Published online Oct 27, 2020. doi: 10.4240/wjgs.v12.i10.407
Peer-review started: July 29, 2020
First decision: August 9, 2020
Revised: August 19, 2020
Accepted: September 14, 2020
Article in press: September 14, 2020
Published online: October 27, 2020
Processing time: 89 Days and 13.9 Hours
Currently, extracellular vesicles and particularly exosomes have gained a lot of research interest due to their unique roles in several biological processes. Noncoding RNAs (microRNAs, long noncoding RNAs and circular RNAs) represent a class of functional RNA with distinct regulatory roles in tumorigenesis and cancer progression. Cholangiocarcinoma is a rare but highly aggressive type of malignancy that is very challenging to diagnose, especially in early stages; surgical resection still represents the sole potentially curative treatment option. Hence, there is an urgent need for the discovery of novel diagnostic and prognostic biomarkers. Hereby, we provide a comprehensive review of the most recent discoveries that focus on exosomal noncoding RNAs in cholangio-carcinoma with the aim to identify new molecular players that could be used as biomarkers and therapeutic targets.
Core Tip: Although there are currently several original research studies investigating the role of noncoding RNAs in cholangiocarcinoma, very few have focused specifically on exosomal noncoding RNA signatures. This is the first review to summarize and report current data regarding exosomal noncoding RNAs in cholangiocarcinoma and discuss their potential future clinical applications.
- Citation: Laschos K, Lampropoulou DI, Aravantinos G, Piperis M, Filippou D, Theodoropoulos G, Gazouli M. Exosomal noncoding RNAs in cholangiocarcinoma: Laboratory noise or hope? World J Gastrointest Surg 2020; 12(10): 407-424
- URL: https://www.wjgnet.com/1948-9366/full/v12/i10/407.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v12.i10.407
Cholangiocarcinoma (CCA) is a type of highly heterogenous group of epithelial malignancies that can originate from any division of the biliary tree. The classification of CCA is based on the anatomic location with regards to the liver and includes three subtypes: Intrahepatic, perihilar (Klatskin tumor) and distal extrahepatic[1]. The heterogenous nature of this type of cancer also corresponds to several epidemiological, biological and clinicopathological features. Intrahepatic CCA is the second most common primary liver cancer after hepatocellular carcinoma, and its prognosis is very poor, mainly depending on the potential and extent of surgical resection[2,3]; the five-year survival rates drop to 2% for cases that are diagnosed with distant metastases[4]. Treatment options for inoperable and/or microscopically positive surgical resection margin (R1) cases, include chemoradiation therapy[5]. However, the response rates still remain very low, and the clinical management of the advanced CCA is lacking a “gold standard” chemotherapy regimen[6].
Several risk factors have been associated with CCA pathogenesis including hepatobiliary disorders such as primary sclerosing cholangitis (PSC), the presence of choledochal cysts, hepatolithiasis, viral hepatitis B and C-induced chronic hepatitis and cirrhosis[7-9]. Moreover, parasite (Opisthorchis viverrini or Clonorchis sinensis)- induced infections[10] as well as genetic factors[11], obesity, smoking and alcohol consumption have also been correlated with increased risk of CCA[9].
CCA is a highly aggressive, rare type of malignancy that is very challenging to diagnose at an early, potentially curable stage. The confirmation of the diagnosis usually results from the combination of: (1) Imaging, such as computed tomography, magnetic resonance imaging, magnetic resonance cholangiopancreatography, endoscopic retrograde cholangiopancreatography and positron emission tomography; (2) Biochemical; and (3) Histological data[12]. Moreover, the assessment of cancer biomarkers, namely carbohydrate antigen 19-9 and carcinoembryonic antigen, is a common routine practice towards the diagnosis, but their usefulness remains controversial due to their low sensitivity and specificity to detect CCA in early stages[13]. Therefore, there is an urgent need for novel diagnostic and therapeutic approaches towards the prompt detection and management of CCA.
Exosomes represent a distinct type of extracellular vesicle (EV) that mediate intercellular communication and have recently emerged as promising biomarkers and therapeutic targets in the cancer field[14,15]. There is also evidence that circulating EVs play an important role in the pathogenesis of liver disease via the modulation of several cell signaling mechanisms[16]. Many studies have shown that there is a different expression of exosomal noncoding RNAs (ncRNAs) in cancer patients compared to healthy subjects and often mirrors the type of cancer. The mechanisms of EV packaging remain unclear; however, several factors that affect the composition of EVs have been described so far. These include: (1) The upregulation of a distinct RNA type in parental cells; and (2) The existence of sorting processes that may be biotype-specific[17]. The content of exosomes consists of a mosaic cargo including proteins, lipids and nucleic acids[18]. Tumor-derived (TD) exosomes can act as transporters of this load; the latest may include cancer-related signaling molecules that can be transferred to other recipient cells via exosome fusion with the target cell membrane. The transported genetic information can subsequently regulate gene expression in the recipient cell, which in turn may trigger several tumor-related processes, such as tumor proliferation, epigenetic reprogramming, invasion and metastasis[19-22].
Hereby, we review the current evidence regarding the roles of exosomal ncRNAs and discuss their diagnostic and therapeutic potential in CCA. For clarity, because the term “EVs” has been ambiguous in the literature and often coincides and/or is being confused with the term “exosomes,” in the present work we will try to include data that refer to primarily exosomal and secondarily EV-derived ncRNAs in CCA.
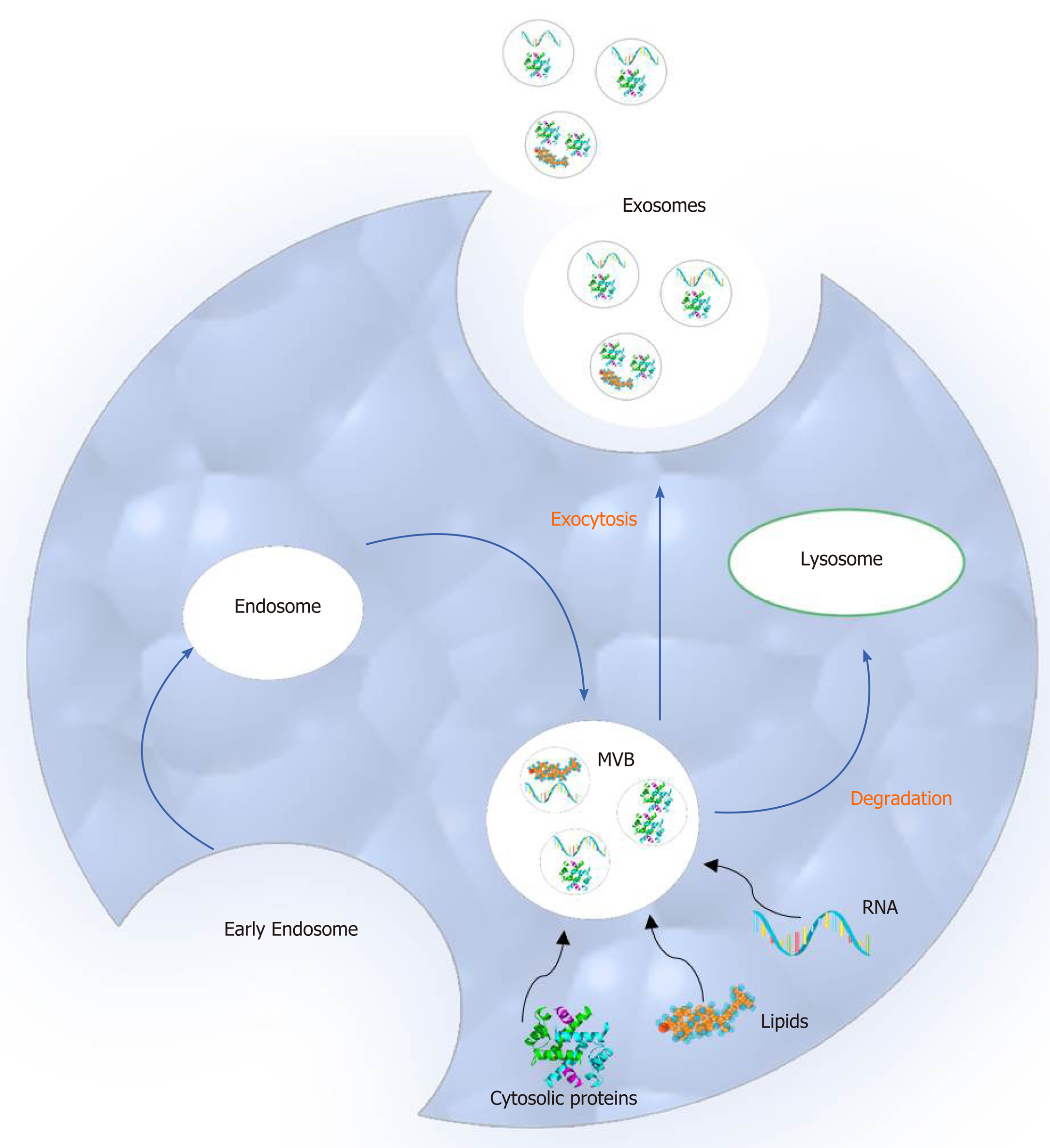
Compared to microvesicles (the second main type of extracellular vesicles), exosomes differ in size and biogenesis pathway. More specifically, exosomes are smaller than microvesicles ranging in diameter from 30 to 100 nm and originate from the endosomal network. Moreover, they are released to the extracellular space by multivesicular bodies (MVBs) following fusion with the cellular membrane. On the other hand, microvesicles are larger in size (50 to 1000 nm in diameter), and they are secreted to the extracellular environment through direct outward budding of the plasma membrane[15-19].
The process of exosome biogenesis includes the following steps: (1) Early endosome creation from the plasma membrane; (2) MVB formation; and (3) Initial formation of exosomes as intraluminal vesicles (ILVs) in the MVBs. These components are either degraded by lysosomes or released as exosomes to the extracellular space after fusion with the plasma membrane. During this process, several biomolecules such as cytosolic proteins, lipids and nucleic acids are incorporated in the MVBs[20] (Figure 1).
Two main mechanisms orchestrate ILV formation and the packaging of bioactive exosomal cargo. The first one depends on the endosomal sorting complexes required for the transport system whereas the second depends on raft-based microdomains, such as tetraspanin-enriched microdomains, tetraspanins and lipids[21].
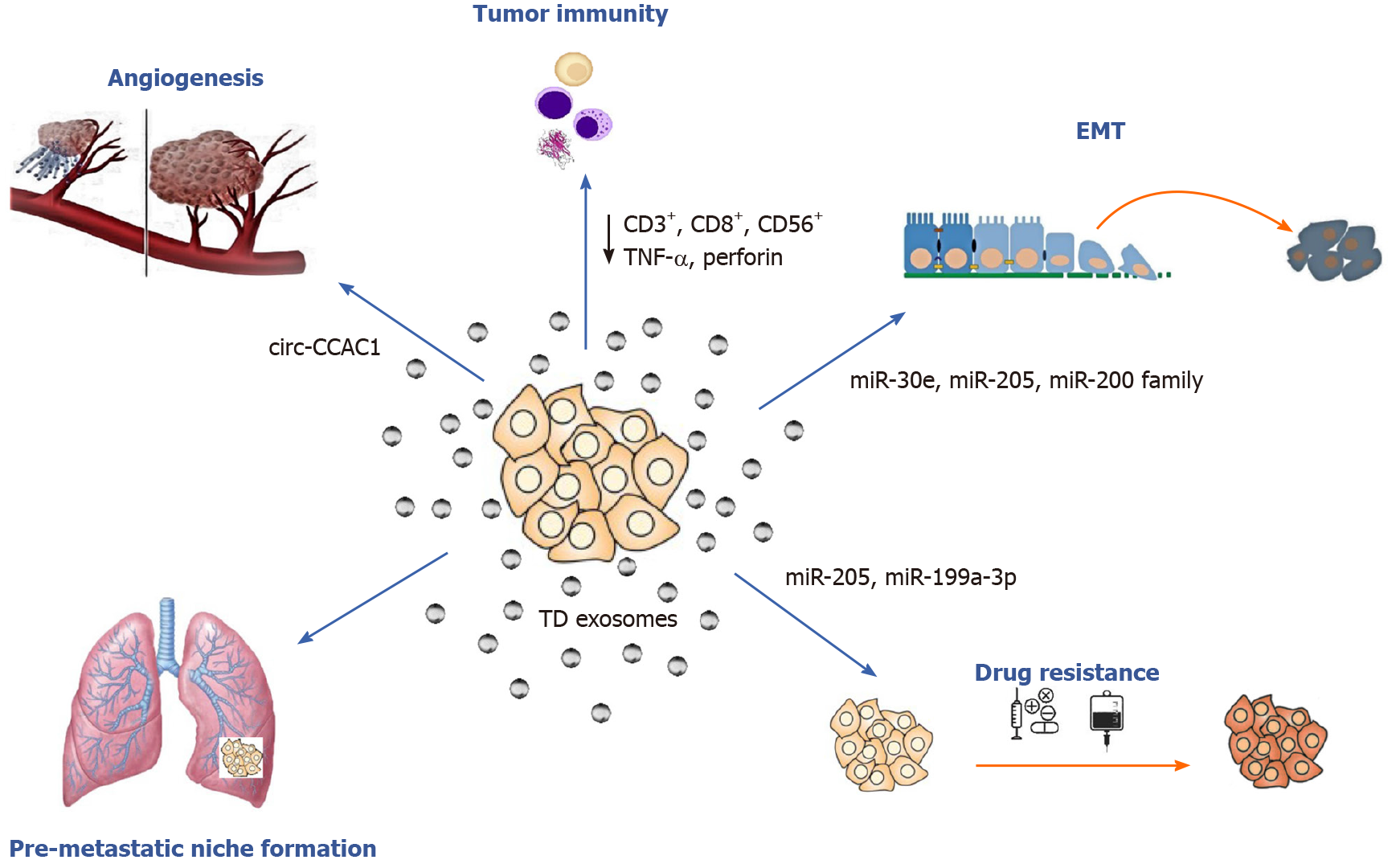
Exosomes and their parental cell-specific cargos can be secreted by all eukaryotic cells, both healthy and tumor, into the extracellular environment. Then they can either enter neighboring recipient cells by endocytosis or travel through biological fluids such as blood, urine and saliva. Exosome uptake from the recipient cell takes place after cellular recognition and internalization. It has been reported that tumor cells can release ten times more exosomes than healthy cells and that TD exosomes exhibit pro-oncogenic properties, such as promoting cell proliferation, epithelial-to-mesenchymal transition (EMT), angiogenesis, metastasis and drug resistance[21,23] (Figure 2). With regard to CCA, bile EV concentrations were found to be significantly increased in patients with CCA suggesting that they could be used for diagnostic testing[24]. In conclusion, exosomes are considered to be crucial mediators of intercellular communication because they can transfer their content and alter biological responses in other cells.
Accumulating evidence shows that exosomal components may play crucial roles in several cancer related processes such as angiogenesis and metastasis colonization. The “pre-metastatic niche” is the microenvironment created by TD exosomes and facilitates metastasis[25]. Recent work has also demonstrated that exosomes promote neoangiogenesis at the pre-metastatic niche by several mechanisms including: (1) Protein activation (i.e., activation of transcription factor 2 and metastasis-associated protein 1)[26]; (2) Vascular permeability promotion via soluble E-cadherin[27]; and (3) The release of proangiogenic factors that promote neovascularization [microRNAs (miRNAs), vascular endothelial growth factor and cytokines][28,29]. Moreover, several mechanisms implicating TD exosomal miRNAs, kinases and other factors affecting the process of metastasis have been identified in current literature[30,31].
Over the last decade several studies have focused on the role of EVs and exosomes in biliary pathophysiology and CCA pathogenesis[16,32]. In 2010, Masyuk et al[33] found that bile exosomes released by normal cholangiocytes directly interacted with primary cilia and inhibited cell proliferation via the ERK signaling pathway. During CCA development, EVs promote the myofibroblast-like transdifferentiation of bone marrow mesenchymal stem cells (MSCs) and thus favor the formation of tumor stroma. Moreover, they stimulated IL-6 production contributing further to CCA growth[34,35]. Chen et al[36] reported that TD exosomes contributed to CCA escape from immune recognition by downregulating CD3+, CD8+, NK (CD56+) cells and by decreasing TNF-α and perforin production.
Additionally, CCA cell-derived EVs are loaded with a unique content that has been associated with tumorigenic effects. Proteomic analysis identified various oncogenic proteins such as epidermal growth factor receptor and integrin beta-4 in CCA vs healthy cholangiocyte-derived EVs[37]. Accordingly, Dutta et al[38], reported that various cancer-related proteins (i.e., large neutral amino acids transporter small subunit 1 (LAT1), 4F2 cell-surface antigen heavy chain, pyruvate kinase) were disclosed in CCA-derived exosomes compared to normal human cholangiocytes (H69), providing evidence for their direct intercellular transport by the exosomes. Recently, the phosphorylation level of exosomal heat shock protein 90 was also found to be significantly related with tumor malignancy in an in vitro model of isogenic CCA cells[39].
Dysregulation of cellular miRNAs in several types of cancer has been a topic of extensive investigation; currently there is emerging evidence that exosomal miRNA expression is also modified, suggesting that it may serve as a potential biomarker for cancer diagnosis and prognosis[40]. In 2013, Huang et al[41] first reported that amongst other exosomal RNA species, miRNAs were the most abundant in human plasma-derived exosomes. miRNAs are short ncRNAs consisting of 21–25 nucleotides that are critically involved in the regulation of gene expression[19]. The exosome-loading process starts after miRNA biogenesis and includes a complex of different components such as mature and pre-miRNAs, other RNA species, proteins and lipids. This cargo mirrors the content of the parent cell and is transferred to the recipient cell via fusion with the plasma membrane. Subsequently, transported miRNAs can play regulatory roles in the recipient cells[42]. Thus, CCA-derived exosomes can act as cancer migration and invasion mediators by transferring oncogenic miRNAs to normal cholangiocytes.
To date, several studies have investigated the role of miRNAs in the initiation and progression of CCA, but only a few have focused specifically on the exosomal miRNA profiling and role (Table 1). Reportedly, miR-205 can act as an oncogene or tumor suppressor[43]; miR-205 overexpression has been implicated in the development and progression of several cancers[44,45]. Okamoto et al[46] reported that miR-205 levels were associated with gemcitabine resistance in HuH28 cell lines and that miR-205 upregulation was related with restoring gemcitabine sensitivity. Interestingly, exosome-derived miR-205 from human CCA cell lines was found to be overexpressed, and knockdown of miR-205-5p expression repressed migration and invasion in CCA cell lines[47]. The same study also supported the role of exosomal miR-200 family members in CCA progression. Consistent with this observation, Shen et al[48] found that the miR-200 family was differentially expressed in peripheral blood-derived exosomes of CCA patients.
| microRNA | Expression | Type of EVs | EV source | Major finding | Potential application | Ref. |
| miR-205 | ↑ | Exosomes | Human CCA cell lines | Downregulation of miR-205-5p decreased migration and invasion in CCA cell lines | Therapy monitoring/therapeutic target | [47] |
| Members of miR-200 family | ||||||
| miR-200c-3p, miR-200b-3p, miR-200a-3p, miR-429 and miR-141-3p | ↑ | Exosomes | Human CCA cell lines | Supported the role of exosomal miR-200 family in CCA progression | Prognostic value | [47] |
| miR-200c-3p, miR-200a/c-3p | ↑ | Exosomes | Peripheral blood samples (36 patients) | (a) miR-200c-3p emerged as a potential diagnostic biomarker; and (b) miR-200a/c-3p emerged as a potential diagnostic and prognostic biomarker | Early diagnostic and prognostic value | [48] |
| miR-199 family | ↓ | Exosomes | Human CCA cell lines | Supported the role of miR-199 family in CCA carcinogenesis | - | [47] |
| miR-214 | ↓ | Exosomes | Human CCA cell lines | Supported the role of miR-214 in CCA carcinogenesis | - | [47] |
| 5 miR-based panel (miR-191, miR-486-3p, miR-1274b, miR-16 and miR-484) | ↑ | EVs | Bile samples (46 CCA vs 50 control patients with PSC, biliary obstruction and bile leak) | (a) The panel displayed a 67% sensitivity and 96% specificity for CCA diagnosis; and (b) tool for differential diagnosis between biliary obstruction of nonmalignant etiologies and CAA | Diagnostic value | [49] |
| miR-30e | ↓ | EVs | Nonmalignant human cell vs CCA cell lines | Encapsulation of miR-30e in EVs could suppress CCA cell invasion and migration by inhibiting EMT | EVs may be used as vehicles for delivery of therapeutic agents | [50] |
| miR-195 | ↓ | EVs | Human liver stellate cell line | Coculture of CCA and stellate cell lines resulted in downregulation of miR-195. EV-mediated miR-195 transfer targeted tumor cells and inhibited proliferation in a rat model. | EVs may be used as vehicles for delivery of therapeutic agents | [52] |
| miR-604 | ↑ | EVs | Serum | Displayed 0.944 diagnostic capacity for CCA | Diagnostic value | [53] |
| miR-551B | ↑ | EVs | Serum | Displayed 0.909 diagnostic capacity for CCA | Diagnostic value | [53] |
| miR-96-5p, miR-151a-5p, miR-191-5p and miR-4732-3p | ↑ | Exosomes | Blood | Stage II CCA patients displayed the highest levels | Diagnostic value in early CCA stages | [58] |
| miR-9-5p | ↑ | Exosomes | Human ICC samples | Significant association with malignancy promotion via ↑IL-6 expression in vCAFs | Prognostic value | [59] |
On the contrary, exosomal miR-199 family members and their clustered miRNA, hsa-miR-214-3p were found to be downregulated in human CCA cell line-derived exosomes, supporting their role in CCA carcinogenesis[47]. Furthermore, a panel of five miRNAs (miR-191, miR-486-3p, miR-1274b, miR-16 and miR-484) were upregulated in bile EVs from CCA patients vs a control group of patients suffering from PSC, biliary obstruction and bile leak. Of note, the study isolation protocol supported that the identified EVs were probably exosomes[49].
It is well known that cancer invasion and metastasis have also been associated with EMT promotion. A recent study investigated the role of EV-miRNAs in regulating EMT process in CCA cells. The authors concluded that miR-30e expression was decreased by TGF-β[50]; the latter has been previously identified as an EMT inducer[51]. More importantly, this study demonstrated that miR-30e encapsulation in EVs could halt CCA cell invasion and migration by inhibiting EMT[50]. Similarly, miR-195 levels were downregulated in cholangiocarcinoma cells, and EV-incorporated miR-195 decreased cancer progression in a rat CCA model[52].
Another recent study demonstrated that miR-551B and miR-604 were significantly upregulated in serum EVs, displaying an optimal diagnostic capacity for CCA[53]. Interestingly, Chang et al[54] had previously reported that decreased miR-551b-3p levels were associated with poor overall survival of CCA patients. It is worth noting that miR-551b-3p expression in cancer varies in the literature. For instance, miR-551b-3p upregulation has been reported in papillary thyroid carcinoma[55] and ovarian cancer[56], whereas gastric cancer has been associated with miR-551b-3p downregulation[57]. Therefore, further research may be required in order to investigate the expression and functional roles of miR-551b-3p in CCA.
Recently, four miRNAs (miR-96-5p, miR-151a-5p, miR-191-5p and miR-4732-3p) were found to be significantly overexpressed in blood-derived exosomes of CCA patients[58], and exosomal miR-9-5p was proposed as a potential prognostic biomarker for intrahepatic cholangiocarcinoma[59]. Mir-9-5p has been previously associated with angiogenesis promotion in cervical cancer[60].
Long ncRNAs (lncRNAs) represent a subclass of ncRNAs (more than 200 nucleotides long) with distinct roles in several biological processes including cell proliferation, differentiation, invasion and metastasis. Several studies have found that lncRNAs can act as crucial mediators of cancer development, including CCA. Recent evidence supports their involvement in CCA progression via the competing endogenous RNA (ceRNA) network[61]. lncRNA levels in secreted exosomes have been suggested to be similar with those detected in plasma[62]. Although some lncRNA-specific loading mechanisms have been described in the literature[63], the exact process that drives the exosomal loading with a specific biological cargo remains unclear.
Many studies have investigated the abnormal expression of specific lncRNAs and their association with CCA development and progression. However, little research has focused on the exosome and/or EV-lncRNA expression. Given that a very recent extensive review summarizes the roles of lncRNAs in CCA[64], we will refer to and discuss current literature evidence based on studies investigating the functional roles of lncRNAs in EVs with an emphasis to exosomes (Table 2).
| lncRNA | Expression | Type of EVs | EV source | Major finding | Potential application | Ref. |
| ENST00000517758.1 and ENST00000588480.1 | ↑ | Exosomes | Bile samples of CCA (n = 35) and biliary obstruction patients (n = 56) | (a) ENST00000588480.1 expression may contribute to tumorigenesis and CCA progression (via p53 signaling pathway); and (b) Combined ENST00000588480.1 and ENST00000517758.1 exhibited higher sensitivity than CA19-9 (82.9% vs 74.3%) | Diagnostic value; Prognostic value/Therapeutic target | [63] |
| MALAT-1 | ↑ | EVs | Serum | Upregulated in EVs from CCA vs PSC patients | Diagnostic value | [53] |
Ge et al[63], reported that two lncRNAs (ENST00000588480.1 and
In summary, despite the emerging role of exosomal lncRNAs as potential cancer biomarkers, to date very few researches have been focused on CCA. Most of the available data cannot determine direct associations between the exosomal lncRNAs and CCA development and progression. Moreover, to our knowledge no evidence exists regarding the sensitivity and specificity of lncRNAs in a clinical setting.
Besides miRNAs and lncRNAs, circular RNAs (circRNAs) represent another subclass of bioactive ncRNAs. Originally, circRNAs were considered an RNA splicing byproduct with negligible functions. Recent findings suggested that exosomal circRNAs can serve as candidate cancer biomarkers due to their high stability in exosomes[68].
CircRNA production originates from pre-mRNA back-splicing of exons, resulting in a single-stranded, closed, circular structure. Emerging evidence shows that they participate in several pathophysiological processes and that their expression is significantly altered during cancer development[69,70]. Conn et al[71] proposed that circRNAs are implicated in the EMT process and thus affect cell migration, invasion and tumor metastasis. Their regulatory role in the transcription process has also been reported[72].
Few studies have investigated the role of circRNAs in CCA tumorigenesis and progression. Cdr1as was found to be upregulated in cholangiocarcinoma tissues, and its expression level was positively correlated with clinicopathological parameters (tumor, node, metastasis stage, lymph node invasion and postsurgery recurrence). The authors also supported that high Cdr1as expression was associated with poor overall survival, highlighting the potential role of this circRNA as a prognostic biomarker[73]. Another study found that hsa_circ_0001649 was downregulated in CCA tissues, and it was associated with tumor size and grade. Moreover, it was suggested that upregulation of hsa_circ_0001649 resulted in tumor suppression both in vivo and in vitro[74]. Increased hsa_circ_0001649 expression was also negatively correlated with tumor progression in hepatocellular carcinoma[75] and in pancreatic ductal adenocarcinoma[76]. Its potential role as a tumor suppressor in gastrointestinal malignancies was further supported by a recent study; upregulation of hsa_circ_0001649 inhibited tumor growth and metastasis in gastric cancer cells[77]. Finally, Xu et al[78] proposed that circ_0005230 inhibited cell apoptosis and promoted cell proliferation and metastasis in CCA cells.
To the best of our knowledge, only two studies have investigated the possible association of exosomal circRNA expression in cholangiocarcinoma so far. According to Wang et al[79], circRNA 0000284 was found to be significantly upregulated in CCA cell lines, tumor tissues and plasma exosomes. Furthermore, exosome-mediated circ-0000284 transfer to neighboring normal cells resulted in tumorigenesis and CCA progression. Hence, circ-0000284 was reported to exhibit autocrine and paracrine actions through exosomal intercellular communication. Recently, circ-CCAC1 was found upregulated in circulating EVs exerting a potential role in CCA diagnosis and prognosis[80].
Piwi-interacting RNAs (piRNAs) represent the largest class of ncRNAs; piRNAs are 26 to 31 nucleotides long and specifically interact with piwi-domain containing proteins[81]. More recent evidence suggests that piRNAs are involved in gene regulation at epigenetic and posttranscriptional levels, emerging as new mediators in the process of carcinogenesis[82,83]. In 2016, Yuan et al[84] noticed that piRNAs were differentially expressed in plasma-derived exosomes of cancer patients compared to healthy subjects. Currently, the majority of published studies have focused on the potential role of piRNAs as diagnostic and prognostic biomarkers in other types of malignancies, such as colorectal cancer and hepatocellular carcinoma[85,86].
On the other hand, very little is known about the roles of piRNAs in CCA. Chen et al[87] reported that piwi-like protein 2 was significantly overexpressed in both hilar CCA tissues and the QBC939 cell line. Based on this observation, Gu et al[88] further investigated exosomal piRNA signatures and found that several piRNAs were differentially expressed in CCA patients compared to healthy individuals. More importantly, the authors suggested that piR-10506469 was significantly overexpressed in plasma-derived exosomes from CCA patients and that piR-10506469 and piR-20548188 were significantly downregulated after surgery, suggesting that these piRNAs may serve as potential diagnostic and prognostic biomarkers.
In terms of epigenetic modifications, miRNA downregulation has often been associated with hypermethylation of their promoters. For instance, epigenetic silencing of tumor suppressor miR-370 in human CCA has been linked to hypermethylation of its promoter by IL-6-dependent overexpression of DNA methyltransferases[89]. Interestingly, An et al[90] suggested that miR-370 silencing in CCA follows Knudson’s “two-hit hypothesis” mechanism via IL-6 mediated maternal to paternal epigenotype switch. Furthermore, CpG island hypermethylation of miR-373 resulted in miR-373 downregulation in hilar cholangiocarcinoma[91]. Accordingly, increased methylation of CpG sites upstream of miR-376c gene was found in intrahepatic CCA cell lines[92]. CCA tumorigenesis and progression has also been associated with Notch pathway activation[93]. Enhancer of zeste homolog 2 (EZH2) and DNA methylation-induced miR-34a silencing resulted in the promotion of CCA cell growth through activation of the Notch pathway[94]. Interferon regulatory factor-1 (IRF-1) has been suggested as a tumor suppressor in CCA[95], and miR-383 has been recently found to directly target IRF-1[96]. These findings suggest that the targeting of IRF-1 by miR-383 may be the molecular basis for IRF-1 downregulation in CCA.
ncRNAs can regulate gene expression at several levels (epigenetic, transcriptional and posttranscriptional). On this basis, the regulatory scenario is enormous and remains under investigation. The exact underlying mechanisms of many ncRNA functions remain unclear. In the following section will focus on the molecular mechanisms of exosomal miRNAs in CCA.
The hypomethylated status of the miR-429 promoter has been correlated with increased miR-429 expression in CCA. Goeppert et al[97] demonstrated that epigenetically dysregulated miR-429 directly targeted cadherin-6 and promoted tumor growth. Exosomal miR-551b levels were upregulated in EVs isolated from CCA patients[53]. LncRNA SMARCC2 acts as a “sponge” RNA promoting the aberrant miR-551b-3p expression in gastric cancer[57]. On the other hand, Chang et al[54] demonstrated that miR-551b-3p directly targeted and decreased CCND1 expression, inhibiting CCA cell cycle progression and proliferation. miR-551b downregulation in breast cancer patients was associated with hypermethylation of its promoter[98]. Exosome-derived miR-205 and members of the miR-200 family were found to be overexpressed in CCA cell lines[47]. It has been previously suggested that these miRNAs cooperatively regulate EMT by targeting the transcriptional repressors ZEB1 and SIP1[99]. Moreover, miR-205 upregulation was associated with enhancing gemcitabine sensitivity in CCA cell lines; however, the authors could not identify possible target genes that could be associated with chemosensitivity[46]. Another study suggested that miR-205 promotes tumor invasion and metastasis in ovarian cancer via suppressing PTEN/SMAD4 expression[44]. MiR-199a-3p has also been implicated in increasing cisplatin sensitivity by inhibiting the mTOR pathway and by downregulating the MDR1 gene[100]. Recently, Zhang et al[101] reported that exosomes can act as carriers of miR-199a-3p in hepatocellular carcinoma. Interestingly, the authors concluded that intravenous injection of exo-miR-199a-3p increased resistance to cisplatin, offering a novel option for cisplatin refractory cancers. KEGG pathway analysis revealed that several CCA-associated, exosomal miRNA (miR-96-5p, miR-151a-5p, miR-191-5p and miR-4732-3p) target genes were enriched in the MAPK signaling pathway, suggesting their role in the process of neurogenesis[58]. miR-9-5p is another important miRNA that was upregulated in CCA-derived exosomes; miR-9-5p expression in intrahepatic cholangiocarcinoma was correlated with IL-6 upregulation in vascular cancer-associated fibroblasts via EZH2 overexpression[59]. Furthermore, Wei et al[60], suggested that it could promote angiogenesis in cervical cancer by targeting suppressor of cytokine signaling 5.
Accumulating evidence has demonstrated that lncRNAs may interact with miRNAs as ceRNAs and regulate gene expression at the posttranscriptional level. Several lncRNAs, such as lncRNA TUG1, lncRNA HULC, lncRNA H19, PVT1 and LINC01296 have been identified as ceRNAs in CCA[64,102]. Furthermore, another oncogenic lncRNA, SPRY4-IT1, interacts with EZH2, lysine specific demethylase 1 and DNA methyltransferase 1 serving as a molecular scaffold[64]. In addition, some lncRNAs, such as SNHG1, directly interact with EZH2 and regulate gene transcription[103].
Exosomal MALAT-1 was upregulated in serum isolated EVs from CCA patients. Mechanistically, MALAT-1 acted as a ceRNA via miR-204-dependent CXCR4 regulation[65]. Furthermore, Wang et al[104] supported that MALAT-1 exerted its oncogenic functions in CCA by activating PI3K/Akt pathway. Zinc fingers and homeoboxes 1 (ZHX1) functions as a transcription repressor binding DNA methyltransferase 3B; ZHX1 was associated with CCA development and metastasis[105]. A recent study supported that suppression of the lncRNA MALAT1/miR-199a/ZHX1 axis inhibited glioblastoma progression[106]. Given that exosomal MALAT-1 and miR-199a roles in CCA have been previously described, future studies could investigate the potential role of MALAT1 in ZHX1 regulation by sponging miR-199a. The lncRNAs, ENST00000588480.1 and ENST00000517758.1 have been associated with promoting CCA development via the p53 signaling pathway[63].
The functions and underlying molecular mechanisms of circRNAs in cancer have gained increasing scientific interest. Some of the proposed mechanisms include their ability to (a) regulate gene transcription by binding to RNA polymerase II; (b) alter protein activity; and c) act as ceRNAs[107].
The oncogenic role of circ_0005230 in CCA was first described by Xu et al[78], who proposed its role as a ceRNA by sponging miR-1238 and miR-1299. Circ_0005230 was previously associated with an unfavorable prognosis of breast cancer patients; reportedly, it can act as a miR-618 sponge and thus enhance CBX8 expression[108]. Increasing evidence supports the role of hsa_circ_0001649 as a tumor suppressor[74-77]. Its expression has been correlated with the ERK and Wnt/β-catenin pathway[77]. Matrix metalloproteinases play crucial roles in several cancer-related processes such as tumor neovascularization and metastasis[109]. Matrix metalloproteinase-9 was significantly regulated by hsa_circ_0001649 expression in CCA cells[74].
Concerning exosomal circRNAs in CCA, circ-0000284 was identified as a ceRNA, directly binding to miRNA-637 and thus stimulating LY6E expression[79]. Finally, the newly identified circ-CCAC1 promoted CCA progression via YY1 upregulation by sponging miR-514a-5p. YY1 is a transcription factor and gene target of miR-514a-5p. The authors suggested positive correlations among circ-CCAC1, YY1 and CAMLG expression levels[80].
Exosomes and their diverse cargos represent a relatively novel and very promising field of investigation in cancer research. Their distinct properties have been valued by scholars, and they have currently become a research hotspot. CCA cells harbor specific ncRNA expression profiles transferred by EVs to neighboring or distant cells. Exosomal secretion into bodily fluids offers a novel, non-invasive liquid biopsy approach by detecting specific ncRNA profiles in serum or urine. Interestingly, although glomerular infiltration seems to affect the number of detectable ncRNAs in urine, some ncRNAs were found to be significantly upregulated in urine EVs isolated from patients with CCA[53]. These findings indicate a potential diagnostic value as noninvasive biomarkers for CCA. Moreover, we suggest that further research should focus on exosomal ncRNA signatures that have been associated with early stage CCA detection[47,48,58,66], especially because late stage disease is not amenable to curative treatments.
Although EV isolation and characterization techniques have been improved in recent years, several concerns still arise regarding the accurate extraction and quantification of exosomal ncRNAs. Thus, caution in interpreting current research results is clearly warranted. With regard to CCA studies, several points should be taken into consideration to explain the conflicting results[49]: The relatively small number of published studies; the low patient sample size; and the different specimen collection, storage and processing procedures. This contradiction may also be attributed to the different ncRNA expression patterns among tissues and fluids of different origin. Moreover, despite exosomes being considered relatively stable[110], there is still a need for developing universal methodologies in order to establish reproducible and reliable data.
Another topic of great importance in CCA management is associated with the role of systemic therapy. Despite insufficient response, chemotherapy remains the mainstay for patients with advanced CCA. Gemcitabine, cisplatin, 5-fluorouracil and oxaliplatin represent the chemotherapy agents that are still included in current therapeutic options[111]. Targeted and immunotherapy agents have also shown some potential in specific patient subpopulations. A comprehensive assessment of interpatient and intratumor heterogeneity is essential in order to understand the underlying drug resistance mechanisms and move towards a more personalized approach. In this context, exosomal ncRNAs may serve as biomarkers associated with response to several therapeutic regimens. There are currently only a few studies that have investigated the potential role of exosomal miRNAs as monitoring biomarkers in cancer treatment. For instance, a recent study suggested that plasma exosomal miR-125b expression may act as a tool for the early detection of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients[112]. Similarly, Wei et al[113] reported that exosomal miR-222-3p played an important role in gemcitabine resistance by targeting SOCS3 in non-small cell lung cancer patients. Another exosomal miRNA, miR-425-3p was also proposed as a potential prognostic marker in cisplatin-resistant non-small cell lung cancer patients[114]. Exosome-transferred miR-199a-3p mimics seemed to reverse cisplatin resistance in hepatocellular carcinoma cells[101]. However, to the best of our knowledge, no study has investigated exosome-derived ncRNA signatures in CCA drug resistance. Therefore, additional research in this field is needed in order to identify new biomarkers.
The high potential of exosomes in CCA diagnosis, prognosis and therapeutic approaches has gained considerable research interest. In line with the observation from Kitdumrongthum et al[47], one of the most promising therapeutic strategies for exosomes is the inhibition of oncogenic ncRNAs. Selectively loaded exosomes could serve as vehicles for the targeted delivery of molecules with antitumor properties (antioncogenic ncRNA agents). To expand this perspective, such molecules may also include ncRNAs that mimic endogenous, tumor suppressing ncRNAs. Indeed, Ota et al[50] suggested that miR-30e encapsulated in EVs may suppress CCA metastasis by inhibiting EMT and thus could serve as a therapeutic agent. Similarly, EV-mediated miR-195 transfer was found to target tumor cells and inhibit CCA cell proliferation in a rat model[52]. An interesting procedure has already been described and includes the isolation and insertion of designated therapeutic biomolecules into exosomes. Then, the modified exosomes are reintroduced to the patient and regulated cellular functions that inhibited tumor growth[115]. From a safety perspective, autologous EV administration has been well tolerated, exhibiting mild inflammatory responses[116].
Moreover, the design and efficacy of exosome-based therapeutic approaches demands comprehensive understanding of exosome pharmacokinetics. So far, the in vivo tracking of exosomes includes fluorescence labeling or radiolabeling methods[117]. The half-life of exosomes and EVs have been reported in the literature[118,119]. It is interesting to mention that exosomes isolated from patient fluids or tissues have longer circulation times due to low immunogenicity[120]. However, there are several factors that affect exosome concentration in systematic circulation and target tissues. Apart from the route of administration and dose, the pharmacokinetic parameters of exosomes depend on individual genetic variations, blood flow, organ volume and clearance[121,122]. Several bioengineering strategies, including polyethylene glycol-based formulations, have been proposed in order to improve the exosome half-life in circulation and target tissues[123]. Further research focusing on tissue-specific analytical methods towards dose individualization is needed due to variations in exosome absorption, distribution, metabolism and elimination.
Considering the above, an optimal drug delivery system should be able to escape immune defense mechanisms, transfer the incorporated cargo selectively to tumor sites and display minimum toxicity to normal tissues. Exosomes, as a natural body product, can avoid phagocytosis, enter target cells and escape degradation by lysosomes with limited immune response[124]. In general, EV tropism depends on the nature of the progenitor cell[125]. Thus, the appropriate exosome selection for engineering is crucial. Indeed, an increasing number of studies have proposed EV modification towards enhancing targeted drug delivery and anticancer efficacy. For instance, paclitaxel-loaded EVs from prostate cancer cells displayed improved efficacy, and the targeted delivery was partially attributed to surface proteins. Moreover, the authors suggested that the use of autologous cancer cell-derived EVs offer an advantage because they are taken up by both parent cells and other cells in the tumor microenvironment[126]. Accordingly, genetic modification of MSCs-derived and dendritic cell-derived exosomes has been a subject of extensive research in several tumor types[127-131]. Various methods for EV engineering have been described; electroporation for miRNA loading and incubation for loading chemotherapy drugs into exosomes are two of the most commonly used techniques[132].
In conclusion, despite that numerous studies have suggested possible therapeutic applications for exosomes, very few clinical trials have been conducted until now. Indeed, our brief search in ClinicalTrials.gov did not reveal any clinical trial using exosomes for CCA treatment. On the other hand, there are very few interventional clinical trials (phase I-II) investigating the role of exosomes as drug-delivery microsystems in the treatment of other types of cancer[133-136]. This may be attributed to the fact that there are still several questions that need to be answered, such as the exact recipient cell uptake processes of exosomes in vivo. Nevertheless, we did identify one prospective observational study aiming to characterize TD exosomal ncRNAs as potential biomarkers in CCA[137]. Finally, it is worth mentioning that one recently added interventional clinical trial aims to investigate exosomal PD-L1 and miRNA expression profiles in non-small cell lung cancer patients receiving immunotherapy[138].
Exosomal ncRNAs represent a fast growing and promising field of current cancer research. CCA-derived exosomes are loaded with unique ncRNA signatures that may serve as important tools for the early diagnosis and prognosis of cholangiocarcinoma. Further, targeted, large-scale research is needed in order to identify new diagnostic and prognostic biomarkers with acceptable sensitivity and specificity in CCA. Although the concept of exosomes as delivery “nanosystems” of antitumor biomolecules is very recent, it provides a very promising prospect in CCA treatment due to the limited treatment options for this type of cancer.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cui Y, Ullah M S-Editor: Wang JL L-Editor: Filipodia P-Editor: Zhang YL
| 1. | Blechacz B, Komuta M, Roskams T, Gores GJ. Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:512-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 533] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 2. | Bergquist A, von Seth E. Epidemiology of cholangiocarcinoma. Best Pract Res Clin Gastroenterol. 2015;29:221-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 290] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 3. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 836] [Article Influence: 104.5] [Reference Citation Analysis (2)] |
| 4. | Sato K, Glaser S, Alvaro D, Meng F, Francis H, Alpini G. Cholangiocarcinoma: novel therapeutic targets. Expert Opin Ther Targets. 2020;24:345-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 5. | Blechacz B. Cholangiocarcinoma: Current Knowledge and New Developments. Gut Liver. 2017;11:13-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 346] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 6. | Macias RI. Cholangiocarcinoma: Biology, Clinical Management, and Pharmacological Perspectives. ISRN Hepatol. 2014;2014:828074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 7. | Feng X, Zheng S, Xia F, Ma K, Wang S, Bie P, Dong J. Classification and management of hepatolithiasis: A high-volume, single-center's experience. Intractable Rare Dis Res. 2012;1:151-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (2)] |
| 8. | Lazaridis KN, LaRusso NF. Primary Sclerosing Cholangitis. N Engl J Med. 2016;375:1161-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 358] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 9. | Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology. 2011;54:173-184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 722] [Cited by in RCA: 689] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 10. | Khan AS, Dageforde LA. Cholangiocarcinoma. Surg Clin North Am. 2019;99:315-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 143] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 11. | Walter D, Hartmann S, Waidmann O. Update on cholangiocarcinoma: potential impact of genomic studies on clinical management. Z Gastroenterol. 2017;55:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 967] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 13. | Macias RIR, Kornek M, Rodrigues PM, Paiva NA, Castro RE, Urban S, Pereira SP, Cadamuro M, Rupp C, Loosen SH, Luedde T, Banales JM. Diagnostic and prognostic biomarkers in cholangiocarcinoma. Liver Int. 2019;39 Suppl 1:108-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 14. | Tkach M, Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1828] [Cited by in RCA: 2569] [Article Influence: 285.4] [Reference Citation Analysis (0)] |
| 15. | Wang JP, Tang YY, Fan CM, Guo C, Zhou YH, Li Z, Li XL, Li Y, Li GY, Xiong W, Zeng ZY, Xiong F. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2018;9:12487-12502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Hirsova P, Ibrahim SH, Verma VK, Morton LA, Shah VH, LaRusso NF, Gores GJ, Malhi H. Extracellular vesicles in liver pathobiology: Small particles with big impact. Hepatology. 2016;64:2219-2233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 187] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 17. | Abels ER, Breakefield XO. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol. 2016;36:301-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 724] [Cited by in RCA: 1236] [Article Influence: 137.3] [Reference Citation Analysis (0)] |
| 18. | Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4900] [Cited by in RCA: 6151] [Article Influence: 512.6] [Reference Citation Analysis (0)] |
| 19. | Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14:14240-14269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 406] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 20. | Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Krämer-Albers EM, Laitinen S, Lässer C, Lener T, Ligeti E, Linē A, Lipps G, Llorente A, Lötvall J, Manček-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-'t Hoen EN, Nyman TA, O'Driscoll L, Olivan M, Oliveira C, Pállinger É, Del Portillo HA, Reventós J, Rigau M, Rohde E, Sammar M, Sánchez-Madrid F, Santarém N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3959] [Cited by in RCA: 4122] [Article Influence: 412.2] [Reference Citation Analysis (0)] |
| 21. | Dilsiz N. Role of exosomes and exosomal microRNAs in cancer. Future Sci OA. 2020;6:FSO465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 22. | Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1505] [Cited by in RCA: 1404] [Article Influence: 234.0] [Reference Citation Analysis (0)] |
| 23. | Fontana S, Giallombardo M, Alessandro R. Technical aspects for the evaluation of exosomes and their content. In: Russo A, Giordano A, Rolfo C, editors. Liquid Biopsy in Cancer Patients. Cham: Humana Press, 2017: 61–70. |
| 24. | Severino V, Dumonceau JM, Delhaye M, Moll S, Annessi-Ramseyer I, Robin X, Frossard JL, Farina A. Extracellular Vesicles in Bile as Markers of Malignant Biliary Stenoses. Gastroenterology 2017; 153: 495-504. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 25. | Yu Z, Zhao S, Ren L, Wang L, Chen Z, Hoffman RM, Zhou J. Pancreatic cancer-derived exosomes promote tumor metastasis and liver pre-metastatic niche formation. Oncotarget. 2017;8:63461-63483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 26. | Yi H, Ye J, Yang XM, Zhang LW, Zhang ZG, Chen YP. High-grade ovarian cancer secreting effective exosomes in tumor angiogenesis. Int J Clin Exp Pathol. 2015;8:5062-5070. [PubMed] |
| 27. | Tang MKS, Yue PYK, Ip PP, Huang RL, Lai HC, Cheung ANY, Tse KY, Ngan HYS, Wong AST. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat Commun. 2018;9:2270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 176] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 28. | Whiteside TL. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv Clin Chem. 2016;74:103-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 572] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 29. | Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-miRNAs as a New Tool for Liquid Biopsy in Lung Cancer. Cancers (Basel). 2019;11:888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 30. | Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci. 2016;37:606-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 386] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 31. | Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell. 2019;49:347-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 898] [Article Influence: 179.6] [Reference Citation Analysis (0)] |
| 32. | Maji S, Matsuda A, Yan IK, Parasramka M, Patel T. Extracellular vesicles in liver diseases. Am J Physiol Gastrointest Liver Physiol. 2017;312:G194-G200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 33. | Masyuk AI, Huang BQ, Ward CJ, Gradilone SA, Banales JM, Masyuk TV, Radtke B, Splinter PL, LaRusso NF. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol. 2010;299:G990-G999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 34. | Haga H, Yan IK, Takahashi K, Wood J, Zubair A, Patel T. Tumour cell-derived extracellular vesicles interact with mesenchymal stem cells to modulate the microenvironment and enhance cholangiocarcinoma growth. J Extracell Vesicles. 2015;4:24900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 35. | Park J, Tadlock L, Gores GJ, Patel T. Inhibition of interleukin 6-mediated mitogen-activated protein kinase activation attenuates growth of a cholangiocarcinoma cell line. Hepatology. 1999;30:1128-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 163] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 36. | Chen JH, Xiang JY, Ding GP, Cao LP. Cholangiocarcinoma-derived exosomes inhibit the antitumor activity of cytokine-induced killer cells by down-regulating the secretion of tumor necrosis factor-α and perforin. J Zhejiang Univ Sci B. 2016;17:537-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, Erice O, Gonzalez E, Jimenez-Agüero R, Lacasta A, Ibarra C, Sanchez-Campos A, Jimeno JP, Lammert F, Milkiewicz P, Marzioni M, Macias RIR, Marin JJG, Patel T, Gores GJ, Martinez I, Elortza F, Falcon-Perez JM, Bujanda L, Banales JM. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. 2017;66:1125-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 225] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 38. | Dutta S, Reamtong O, Panvongsa W, Kitdumrongthum S, Janpipatkul K, Sangvanich P, Piyachaturawat P, Chairoungdua A. Proteomics profiling of cholangiocarcinoma exosomes: A potential role of oncogenic protein transferring in cancer progression. Biochim Biophys Acta. 2015;1852:1989-1999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 39. | Weeraphan C, Phongdara A, Chaiyawat P, Diskul-Na-Ayudthaya P, Chokchaichamnankit D, Verathamjamras C, Netsirisawan P, Yingchutrakul Y, Roytrakul S, Champattanachai V, Svasti J, Srisomsap C. Phosphoproteome Profiling of Isogenic Cancer Cell-Derived Exosome Reveals HSP90 as a Potential Marker for Human Cholangiocarcinoma. Proteomics. 2019;19:e1800159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 40. | Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126:1163-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 269] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 41. | Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 765] [Cited by in RCA: 808] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 42. | Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 305] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 43. | Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZ, Ren XB, Cao S. MiR-205 in cancer: an angel or a devil? Eur J Cell Biol. 2013;92:54-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 44. | Li J, Hu K, Gong G, Zhu D, Wang Y, Liu H, Wu X. Upregulation of MiR-205 transcriptionally suppresses SMAD4 and PTEN and contributes to human ovarian cancer progression. Sci Rep. 2017;7:41330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 45. | Su N, Qiu H, Chen Y, Yang T, Yan Q, Wan X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol Rep. 2013;29:2297-2302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 46. | Okamoto K, Miyoshi K, Murawaki Y. miR-29b, miR-205 and miR-221 enhance chemosensitivity to gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS One. 2013;8:e77623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 47. | Kitdumrongthum S, Metheetrairut C, Charoensawan V, Ounjai P, Janpipatkul K, Panvongsa W, Weerachayaphorn J, Piyachaturawat P, Chairoungdua A. Dysregulated microRNA expression profiles in cholangiocarcinoma cell-derived exosomes. Life Sci. 2018;210:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 48. | Shen L, Chen G, Xia Q, Shao S, Fang H. Exosomal miR-200 family as serum biomarkers for early detection and prognostic prediction of cholangiocarcinoma. Int J Clin Exp Pathol. 2019;12:3870-3876. [PubMed] |
| 49. | Li L, Masica D, Ishida M, Tomuleasa C, Umegaki S, Kalloo AN, Georgiades C, Singh VK, Khashab M, Amateau S, Li Z, Okolo P, Lennon AM, Saxena P, Geschwind JF, Schlachter T, Hong K, Pawlik TM, Canto M, Law J, Sharaiha R, Weiss CR, Thuluvath P, Goggins M, Shin EJ, Peng H, Kumbhari V, Hutfless S, Zhou L, Mezey E, Meltzer SJ, Karchin R, Selaru FM. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60:896-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 180] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 50. | Ota Y, Takahashi K, Otake S, Tamaki Y, Okada M, Aso K, Makino Y, Fujii S, Ota T, Haneda M. Extracellular vesicle-encapsulated miR-30e suppresses cholangiocarcinoma cell invasion and migration via inhibiting epithelial-mesenchymal transition. Oncotarget. 2018;9:16400-16417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 51. | Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 2167] [Article Influence: 135.4] [Reference Citation Analysis (0)] |
| 52. | Li L, Piontek K, Ishida M, Fausther M, Dranoff JA, Fu R, Mezey E, Gould SJ, Fordjour FK, Meltzer SJ, Sirica AE, Selaru FM. Extracellular vesicles carry microRNA-195 to intrahepatic cholangiocarcinoma and improve survival in a rat model. Hepatology. 2017;65:501-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 53. | Lapitz A, Arbelaiz A, O'Rourke CJ, Lavin JL, Casta A, Ibarra C, Jimeno JP, Santos-Laso A, Izquierdo-Sanchez L, Krawczyk M, Perugorria MJ, Jimenez-Aguero R, Sanchez-Campos A, Riaño I, Gónzalez E, Lammert F, Marzioni M, Macias RIR, Marin JJG, Karlsen TH, Bujanda L, Falcón-Pérez JM, Andersen JB, Aransay AM, Rodrigues PM, Banales JM. Patients with Cholangiocarcinoma Present Specific RNA Profiles in Serum and Urine Extracellular Vesicles Mirroring the Tumor Expression: Novel Liquid Biopsy Biomarkers for Disease Diagnosis. Cells. 2020;9:721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 54. | Chang W, Wang Y, Li W, Shi L, Geng Z. MicroRNA-551b-3p inhibits tumour growth of human cholangiocarcinoma by targeting Cyclin D1. J Cell Mol Med. 2019;23:4945-4954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 55. | Swierniak M, Wojcicka A, Czetwertynska M, Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M, Koperski L, de la Chapelle A, Jazdzewski K. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98:E1401-E1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 113] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 56. | Wei Z, Liu Y, Wang Y, Zhang Y, Luo Q, Man X, Wei F, Yu X. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med Oncol. 2016;33:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 57. | Yuan H, Chen Z, Bai S, Wei H, Wang Y, Ji R, Guo Q, Li Q, Ye Y, Wu J, Zhou Y, Qiao L. Molecular mechanisms of lncRNA SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett. 2018;418:84-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 58. | Xue XY, Liu YX, Wang C, Gu XJ, Xue ZQ, Zang XL, Ma XD, Deng H, Liu R, Pan L, Liu SH. Identification of exosomal miRNAs as diagnostic biomarkers for cholangiocarcinoma and gallbladder carcinoma. Signal Transduct Target Ther. 2020;5:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 59. | Zhang M, Yang H, Wan L, Wang Z, Wang H, Ge C, Liu Y, Hao Y, Zhang D, Shi G, Gong Y, Ni Y, Wang C, Zhang Y, Xi J, Wang S, Shi L, Zhang L, Yue W, Pei X, Liu B, Yan X. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol. 2020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 356] [Article Influence: 71.2] [Reference Citation Analysis (0)] |
| 60. | Wei YQ, Jiao XL, Zhang SY, Xu Y, Li S, Kong BH. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci. 2019;23:7314-7326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 61. | Li G, Liu T, Zhang B, Chen W, Ding Z. Genome-wide identification of a competing endogenous RNA network in cholangiocarcinoma. J Cell Biochem. 2019;120:18995-19003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 62. | Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 319] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 63. | Ge X, Wang Y, Nie J, Li Q, Tang L, Deng X, Wang F, Xu B, Wu X, Zhang X, You Q, Miao L. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget. 2017;8:69995-70005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 64. | Jiang F, Ling X. The Advancement of Long Non-Coding RNAs in Cholangiocarcinoma Development. J Cancer. 2019;10:2407-2414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 65. | Tan X, Huang Z, Li X. Long Non-Coding RNA MALAT1 Interacts With miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. J Cell Biochem. 2017;118:3643-3653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 66. | Shi J, Li X, Zhang F, Kong L, Zhang X, Cheng Y, Guan Q, Cao X, Zhu W, Ou K, Chen Q, Hu S. The Plasma LncRNA Acting as Fingerprint in Hilar Cholangiocarcinoma. Cell Physiol Biochem. 2018;49:1694-1702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 67. | Tang Z, Wei G, Zhang L, Xu Z. Signature microRNAs and long noncoding RNAs in laryngeal cancer recurrence identified using a competing endogenous RNA network. Mol Med Rep. 2019;19:4806-4818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 68. | Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1303] [Cited by in RCA: 1749] [Article Influence: 174.9] [Reference Citation Analysis (0)] |
| 69. | Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 1295] [Article Influence: 143.9] [Reference Citation Analysis (0)] |
| 70. | Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 518] [Cited by in RCA: 609] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 71. | Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1218] [Cited by in RCA: 1616] [Article Influence: 161.6] [Reference Citation Analysis (0)] |
| 72. | Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1619] [Cited by in RCA: 2219] [Article Influence: 221.9] [Reference Citation Analysis (0)] |
| 73. | Jiang XM, Li ZL, Li JL, Xu Y, Leng KM, Cui YF, Sun DJ. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. Eur Rev Med Pharmacol Sci. 2018;22:365-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 39] [Reference Citation Analysis (0)] |
| 74. | Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L, Cui Y, Jiang X. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 75. | Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 367] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 76. | Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 77. | Sun H, Wang Q, Yuan G, Quan J, Dong D, Lun Y, Sun B. Hsa_circ_0001649 restrains gastric carcinoma growth and metastasis by downregulation of miR-20a. J Clin Lab Anal. 2020;34:e23235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 78. | Xu Y, Yao Y, Liu Y, Wang Z, Hu Z, Su Z, Li C, Wang H, Jiang X, Kang P, Sun D, Zhong X, Cui Y. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY). 2019;11:1907-1917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 79. | Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, Sun Y, Su Y. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci (Lond). 2019;133:1935-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 80. | Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, Shi G, Ji D, Huang P, Zheng W, Li Z, Li J, Huang L, Yu L, Zhou Y, Jiang X, Wang H, Li C, Su Z, Tai S, Zhong X, Wang Z, Cui Y. A novel circular RNA, circ-CCAC1, contributes to CCA progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 81. | Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709-1714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 623] [Cited by in RCA: 672] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 82. | Fagegaltier D, Falciatori I, Czech B, Castel S, Perrimon N, Simcox A, Hannon GJ. Oncogenic transformation of Drosophila somatic cells induces a functional piRNA pathway. Genes Dev. 2016;30:1623-1635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 83. | Wang Z, Liu N, Shi S, Liu S, Lin H. The Role of PIWIL4, an Argonaute Family Protein, in Breast Cancer. J Biol Chem. 2016;291:10646-10658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 84. | Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, Tsai S, Kohli M, Boardman L, Patel T, Wang L. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 210] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 85. | Weng W, Liu N, Toiyama Y, Kusunoki M, Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y, Goel A. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. 2018;17:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 86. | Tan C, Cao J, Chen L, Xi X, Wang S, Zhu Y, Yang L, Ma L, Wang D, Yin J, Zhang T, John Lu Z. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin Chem. 2019;65:905-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 87. | Chen YJ, Xiong XF, Wen SQ, Tian L, Cheng WL, Qi YQ. Expression and clinical significance of PIWIL2 in hilar cholangiocarcinoma tissues and cell lines. Genet Mol Res. 2015;14:7053-7061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 88. | Gu X, Wang C, Deng H, Qing C, Liu R, Liu S, Xue X. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim Biophys Sin (Shanghai). 2020;52:475-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 89. | Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27:378-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 183] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 90. | An F, Yamanaka S, Allen S, Roberts LR, Gores GJ, Pawlik TM, Xie Q, Ishida M, Mezey E, Ferguson-Smith AC, Mori Y, Selaru FM. Silencing of miR-370 in human cholangiocarcinoma by allelic loss and interleukin-6 induced maternal to paternal epigenotype switch. PLoS One. 2012;7:e45606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 91. | Chen YJ, Luo J, Yang GY, Yang K, Wen SQ, Zou SQ. Mutual regulation between microRNA-373 and methyl-CpG-binding domain protein 2 in hilar cholangiocarcinoma. World J Gastroenterol. 2012;18:3849-3861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 92. | Iwaki J, Kikuchi K, Mizuguchi Y, Kawahigashi Y, Yoshida H, Uchida E, Takizawa T. MiR-376c down-regulation accelerates EGF-dependent migration by targeting GRB2 in the HuCCT1 human intrahepatic cholangiocarcinoma cell line. PLoS One. 2013;8:e69496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 93. | Razumilava N, Gores GJ. Notch-driven carcinogenesis: the merging of hepatocellular cancer and cholangiocarcinoma into a common molecular liver cancer subtype. J Hepatol. 2013;58:1244-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 94. | Kwon H, Song K, Han C, Zhang J, Lu L, Chen W, Wu T. Epigenetic Silencing of miRNA-34a in Human Cholangiocarcinoma via EZH2 and DNA Methylation: Impact on Regulation of Notch Pathway. Am J Pathol. 2017;187:2288-2299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 95. | Wan P, Zhang J, Du Q, Geller DA. The clinical significance and biological function of interferon regulatory factor 1 in cholangiocarcinoma. Biomed Pharmacother. 2018;97:771-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 96. | Wan P, Chi X, Du Q, Luo J, Cui X, Dong K, Bing Y, Heres C, Geller DA. miR-383 promotes cholangiocarcinoma cell proliferation, migration, and invasion through targeting IRF1. J Cell Biochem. 2018;119:9720-9729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 97. | Goeppert B, Ernst C, Baer C, Roessler S, Renner M, Mehrabi A, Hafezi M, Pathil A, Warth A, Stenzinger A, Weichert W, Bähr M, Will R, Schirmacher P, Plass C, Weichenhan D. Cadherin-6 is a putative tumor suppressor and target of epigenetically dysregulated miR-429 in cholangiocarcinoma. Epigenetics. 2016;11:780-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 98. | Oltra SS, Peña-Chilet M, Vidal-Tomas V, Flower K, Martinez MT, Alonso E, Burgues O, Lluch A, Flanagan JM, Ribas G. Methylation deregulation of miRNA promoters identifies miR124-2 as a survival biomarker in Breast Cancer in very young women. Sci Rep. 2018;8:14373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 99. | Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2843] [Cited by in RCA: 3080] [Article Influence: 181.2] [Reference Citation Analysis (0)] |
| 100. | Li Q, Xia X, Ji J, Ma J, Tao L, Mo L, Chen W. MiR-199a-3p enhances cisplatin sensitivity of cholangiocarcinoma cells by inhibiting mTOR signaling pathway and expression of MDR1. Oncotarget. 2017;8:33621-33630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 101. | Zhang K, Shao CX, Zhu JD, Lv XL, Tu CY, Jiang C, Shang MJ. Exosomes function as nanoparticles to transfer miR-199a-3p to reverse chemoresistance to cisplatin in hepatocellular carcinoma. Biosci Rep. 2020;40:BSR20194026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 102. | Xu F, Zhao Y, Qin G, Huan Y, Li L, Gao W. Comprehensive analysis of competing endogenous RNA networks associated with cholangiocarcinoma. Exp Ther Med. 2019;18:4103-4112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 103. | Yu Y, Zhang M, Wang N, Li Q, Yang J, Yan S, He X, Ji G, Miao L. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell Death Dis. 2018;9:746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 104. | Wang C, Mao ZP, Wang L, Wu GH, Zhang FH, Wang DY, Shi JL. Long non-coding RNA MALAT1 promotes cholangiocarcinoma cell proliferation and invasion by activating PI3K/Akt pathway. Neoplasma. 2017;64:725-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 105. | Kwon RJ, Han ME, Kim JY, Liu L, Kim YH, Jung JS, Oh SO. ZHX1 Promotes the Proliferation, Migration and Invasion of Cholangiocarcinoma Cells. PLoS One. 2016;11:e0165516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 106. | Liao K, Lin Y, Gao W, Xiao Z, Medina R, Dmitriev P, Cui J, Zhuang Z, Zhao X, Qiu Y, Zhang X, Ge J, Guo L. Blocking lncRNA MALAT1/miR-199a/ZHX1 Axis Inhibits Glioblastoma Proliferation and Progression. Mol Ther Nucleic Acids. 2019;18:388-399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 107. | Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and Exosomes: A Mysterious Frontier for Human Cancer. Mol Ther Nucleic Acids. 2020;19:384-392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 108. | Xu Y, Yao Y, Leng K, Ji D, Qu L, Liu Y, Cui Y. Increased Expression of Circular RNA circ_0005230 Indicates Dismal Prognosis in Breast Cancer and Regulates Cell Proliferation and Invasion via miR-618/ CBX8 Signal Pathway. Cell Physiol Biochem. 2018;51:1710-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 109. | Quintero-Fabián S, Arreola R, Becerril-Villanueva E, Torres-Romero JC, Arana-Argáez V, Lara-Riegos J, Ramírez-Camacho MA, Alvarez-Sánchez ME. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front Oncol. 2019;9:1370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 627] [Article Influence: 104.5] [Reference Citation Analysis (0)] |
| 110. | Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 223] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 111. | Benson AB 3rd, D'Angelica MI, Abbott DE, Abrams TA, Alberts SR, Saenz DA, Are C, Brown DB, Chang DT, Covey AM, Hawkins W, Iyer R, Jacob R, Karachristos A, Kelley RK, Kim R, Palta M, Park JO, Sahai V, Schefter T, Schmidt C, Sicklick JK, Singh G, Sohal D, Stein S, Tian GG, Vauthey JN, Venook AP, Zhu AX, Hoffmann KG, Darlow S. NCCN Guidelines Insights: Hepatobiliary Cancers, Version 1.2017. J Natl Compr Canc Netw. 2017;15:563-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 254] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 112. | Yagi T, Iinuma H, Hayama T, Matsuda K, Nozawa K, Tsukamoto M, Shimada R, Akahane T, Tsuchiya T, Ozawa T, Hashiguchi Y. Plasma exosomal microRNA-125b as a monitoring biomarker of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients. Mol Clin Oncol. 2019;11:416-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 113. | Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, Ding L, Zhang Y, Zhang L, Li N, Li Y, Liu Y. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 203] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 114. | Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan M, Xue W, Wang Y, Li W, Shu Y, Xu Q, Shen Y. Exosomal Transfer Of Cisplatin-Induced miR-425-3p Confers Cisplatin Resistance In NSCLC Through Activating Autophagy. Int J Nanomedicine. 2019;14:8121-8132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 93] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 115. | Samanta S, Rajasingh S, Drosos N, Zhou Z, Dawn B, Rajasingh J. Exosomes: new molecular targets of diseases. Acta Pharmacol Sin. 2018;39:501-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 315] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 116. | Escudier B, Dorval T, Chaput N, André F, Caby MP, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, Boccaccio C, Bonnerot C, Dhellin O, Movassagh M, Piperno S, Robert C, Serra V, Valente N, Le Pecq JB, Spatz A, Lantz O, Tursz T, Angevin E, Zitvogel L. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med. 2005;3:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 763] [Cited by in RCA: 1006] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 117. | Kim DH, Kothandan VK, Kim HW, Kim KS, Kim JY, Cho HJ, Lee YK, Lee DE, Hwang SR. Noninvasive Assessment of Exosome Pharmacokinetics In Vivo: A Review. Pharmaceutics. 2019;11:649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 118. | Pocsfalvi G, Stanly C, Vilasi A, Fiume I, Capasso G, Turiák L, Buzas EI, Vékey K. Mass spectrometry of extracellular vesicles. Mass Spectrom Rev. 2016;35:3-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 119. | Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire C, Chen JW, Tannous BA, Breakefield XO. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014;8:483-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 686] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 120. | Liu C, Su C. Design strategies and application progress of therapeutic exosomes. Theranostics. 2019;9:1015-1028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 307] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 121. | Gerlowski LE, Jain RK. Physiologically based pharmacokinetic modeling: principles and applications. J Pharm Sci. 1983;72:1103-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 389] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 122. | Rodrigues D, Rowland A. From Endogenous Compounds as Biomarkers to Plasma-Derived Nanovesicles as Liquid Biopsy; Has the Golden Age of Translational Pharmacokinetics-Absorption, Distribution, Metabolism, Excretion-Drug-Drug Interaction Science Finally Arrived? Clin Pharmacol Ther. 2019;105:1407-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 123. | Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen I, Klyachko NL, Kabanov AV, Batrakova EV. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine. 2018;14:195-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 498] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 124. | Bunggulawa EJ, Wang W, Yin T, Wang N, Durkan C, Wang Y, Wang G. Recent advancements in the use of exosomes as drug delivery systems. J Nanobiotechnology. 2018;16:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 391] [Cited by in RCA: 419] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 125. | Di Rocco G, Baldari S, Toietta G. Towards Therapeutic Delivery of Extracellular Vesicles: Strategies for In Vivo Tracking and Biodistribution Analysis. Stem Cells Int. 2016;2016:5029619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 126. | Saari H, Lázaro-Ibáñez E, Viitala T, Vuorimaa-Laukkanen E, Siljander P, Yliperttula M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J Control Release. 2015;220:727-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 473] [Article Influence: 47.3] [Reference Citation Analysis (1)] |
| 127. | Reza AMMT, Choi YJ, Yasuda H, Kim JH. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci Rep. 2016;6:38498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 174] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 128. | Ko SF, Yip HK, Zhen YY, Lee CC, Lee CC, Huang CC, Ng SH, Lin JW. Adipose-Derived Mesenchymal Stem Cell Exosomes Suppress Hepatocellular Carcinoma Growth in a Rat Model: Apparent Diffusion Coefficient, Natural Killer T-Cell Responses, and Histopathological Features. Stem Cells Int. 2015;2015:853506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 128] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 129. | Katakowski M, Buller B, Zheng X, Lu Y, Rogers T, Osobamiro O, Shu W, Jiang F, Chopp M. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335:201-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 589] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 130. | Pitt JM, André F, Amigorena S, Soria JC, Eggermont A, Kroemer G, Zitvogel L. Dendritic cell-derived exosomes for cancer therapy. J Clin Invest. 2016;126:1224-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 472] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 131. | Viaud S, Terme M, Flament C, Taieb J, André F, Novault S, Escudier B, Robert C, Caillat-Zucman S, Tursz T, Zitvogel L, Chaput N. Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: a role for NKG2D ligands and IL-15Ralpha. PLoS One. 2009;4:e4942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 354] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 132. | You B, Xu W, Zhang B. Engineering exosomes: a new direction for anticancer treatment. Am J Cancer Res. 2018;8:1332-1342. [PubMed] |
| 133. | Roussy G. Trial of a Vaccination With Tumor Antigen-loaded Dendritic Cell-derived Exosomes (CSET 1437). In: Clinical.Trials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT01159288 ClinicalTrials.gov Identifier: NCT01159288. |
| 134. | Redman R. Edible Plant Exosome Ability to Prevent Oral Mucositis Associated With Chemoradiation Treatment of Head and Neck Cancer. In: Clinical.Trials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT01668849 ClinicalTrials.gov Identifier: NCT01668849. |
| 135. | M.D. Anderson Cancer Center. iExosomes in Treating Participants With Metastatic Pancreas Cancer With KrasG12D Mutation. In: Clinical.Trials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT03608631 ClinicalTrials.gov Identifier: NCT03608631. |
| 136. | Miller D. Study Investigating the Ability of Plant Exosomes to Deliver Curcumin to Normal and Colon Cancer Tissue. In: Clinical.Trials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT01294072 ClinicalTrials.gov Identifier: NCT01294072. |
| 137. | The Second Hospital of Nanjing Medical University. ncRNAs in Exosomes of Cholangiocarcinoma. In: Clinical.Trials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT03102268 ClinicalTrials.gov Identifier: NCT03102268. |
| 138. | Wang J. Prediction of Immunotherapeutic Effect of Advanced Non-small Cell Lung Cancer. In: Clinical.Trials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT04427475 ClinicalTrials.gov Identifier: NCT04427475. |