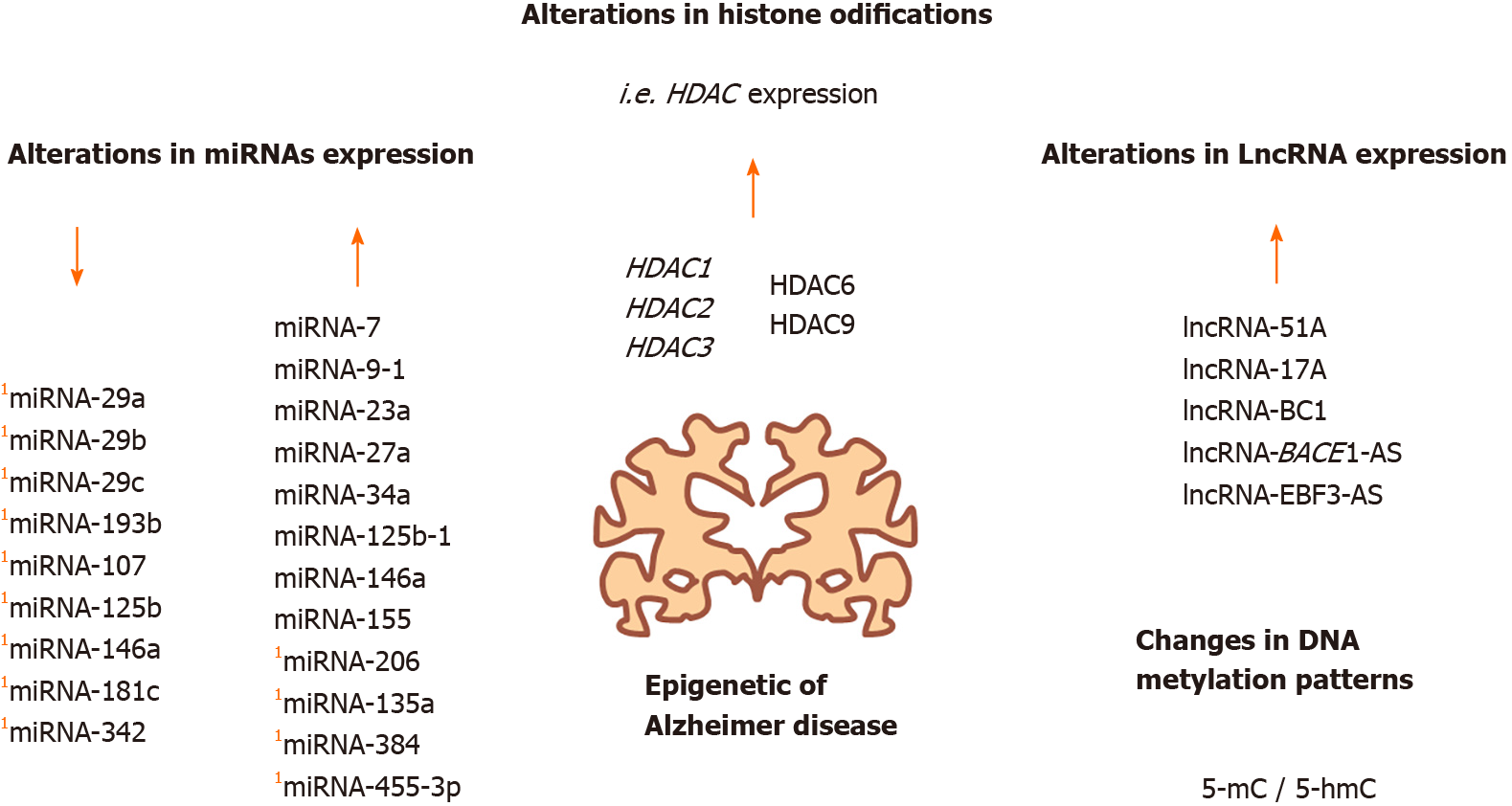
With transgenic mice as an AD model, it has been determined that HDAC2 is primarily expressed in the hippocampus and prefrontal cortex and reduces the density of dendritic spines, the number of synapses, synaptic plasticity, and memory formation in comparison with wild-type mice[55,56]. Additionally, by studying the neurodegeneration process in the brain of CK-p25 mice, researchers detected high levels of HDAC2 and reduced histone acetylation for genes related to learning and memory, as well as the inhibition of genes related to neuroplasticity[57]. HDAC1/2 expression abated in microglia from AD transgenic mice reduced amyloid load, improving cognitive function[58]. Furthermore, in the study of monozygotic twins (previously mentioned), HDAC2, and HDAC9 expression levels in peripheral blood cells were higher in the AD twin than in the healthy twin[59]. Also, increased HDAC3 expression is associated with a decreased memory in the brains of AD mouse models, whereas the loss of HDAC3 expression, experimentally induced in the dorsal hippocampus, appears to improve memory[60-62] (Figure 2).
In addition, HDAC6 is increased in the cortex and hippocampus of AD patients, and it colocalizes with the tau protein in the hippocampus, whereas a reduction in the levels of tau is observed when HDAC6 levels are decreased[63]. It was reported that reduced HDAC6 levels might improve cognitive activity in double transgenic mice (APPPS 1-21 /HDAC6–/–)[64]. In contrast, in one report that used HDAC4 knock-out mice suggested that the lack of HDAC4 reduces learning and memory. Thus, some HDACs are overexpressed in AD patients, whereas HDAC4 seems to decrease synaptic plasticity and memory formation[65].
The acetylation of tau promotes pathological tau aggregation, but SIRT1 can deacetylate tau. Nonetheless, SIRT1 expression levels are decreased in the cortex in AD[66-68]. Additionally, several AD mouse models have been treated with HDAC inhibitors such as sodium butyrate, trichostatin A, and valproic acid and have shown improvements in learning and memory, some of which result from reduced β-amyloid levels[69,70]. Some studies on histone modifications have also been reported. For example, a reduction in H3K18ac and H3K23ac has been identified in the AD brains[71]. Other potentially relevant modifications are H3K27me3 and H3K4me3 modifications, which are enriched in DNA-hypermethylated regions and are associated with aging and AD[72].
Several non-coding RNAs are implicated in the differentiation, connections, and functions of the neurons, as well as in neurodegenerative processes, participating in proteostatic mechanisms, mitochondrial dysfunction, apoptosis, and neurotrophic factor reduction in the neurons and glial cells[73]. One study reported that intracerebroventricular injection of β-amyloid 1–42 resulted in an AD pattern and deregulation of non-coding RNAs in the hippocampus region[74] that included miRNAs and lncRNAs. Interestingly, it has been proposed a putative role as blood-based biomarkers for some miRNAs in AD[75]. For instance, the expression of several miRNAs such as miRNA-29a, miRNA-29b, and miRNA-29c was reduced with an increase in BACE1 (β-secretase 1) expression, which is essential for β-amyloid production (Figure 3). This deregulation has been detected in the brain and peripheral blood of AD patients[76-78]. In contrast, increased levels of miRNA-7, miRNA-9-1, miRNA-23a/miRNA-27a, miRNA-34a, miRNA-125b-1, miRNA-146a, and miRNA-155 have been observed in postmortem AD neocortex samples in comparison with healthy controls[79]. Furthermore, increased miRNA-135a and miRNA-384 Levels and decreased miRNA-193b levels have been found in the serum of AD patients compared with healthy controls[80]. Moreover, the upregulation of miRNA-200b and miRNA-200c was detected in Tg2576 transgenic mice. However, the exogenous overexpression of miRNA-200b and miRNA-200c reduced β-amyloid secretion in in vivo and in vitro experiments[81]. Also, it has also been reported that miRNA-107, miRNA-125b, miRNA-146a, miRNA-181c, miRNA-29b, and miRNA-342 Levels are lower in blood cells from AD patients than in blood from healthy patients[82].
Additionally, miRNA-206 is highly expressed in APP/PSEN1 transgenic mice, mainly in plasma, CSF, and hippocampal regions, correlating with a downregulation of BDNF, and this phenomenon has also been observed in AD patients[83-85]. In another AD model (Tg2576 AD), as well as the brain samples from AD patients, the miRNA-206 expression is also increased and negatively regulates the expression of BDNF at the transcriptional level, which affects synaptic plasticity and memory[84,86]. Moreover, miRNA-206 can be detected in early dementia patients through biopsy of olfactory epithelia[83]. AM206, the antagomir of miRNA-206, prevented the pathogenic effect of β-amyloid 1-42 and increased the levels of BDNF, synaptic density, and neurogenesis after intranasal administration[86,87]. Hence, miRNA-20 is considered as a reliable biomarker for AD. Another of the more reliable biomarkers that have been proposed is miRNA-455-3p since its expression is upregulated in serum samples and brain tissues from AD patients, and these results are also corroborated in transgenic mice and AD cell lines (skin fibroblasts and lymphoblast cells). Thereby, miRNA-455-3p is also suggested as a potential peripheral biomarker for this disease[88]. Thus, miRNAs appear to be relevant indicators of AD progression, and the detection of these miRNAs in the blood may be a powerful tool for this disease (Figure 3).
Similarly, microarrays and RNA-seq studies have found significant changes in lncRNA expression in the AD brain compared with control brain samples[89,90]. For instance, an increase in lncRNA-51A has been reported in AD; lncRNA-51A is known by modulating the splicing of sortilin-related receptor 1 (an important gene for traffic and recycling of the β-amyloid precursor), reducing the synthesis of sortilin-related receptor 1 variant A. Consequently, APP processing is altered, and β-amyloid production is increased, which promotes AD progression[91]. As another example, lncRNA-17A levels are also increased in AD and regulate the alternative splicing of the GABAB receptor; moreover, this lncRNA promotes β-amyloid secretion in response to inflammatory signals[92]. A previous study identified an antisense lncRNA for BACE1, which was termed lncRNA-BACE1-AS. Through in vivo and in vitro assays, lncRNA-BACE1-AS was shown to confer stability to BACE1 mRNA, which increases β-amyloid production and AD development. Furthermore, β-amyloid 1-42 overexpression and stressing factors appear to increase lncRNA-BACE1-AS levels, resulting in amyloid protein aggregation. In both AD patients and a murine AD model (APP 695SWE/IND; TgCRND8 or Tg19959), lncRNA-BACE1-AS expression is augmented[93]. Moreover, it has been observed that lncRNA-BC1 is highly expressed in brains from Tg2576-APPswe mice, another mouse model used to study AD. This lncRNA promotes the translation of APP mRNA, which increases the production and aggregation of β-amyloid peptide[94]. Levels of lncRNA- early B cell factor 3 (EBF3)-AS, i.e., an antisense lncRNA for EBF3, are increased in the hippocampus of APP/PSI mouse model for AD. The authors of this study proposed that lncRNA-EBF3-AS may induce EBF3 expression to stimulate neuronal apoptosis under AD conditions[95].
Despite the complexity involved in understanding and treating AD, epigenetic mechanisms have emerged as potential elements of this disease. To date, studies have reported several modifications in the epigenome of brain cells under AD conditions respect to normal conditions. Importantly, some of these changes have been detected in peripheral blood cells, rendering these changes as promising biomarkers for this disease. The application of HDAC inhibitors has demonstrated beneficial results for several AD mouse models[96-105], impacting β-amyloid levels, tau phosphorylation, and hippocampus dendritic spine restoration, improving learning and memory. Thus, the modulation of the epigenetic modifications in AD, and the identification of epigenetic determinants for healthy aging and those for pathological neurodegeneration requires to be deeply studied (Figure 4).
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chen BH S-Editor: Zhang L L-Editor: A P-Editor: Li X
| 1. | Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer's disease. Nat Rev Dis Primers. 2015;1:15056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 1199] [Article Influence: 119.9] [Reference Citation Analysis (0)] |
| 2. | Stoccoro A, Coppedè F. Role of epigenetics in Alzheimer's disease pathogenesis. Neurodegener Dis Manag. 2018;8:181-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 3. | Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Rimmler JB, Locke PA, Conneally PM, Schmader KE. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1239] [Cited by in RCA: 1271] [Article Influence: 41.0] [Reference Citation Analysis (1)] |
| 4. | Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5968] [Cited by in RCA: 6435] [Article Influence: 201.1] [Reference Citation Analysis (0)] |
| 5. | Li R, Lindholm K, Yang LB, Yue X, Citron M, Yan R, Beach T, Sue L, Sabbagh M, Cai H, Wong P, Price D, Shen Y. Amyloid beta peptide load is correlated with increased beta-secretase activity in sporadic Alzheimer's disease patients. Proc Natl Acad Sci. 2004;101:3632-3637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 407] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 6. | Vlassenko AG, Benzinger TL, Morris JC. PET amyloid-beta imaging in preclinical Alzheimer's disease. Biochim Biophys Acta. 2012;1822:370-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 7. | Xin SH, Tan L, Cao X, Yu JT, Tan L. Clearance of Amyloid Beta and Tau in Alzheimer's Disease: from Mechanisms to Therapy. Neurotox Res. 2018;34:733-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 144] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 8. | Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, Brunden KR, Wilson DA, Landreth GE. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 841] [Article Influence: 64.7] [Reference Citation Analysis (0)] |
| 9. | He Z, Guo JL, McBride JD, Narasimhan S, Kim H, Changolkar L, Zhang B, Gathagan RJ, Yue C, Dengler C, Stieber A, Nitla M, Coulter DA, Abel T, Brunden KR, Trojanowski JQ, Lee VM. Amyloid-β plaques enhance Alzheimer's brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat Med. 2018;24:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 479] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 10. | Querol-Vilaseca M, Colom-Cadena M, Pegueroles J, Nuñez-Llaves R, Luque-Cabecerans J, Muñoz-Llahuna L, Andilla J, Belbin O, Spires-Jones TL, Gelpi E, Clarimon J, Loza-Alvarez P, Fortea J, Lleó A. Nanoscale structure of amyloid-β plaques in Alzheimer's disease. Sci Rep. 2019;9:5181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 11. | Sadleir KR, Kandalepas PC, Buggia-Prévot V, Nicholson DA, Thinakaran G, Vassar R. Presynaptic dystrophic neurites surrounding amyloid plaques are sites of microtubule disruption, BACE1 elevation, and increased Aβ generation in Alzheimer's disease. Acta Neuropathol. 2016;132:235-256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 206] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 12. | Selkoe D, Mandelkow E, Holtzman D. Deciphering Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a011460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 13. | Chen Z, Li S, Subramaniam S, Shyy JY, Chien S. Epigenetic Regulation: A New Frontier for Biomedical Engineers. Annu Rev Biomed Eng. 2017;19:195-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 14. | Längst G, Manelyte L. Chromatin Remodelers: From Function to Dysfunction. Genes (Basel). 2015;6:299-324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 15. | Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016;32:42-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 578] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 16. | Kemme CA, Marquez R, Luu RH, Iwahara J. Potential role of DNA methylation as a facilitator of target search processes for transcription factors through interplay with methyl-CpG-binding proteins. Nucleic Acids Res. 2017;45:7751-7759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 319] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 18. | Michaeli Y, Shahal T, Torchinsky D, Grunwald A, Hoch R, Ebenstein Y. Optical detection of epigenetic marks: sensitive quantification and direct imaging of individual hydroxymethylcytosine bases. Chem Commun (Camb). 2013;49:8599-8601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 290] [Cited by in RCA: 355] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 20. | Makarova JA, Shkurnikov MU, Wicklein D, Lange T, Samatov TR, Turchinovich AA, Tonevitsky AG. Intracellular and extracellular microRNA: An update on localization and biological role. Prog Histochem Cytochem. 2016;51:33-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 171] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 21. | Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 965] [Cited by in RCA: 1156] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 22. | Tao BB, Liu XQ, Zhang W, Li S, Dong D, Xiao M, Zhong J. Evidence for the association of chromatin and microRNA regulation in the human genome. Oncotarget. 2017;8:70958-70966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Andersen RE, Lim DA. Forging our understanding of lncRNAs in the brain. Cell Tissue Res. 2018;371:55-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 24. | Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, Kawaji H, Kasukawa T, Itoh M, Burroughs AM, Noma S, Djebali S, Alam T, Medvedeva YA, Testa AC, Lipovich L, Yip CW, Abugessaisa I, Mendez M, Hasegawa A, Tang D, Lassmann T, Heutink P, Babina M, Wells CA, Kojima S, Nakamura Y, Suzuki H, Daub CO, de Hoon MJ, Arner E, Hayashizaki Y, Carninci P, Forrest AR. An atlas of human long non-coding RNAs with accurate 5' ends. Nature. 2017;543:199-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 673] [Cited by in RCA: 731] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 25. | Hu G, Niu F, Humburg BA, Liao K, Bendi S, Callen S, Fox HS, Buch S. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget. 2018;9:18648-18663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 132] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 26. | Soreq L; UK Brain Expression Consortium; North American Brain Expression Consortium, Rose J, Soreq E, Hardy J, Trabzuni D, Cookson MR, Smith C, Ryten M, Patani R, Ule J. Major Shifts in Glial Regional Identity Are a Transcriptional Hallmark of Human Brain Aging. Cell Rep. 2017;18:557-570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 306] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 27. | Kritsilis M, V Rizou S, Koutsoudaki PN, Evangelou K, Gorgoulis VG, Papadopoulos D. Ageing, Cellular Senescence and Neurodegenerative Disease. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 280] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 28. | Martínez-Zamudio RI, Robinson L, Roux PF, Bischof O. SnapShot: Cellular Senescence in Pathophysiology. Cell. 2017;170:1044-1044.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 29. | Martínez-Zamudio RI, Robinson L, Roux PF, Bischof O. SnapShot: Cellular Senescence Pathways. Cell. 2017;170:816-816.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 30. | Nacarelli T, Liu P, Zhang R. Epigenetic Basis of Cellular Senescence and Its Implications in Aging. Genes (Basel). 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 31. | Sidler C, Kovalchuk O, Kovalchuk I. Epigenetic Regulation of Cellular Senescence and Aging. Front Genet. 2017;8:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 32. | Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F, Toledo JB, Gosai SJ, Gregory BD, Torres C, Trojanowski JQ, Wang LS, Johnson FB, Bonini NM, Berger SL. Publisher Correction: Dysregulation of the epigenetic landscape of normal aging in Alzheimer's disease. Nat Neurosci. 2018;21:1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 33. | Christopher MA, Kyle SM, Katz DJ. Neuroepigenetic mechanisms in disease. Epigenetics Chromatin. 2017;10:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, Bennett DA, Colaiácovo MP, Yankner BA. REST and stress resistance in ageing and Alzheimer's disease. Nature. 2014;507:448-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 480] [Cited by in RCA: 573] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 35. | Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS One. 2009;4:e6617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 199] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 36. | Cadena-del-Castillo C, Valdes-Quezada C, Carmona-Aldana F, Arias C, Bermúdez-Rattoni F, Recillas-Targa F. Age-dependent increment of hydroxymethylation in the brain cortex in the triple-transgenic mouse model of Alzheimer's disease. J Alzheimers Dis. 2014;41:845-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 37. | Chouliaras L, Mastroeni D, Delvaux E, Grover A, Kenis G, Hof PR, Steinbusch HW, Coleman PD, Rutten BP, van den Hove DL. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol Aging. 2013;34:2091-2099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 315] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 38. | Condliffe D, Wong A, Troakes C, Proitsi P, Patel Y, Chouliaras L, Fernandes C, Cooper J, Lovestone S, Schalkwyk L, Mill J, Lunnon K. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer's disease brain. Neurobiol Aging. 2014;35:1850-1854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 39. | Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer's disease: decrements in DNA methylation. Neurobiol Aging. 2010;31:2025-2037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 269] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 40. | Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region -224 approximately -101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999;70:288-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 128] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 41. | Hou Y, Chen H, He Q, Jiang W, Luo T, Duan J, Mu N, He Y, Wang H. Changes in methylation patterns of multiple genes from peripheral blood leucocytes of Alzheimer's disease patients. Acta Neuropsychiatr. 2013;25:66-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K, Tamaoka A, Murayama S, Saido T, Tsuji S. Altered CpG methylation in sporadic Alzheimer's disease is associated with APP and MAPT dysregulation. Hum Mol Genet. 2014;23:648-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 43. | West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer's disease patient. J Mol Neurosci. 1995;6:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 143] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 44. | Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68:880-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 45. | Brohede J, Rinde M, Winblad B, Graff C. A DNA methylation study of the amyloid precursor protein gene in several brain regions from patients with familial Alzheimer disease. J Neurogenet. 2010;24:179-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Mano T, Nagata K, Nonaka T, Tarutani A, Imamura T, Hashimoto T, Bannai T, Koshi-Mano K, Tsuchida T, Ohtomo R, Takahashi-Fujigasaki J, Yamashita S, Ohyagi Y, Yamasaki R, Tsuji S, Tamaoka A, Ikeuchi T, Saido TC, Iwatsubo T, Ushijima T, Murayama S, Hasegawa M, Iwata A. Neuron-specific methylome analysis reveals epigenetic regulation and tau-related dysfunction of BRCA1 in Alzheimer's disease. Proc Natl Acad Sci. 2017;114:E9645-E9654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 47. | Celarain N, Sánchez-Ruiz de Gordoa J, Zelaya MV, Roldán M, Larumbe R, Pulido L, Echavarri C, Mendioroz M. TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer's disease hippocampus. Clin Epigenetics. 2016;8:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 48. | Ozaki Y, Yoshino Y, Yamazaki K, Sao T, Mori Y, Ochi S, Yoshida T, Mori T, Iga JI, Ueno SI. DNA methylation changes at TREM2 intron 1 and TREM2 mRNA expression in patients with Alzheimer's disease. J Psychiatr Res. 2017;92:74-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 49. | Smith AR, Smith RG, Condliffe D, Hannon E, Schalkwyk L, Mill J, Lunnon K. Increased DNA methylation near TREM2 is consistently seen in the superior temporal gyrus in Alzheimer's disease brain. Neurobiol Aging. 2016;47:35-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 50. | Blanco-Luquin I, Altuna M, Sánchez-Ruiz de Gordoa J, Urdánoz-Casado A, Roldán M, Cámara M, Zelaya V, Erro ME, Echavarri C, Mendioroz M. PLD3 epigenetic changes in the hippocampus of Alzheimer's disease. Clin Epigenetics. 2018;10:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 51. | Chang L, Wang Y, Ji H, Dai D, Xu X, Jiang D, Hong Q, Ye H, Zhang X, Zhou X, Liu Y, Li J, Chen Z, Li Y, Zhou D, Zhuo R, Zhang Y, Yin H, Mao C, Duan S, Wang Q. Elevation of peripheral BDNF promoter methylation links to the risk of Alzheimer's disease. PLoS One. 2014;9:e110773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 52. | Nagata T, Kobayashi N, Ishii J, Shinagawa S, Nakayama R, Shibata N, Kuerban B, Ohnuma T, Kondo K, Arai H, Yamada H, Nakayama K. Association between DNA Methylation of the BDNF Promoter Region and Clinical Presentation in Alzheimer's Disease. Dement Geriatr Cogn Dis Extra. 2015;5:64-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 53. | Xie B, Xu Y, Liu Z, Liu W, Jiang L, Zhang R, Cui D, Zhang Q, Xu S. Elevation of Peripheral BDNF Promoter Methylation Predicts Conversion from Amnestic Mild Cognitive Impairment to Alzheimer's Disease: A 5-Year Longitudinal Study. J Alzheimers Dis. 2017;56:391-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 54. | Mercorio R, Pergoli L, Galimberti D, Favero C, Carugno M, Dalla Valle E, Barretta F, Cortini F, Scarpini E, Valentina VB, Pesatori AC. PICALM Gene Methylation in Blood of Alzheimer's Disease Patients Is Associated with Cognitive Decline. J Alzheimers Dis. 2018;65:283-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 55. | Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1350] [Cited by in RCA: 1248] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 56. | Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545-40559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 804] [Cited by in RCA: 860] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 57. | Gräff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, Su SC, Samiei A, Joseph N, Haggarty SJ, Delalle I, Tsai LH. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222-226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 559] [Cited by in RCA: 645] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 58. | Datta M, Staszewski O, Raschi E, Frosch M, Hagemeyer N, Tay TL, Blank T, Kreutzfeldt M, Merkler D, Ziegler-Waldkirch S, Matthias P, Meyer-Luehmann M, Prinz M. Histone Deacetylases 1 and 2 Regulate Microglia Function during Development, Homeostasis, and Neurodegeneration in a Context-Dependent Manner. Immunity. 2018;48:514-529.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 59. | D'Addario C, Candia SB, Arosio B, Di Bartolomeo M, Abbate C, Casè A, Candeletti S, Romualdi P, Damanti S, Maccarrone M, Bergamaschini L, Mari D. Transcriptional and epigenetic phenomena in peripheral blood cells of monozygotic twins discordant for alzheimer's disease, a case report. J Neurol Sci. 2017;372:211-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 60. | Janczura KJ, Volmar CH, Sartor GC, Rao SJ, Ricciardi NR, Lambert G, Brothers SP, Wahlestedt C. Inhibition of HDAC3 reverses Alzheimer's disease-related pathologies in vitro and in the 3xTg-AD mouse model. Proc Natl Acad Sci. 2018;115:E11148-E11157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 61. | McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 444] [Cited by in RCA: 404] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 62. | Zhu X, Wang S, Yu L, Jin J, Ye X, Liu Y, Xu Y. HDAC3 negatively regulates spatial memory in a mouse model of Alzheimer's disease. Aging Cell. 2017;16:1073-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 63. | Ding H, Dolan PJ, Johnson GV. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J Neurochem. 2008;106:2119-2130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 309] [Cited by in RCA: 282] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 64. | Govindarajan N, Rao P, Burkhardt S, Sananbenesi F, Schlüter OM, Bradke F, Lu J, Fischer A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer's disease. EMBO Mol Med. 2013;5:52-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 258] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 65. | Kim MS, Akhtar MW, Adachi M, Mahgoub M, Bassel-Duby R, Kavalali ET, Olson EN, Monteggia LM. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J Neurosci. 2012;32:10879-10886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 182] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 66. | Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, Lee VM. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun. 2011;2:252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 566] [Cited by in RCA: 519] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 67. | Gray SG, Ekström TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 389] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 68. | Julien C, Tremblay C, Emond V, Lebbadi M, Salem N, Bennett DA, Calon F. Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J Neuropathol Exp Neurol. 2009;68:48-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 371] [Cited by in RCA: 353] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 69. | Coppedè F. The potential of epigenetic therapies in neurodegenerative diseases. Front Genet. 2014;5:220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 70. | Sung YM, Lee T, Yoon H, DiBattista AM, Song JM, Sohn Y, Moffat EI, Turner RS, Jung M, Kim J, Hoe HS. Mercaptoacetamide-based class II HDAC inhibitor lowers Aβ levels and improves learning and memory in a mouse model of Alzheimer's disease. Exp Neurol. 2013;239:192-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 71. | Zhang K, Schrag M, Crofton A, Trivedi R, Vinters H, Kirsch W. Targeted proteomics for quantification of histone acetylation in Alzheimer's disease. Proteomics. 2012;12:1261-1268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 72. | Watson CT, Roussos P, Garg P, Ho DJ, Azam N, Katsel PL, Haroutunian V, Sharp AJ. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer's disease. Genome Med. 2016;8:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 136] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 73. | Wu YY, Kuo HC. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J Biomed Sci. 2020;27:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 167] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 74. | Wang Z, Xu P, Chen B, Zhang Z, Zhang C, Zhan Q, Huang S, Xia ZA, Peng W. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer's disease-like rats using microarray analysis. Aging (Albany NY). 2018;10:775-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 75. | Htike TT, Mishra S, Kumar S, Padmanabhan P, Gulyás B. Peripheral Biomarkers for Early Detection of Alzheimer's and Parkinson's Diseases. Mol Neurobiol. 2019;56:2256-2277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 76. | Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci. 2008;105:6415-6420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 930] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 77. | Lei X, Lei L, Zhang Z, Zhang Z, Cheng Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer's disease. Int J Clin Exp Pathol. 2015;8:1565-1574. [PubMed] |
| 78. | Yang G, Song Y, Zhou X, Deng Y, Liu T, Weng G, Yu D, Pan S. MicroRNA-29c targets β-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol Med Rep. 2015;12:3081-3088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 79. | Pogue AI, Lukiw WJ. Up-regulated Pro-inflammatory MicroRNAs (miRNAs) in Alzheimer's disease (AD) and Age-Related Macular Degeneration (AMD). Cell Mol Neurobiol. 2018;38:1021-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 80. | Yang TT, Liu CG, Gao SC, Zhang Y, Wang PC. The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer's Disease Biomarkers. Biomed Environ Sci. 2018;31:87-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 104] [Reference Citation Analysis (0)] |
| 81. | Higaki S, Muramatsu M, Matsuda A, Matsumoto K, Satoh JI, Michikawa M, Niida S. Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer's disease models. PLoS One. 2018;13:e0196929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 82. | Fransquet PD, Ryan J. Micro RNA as a potential blood-based epigenetic biomarker for Alzheimer's disease. Clin Biochem. 2018;58:5-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 83. | Moon J, Lee ST, Kong IG, Byun JI, Sunwoo JS, Shin JW, Shim JY, Park JH, Jeon D, Jung KH, Jung KY, Kim DY, Lee SK, Kim M, Chu K. Early diagnosis of Alzheimer's disease from elevated olfactory mucosal miR-206 Level. Sci Rep. 2016;6:20364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 84. | Silvestro S, Bramanti P, Mazzon E. Role of miRNAs in Alzheimer's Disease and Possible Fields of Application. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 85. | Tian N, Cao Z, Zhang Y. MiR-206 decreases brain-derived neurotrophic factor levels in a transgenic mouse model of Alzheimer's disease. Neurosci Bull. 2014;30:191-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 86. | Lee ST, Chu K, Jung KH, Kim JH, Huh JY, Yoon H, Park DK, Lim JY, Kim JM, Jeon D, Ryu H, Lee SK, Kim M, Roh JK. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72:269-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 259] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 87. | Satoh J. Molecular network of microRNA targets in Alzheimer's disease brains. Exp Neurol. 2012;235:436-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 88. | Kumar S, Reddy PH. MicroRNA-455-3p as a Potential Biomarker for Alzheimer's Disease: An Update. Front Aging Neurosci. 2018;10:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 89. | Magistri M, Velmeshev D, Makhmutova M, Faghihi MA. Transcriptomics Profiling of Alzheimer's Disease Reveal Neurovascular Defects, Altered Amyloid-β Homeostasis, and Deregulated Expression of Long Noncoding RNAs. J Alzheimers Dis. 2015;48:647-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 90. | Zhou X, Xu J. Identification of Alzheimer's disease-associated long noncoding RNAs. Neurobiol Aging. 2015;36:2925-2931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 91. | Ciarlo E, Massone S, Penna I, Nizzari M, Gigoni A, Dieci G, Russo C, Florio T, Cancedda R, Pagano A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer's disease brain samples. Dis Model Mech. 2013;6:424-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 92. | Massone S, Vassallo I, Fiorino G, Castelnuovo M, Barbieri F, Borghi R, Tabaton M, Robello M, Gatta E, Russo C, Florio T, Dieci G, Cancedda R, Pagano A. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41:308-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 93. | Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723-730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1201] [Cited by in RCA: 1136] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 94. | Zhang T, Pang P, Fang Z, Guo Y, Li H, Li X, Tian T, Yang X, Chen W, Shu S, Tang N, Wu J, Zhu H, Pei L, Liu D, Tian Q, Wang J, Wang L, Zhu LQ, Lu Y. Expression of BC1 Impairs Spatial Learning and Memory in Alzheimer's Disease Via APP Translation. Mol Neurobiol. 2018;55:6007-6020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 95. | Gu C, Chen C, Wu R, Dong T, Hu X, Yao Y, Zhang Y. Long Noncoding RNA EBF3-AS Promotes Neuron Apoptosis in Alzheimer's Disease. DNA Cell Biol. 2018;37:220-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 96. | Oddo S, Caccamo A, Kitazawa M, Tseng BP, LaFerla FM. Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer's disease. Neurobiol Aging. 2003;24:1063-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 713] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 97. | Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, LaFerla FM. Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2898] [Cited by in RCA: 3136] [Article Influence: 142.5] [Reference Citation Analysis (0)] |
| 98. | Cruz JC, Kim D, Moy LY, Dobbin MM, Sun X, Bronson RT, Tsai LH. p25/cyclin-dependent kinase 5 induces production and intraneuronal accumulation of amyloid beta in vivo. J Neurosci. 2006;26:10536-10541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 160] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 99. | Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40:471-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 464] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 100. | Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer's disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;26:187-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 292] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 101. | Radde R, Bolmont T, Kaeser SA, Coomaraswamy J, Lindau D, Stoltze L, Calhoun ME, Jäggi F, Wolburg H, Gengler S, Haass C, Ghetti B, Czech C, Hölscher C, Mathews PM, Jucker M. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7:940-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 623] [Cited by in RCA: 771] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 102. | Chishti MA, Yang DS, Janus C, Phinney AL, Horne P, Pearson J, Strome R, Zuker N, Loukides J, French J, Turner S, Lozza G, Grilli M, Kunicki S, Morissette C, Paquette J, Gervais F, Bergeron C, Fraser PE, Carlson GA, George-Hyslop PS, Westaway D. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem. 2001;276:21562-21570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 700] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 103. | Hyde LA, Kazdoba TM, Grilli M, Lozza G, Brusa R, Zhang Q, Wong GT, McCool MF, Zhang L, Parker EM, Higgins GA. Age-progressing cognitive impairments and neuropathology in transgenic CRND8 mice. Behav Brain Res. 2005;160:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 104. | Li F, Calingasan NY, Yu F, Mauck WM, Toidze M, Almeida CG, Takahashi RH, Carlson GA, Flint Beal M, Lin MT, Gouras GK. Increased plaque burden in brains of APP mutant MnSOD heterozygous knockout mice. J Neurochem. 2004;89:1308-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 216] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 105. | Hall AM, Roberson ED. Mouse models of Alzheimer's disease. Brain Res Bull. 2012;88:3-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 229] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 106. | Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1028] [Cited by in RCA: 1250] [Article Influence: 56.8] [Reference Citation Analysis (0)] |