Published online Apr 15, 2017. doi: 10.4239/wjd.v8.i4.130
Peer-review started: November 24, 2016
First decision: January 16, 2017
Revised: January 27, 2017
Accepted: February 18, 2017
Article in press: February 20, 2017
Published online: April 15, 2017
Processing time: 141 Days and 12.3 Hours
Natural killer T cells (NKT cells) are innate-like T cells that acquire effector functions while developing in the thymus, polarize into three distinct functional subsets viz. NKT1, NKT2 and NKT17 cells that produce interferon (IFN)-γ, interleukin (IL)-4 and IL-17, respectively. However, there has been no unique surface markers that define each subsets, forcing investigators to use intracellular staining of transcription factors and cytokines in combination of surface markers to distinguish among these subsets. Intracellular staining, however, causes apoptosis and prevents subsequent utilization of NKT cells in functional in vitro and in vivo assays that require viable cells. This limitation has significantly impeded understanding the specific properties of each subset and their interactions with each other. Therefore, there has been fervent efforts to find a specific markers for each NKT cell subset. We have recently identified that syndecan-1 (SDC-1; CD138) as a specific surface marker of NKT17 cells. This discovery now allows visualization of NKT17 in situ and study of their peripheral tissue distribution, characteristics of their TCR and viable sorting for in vitro and in vivo analysis. In addition, it lays the ground working for investigating significance of SDC-1 expression on this particular subset in regulating their roles in host defense and glucose metabolism.
Core tip: Discrete subsets of innate-like Natural killer T (NKT) cells differentially produce three of the most potent and polarizing cytokines, interferon-γ (NKT1), interleukin (IL)-4 (NKT2) and IL-17 (NKT17). But very little is known about how the relationship among the functional subsets of NKT cells is regulated. A major obstacle was the absence of specific single surface markers that reliably identify each subset. Here we highlight our discovery of syndecan-1 as a specific marker of NKT17 subset and its significance for understanding the role of NKT17 in glucose metabolism and autoimmunity.
- Citation: Jaiswal AK, Sadasivam M, Hamad ARA. Syndecan-1-coating of interleukin-17-producing natural killer T cells provides a specific method for their visualization and analysis. World J Diabetes 2017; 8(4): 130-134
- URL: https://www.wjgnet.com/1948-9358/full/v8/i4/130.htm
- DOI: https://dx.doi.org/10.4239/wjd.v8.i4.130
We have recently published that syndecan-1 (SDC-1; CD138) is specifically expressed on the interleukin (IL)-17-producing subset of natural killer cells (NKT17) cells[1]. Briefly, we have previously shown that SDC-1 is expressed on double-negative T cells (DN T cells) that accumulate in lpr and gld mice[2]. After that we sought to know if SDC-1 express in innate cells and detected SDC-1 in a subset of NKT cells. We sorted and analyzed NKT cells subsets by genome-wide gene profiling using microarrays and identified SDC-1 is specifically expressed on the IL-17-producing subset of NKT17 cells[1]. Using SDC-1 expression on NKT17 cells, we visualized their development in the thymus, analyzed their tissue distribution. In addition, we sorted NKT17 cells, out distinguish them from interferon (IFN)-γ-producing NKT1 cells, we sorted each subset and study their characteristics in vitro. In this article, we briefly review SDC-1 expression on immune cells and highlight our results and speculate on the potential role of sdc1 in regulating homeostasis of NKT cells and the implication for glucose homeostasis and body fat development.
SDC-1 is a heparan sulfate proteoglycans that is predominantly expressed on epithelial cells[3,4]. It is composed of a short conserved cytoplasmic domain, a transmembrane domain, and a long variable ectodomain carrying heparan sulfate (HS) glycosaminoglycan chains[5]. Sometimes, SDC-1 used chondroitin/dermatan sulfate beside or instead of HS chains[6]. SDC-1 mediates its functions primarily by using HS chains to bind different ligands[7,8]. These include various growth factors such as fibroblast growth factors, Wnt, vascular endothelial growth factor, hepatocyte growth factor, cell matrix proteins, growth factors, cytokines, and chemokines[3,4] and their receptors[9-12]. Ligand binding to HS is regulated at the cell surface by two sulfatases (SULF-1 and SULF-2) and heparanases[13]. Number, position, and orientation of each sulfate group on HS chains play a role in dictating the ability of SDC-1 to bind ligands and initiate downstream signaling events[14-19]. These regulatory sequences have been proposed to act with both autocrine and paracrine mechanisms and represent potential novel targets for therapeutic interventions, particularly against cancer[12]. In addition, recent discoveries indicate that SDC-1 core proteins also has biological functions and can modulate cell behavior independent of HS. In contrast, the transmembrane and cytoplasmic domains of SDC-1 do not have intrinsic kinase or catalytic activity, but yet play important roles in signal transduction pathway by multimerization and/or interaction with other intracellular components, like GTPases or kinases[20]. This often happens in lipid rafts, which are enriched in glycosphingolipids and cholesterol[21] and essential for receptor binding and signal transduction from the cell surface into the cell. In addition, the short cytoplasmic domain of SDC-1 interacts with a number of cytosolic proteins and plays a role in endocytosis.
Using these various mechanisms, SDC-1 regulates multiple cellular functions, including cell proliferation, differentiation, and survival of adherent cells and tumors. Expression of SDC-1 is dysregulated in a number of cancers, including head and neck, ovarian, breast, and colorectal carcinomas[22]. In addition, SDC-1 has been implicated in regulating whole body energy metabolism in Drosophila[23] and body fat in mice[24]. Role of SDC-1 in cancers, infectious diseases, obesity, wound healing, and angiogenesis were reviewed recently[9,22,25] and hence will not be discussed in depth here.
While ubiquitously expressed on epithelia and other adherent cells, expression of SDC-1 by the immune cells is limited to few cells as discussed below.
Expression in plasma and B cells: SDC-1 is a well known marker of plasma cells[3] and it has been reported on pre-B cells[26]. Other than that, SDC-1 is not known to be widely expressed among various normal immune cell types. SDC-1, however, is commonly expressed by myeloma cells and lymhopid malignancies and it has been implicated in survival, proliferation and metastasis of tumors[27]. But the exact roles of SDC-1 in the development and function of B cells and plasma cell remain poorly understood.
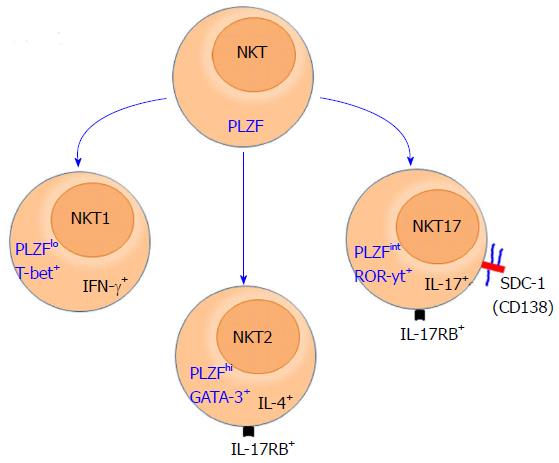
Specific expression of SDC-1 on NKT17 cells: Invariant NKT cells are highly conserved innate-like T cells that, unlike conventional T cells, are restricted to CD1d molecules and recognize glycolipids as antigens[28]. NKT cells acquire their effector functions while developing in the thymus[29] and differentiate into three distinct subsets that produce IFN-γ, IL-4 or IL-17 cytokine. These subsets were labelled in a manner typical to that of T helper cells (Th)1, (Th)2, and (Th17) cells[29,30]. Hence, the IFN-γ-producing subset is referred to as NKT1, the IL-4 producing subset as NKT2, and the subset that produces IL-17 as the NKT17 subset. Due to their innate nature, NKT cells rapidly produce copius amounts of these cytokines upon stimulation, thereby playing critical roles in the initiation and shaping of adaptive immune responses[29,31,32]. These cytokines are highly potent and capable of polarizing adaptive immune responses into Th1, Th2 or Th17 type. Furthermore, because of the ability of these cytokines to inhibit each other function, the overall physiological functions of NKT cells and how the opposing functions of the three subsets are reconciled under physiological and pathological condition remain a mystery. Lack of progress in solving this paradox is rooted in the absence of reliable surface markers that identify and distinguish subsets of NKT cells from one another.
Currently, distinguishing among the NKT subsets is made using intracellular staining for signature transcription factors that control production of IFN-γ (Tbet), IL-4 (PLZF and GATA3), and IL-17 [retinoic acid-related orphan receptor γt (RORγt)][4,33]. Additionally, NKT subsets are identified based on intracellular staining for their signature cytokine, IFN-γ (NKT1), IL-4 (NKT2) and IL-17 (NKT17). Otherwise, It has been difficult to definitively distinguish among NKT cell subsets. Intracellular staining, however, requires fixation and permeabilization, which is a serious limitation that abroagtes the ability of investigators to do in vitro functional analysis using purified invidudual subsets. It has also impeded in vivo tracking and characterization of individual NKT cell subsets and full appreciation of the pathophysiologic functions of each subset. To avoid this problem, a combination of surface markers are currently used for this purpose, but they have their own shortcomings. For example, NKT17 cells can be identified based on low expression of NK1.1 and CD4, and high expression of CCR6 and IL17RB[34]. However, IL-17RB is also expressed by NKT2 cells and expression of NK1.1 is a strain-dependent and absent in most mouse strains[35]. Our recent identification of SDC-1 as a specific marker of NKT17 cells overcome this challenge at least for this subset[1] (Figure 1). This finding has been confirmed by three independent studies[36-38]. This discovery now allows visualization of NKT17 in the thymus and their peripheral tissue distribution, which is leading to novel insights into NKT cell biology.
IL-17 is a potent proinflammatory cytokines that is required in host defense against infections[39] and been implicated in pathogenesis of asthma[40], and autoimmune diseases such as type 1 diabetes[41-43] and regulation of body fat[44]. In addition, IL-17 has been reported to modulate both adipogenesis and functions of adipocytes and glucose metabolism in mice[44,45]. Both IL-17AKO and IL-17RAKO mice has been reported to gain in weight due to the accumulation of visceral fat[44], suggesting involvement of IL-17 in maintaining body fat. NKT17 cells represent about 20% of NKT cells in the thymus[1] and approximately 2%-10% of total NKT cells in secondary lymphoid organs. NKT17 can secrete large amounts of IL-17 in response to various stimuli, such as infections, allergens, tissue injury and metabolic disorders[46,47].
Interestingly, NKT17 cells preferentially reside in visceral adipose tissue in mice[1] and their local and systemic frequencies are reduced in obese patients, suggesting their involvement in inflammation during obesity[48]. In addition, it has been reported that NKT17 could play a pathogenic role in the pathophysiology of diabetes[41]. Therefore, we speculate that studies addressing the roles of SDC-1 expressing NKT17 cell may provide an alternative approach to understanding its role in fat metabolism and glucose homeostasis. Thus, the findings by our group and subsequently other groups that NKT17 cells are identifiable by surface experssion SDC-1 is crucial for clear understanding of their biology and regulation and their physiologic role in thes steady state and disease condition[1,27,38]. For example, SDC-1 provides a unique opportunity for tracking and analysis of NKT17 cells in vivo and for sorting viable NKT17 for various in vitro functional studies and adoptive transfer experiments. In this regards, our findings of great responsiveness of NKT17 than do NKT1 cells is consistent with their preferential localization of NKT17 in white adipose tissue (WAT) and suggest special link to WAT.
The discovery of SDC-1 as specific marker for NKT17 cells laid the foundation for understanding the biology of NKT17 cells and their pathophysiologic functions. In addition, it will be helpful in uncovering specific markers for NKT1 and NKT2 by excluding NKT17 cells and sorting of pure NKT1 and NKT2 cells for gene expression profiling. Future studies are expected to develop into understanding the significance of selective expression of SDC-1 by NKT17 cells and generating new information into the role of SDC-1 in the immune cells, which can lead to develoment of new strategies for manipulating individual subsets of NKT cells for therapeutic purposes.
We thank all members in our lab for their contribution in preparation of this manuscript.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Wakao H, Zhao J S- Editor: Qiu S L- Editor: Ji FF E- Editor: Wu HL
| 1. | Dai H, Rahman A, Saxena A, Jaiswal AK, Mohamood A, Ramirez L, Noel S, Rabb H, Jie C, Hamad AR. Syndecan-1 identifies and controls the frequency of IL-17-producing naïve natural killer T (NKT17) cells in mice. Eur J Immunol. 2015;45:3045-3051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 2. | Mohamood AS, Bargatze D, Xiao Z, Jie C, Yagita H, Ruben D, Watson J, Chakravarti S, Schneck JP, Hamad AR. Fas-mediated apoptosis regulates the composition of peripheral alphabeta T cell repertoire by constitutively purging out double negative T cells. PLoS One. 2008;3:e3465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2114] [Cited by in RCA: 2118] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 4. | Khotskaya YB, Dai Y, Ritchie JP, MacLeod V, Yang Y, Zinn K, Sanderson RD. Syndecan-1 is required for robust growth, vascularization, and metastasis of myeloma tumors in vivo. J Biol Chem. 2009;284:26085-26095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Couchman JR. Syndecans: proteoglycan regulators of cell-surface microdomains? Nat Rev Mol Cell Biol. 2003;4:926-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 345] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 6. | Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 622] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 7. | Carey DJ. Syndecans: multifunctional cell-surface co-receptors. Biochem J. 1997;327:1-16. [PubMed] |
| 8. | Tumova S, Woods A, Couchman JR. Heparan sulfate proteoglycans on the cell surface: versatile coordinators of cellular functions. Int J Biochem Cell Biol. 2000;32:269-288. [PubMed] |
| 9. | Purushothaman A, Uyama T, Kobayashi F, Yamada S, Sugahara K, Rapraeger AC, Sanderson RD. Heparanase-enhanced shedding of syndecan-1 by myeloma cells promotes endothelial invasion and angiogenesis. Blood. 2010;115:2449-2457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 193] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 10. | Alexander CM, Reichsman F, Hinkes MT, Lincecum J, Becker KA, Cumberledge S, Bernfield M. Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat Genet. 2000;25:329-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 286] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 11. | Seidel C, Børset M, Hjertner O, Cao D, Abildgaard N, Hjorth-Hansen H, Sanderson RD, Waage A, Sundan A. High levels of soluble syndecan-1 in myeloma-derived bone marrow: modulation of hepatocyte growth factor activity. Blood. 2000;96:3139-3146. [PubMed] |
| 12. | De Rossi G, Whiteford JR. Novel insight into the biological functions of syndecan ectodomain core proteins. Biofactors. 2013;39:374-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Adhikari N, Carlson M, Lerman B, Hall JL. Changes in expression of proteoglycan core proteins and heparan sulfate enzymes in the developing and adult murine aorta. J Cardiovasc Transl Res. 2011;4:313-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | Wu X, Kan M, Wang F, Jin C, Yu C, McKeehan WL. A rare premalignant prostate tumor epithelial cell syndecan-1 forms a fibroblast growth factor-binding complex with progression-promoting ectopic fibroblast growth factor receptor 1. Cancer Res. 2001;61:5295-5302. [PubMed] |
| 15. | Derksen PW, de Gorter DJ, Meijer HP, Bende RJ, van Dijk M, Lokhorst HM, Bloem AC, Spaargaren M, Pals ST. The hepatocyte growth factor/Met pathway controls proliferation and apoptosis in multiple myeloma. Leukemia. 2003;17:764-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci USA. 2004;101:4158-4163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Szatmári T, Mundt F, Heidari-Hamedani G, Zong F, Ferolla E, Alexeyenko A, Hjerpe A, Dobra K. Novel genes and pathways modulated by syndecan-1: implications for the proliferation and cell-cycle regulation of malignant mesothelioma cells. PLoS One. 2012;7:e48091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 18. | Pap Z, Pávai Z, Dénes L, Kovalszky I, Jung J. An immunohistochemical study of colon adenomas and carcinomas: E-cadherin, Syndecan-1, Ets-1. Pathol Oncol Res. 2009;15:579-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Kato M, Wang H, Bernfield M, Gallagher JT, Turnbull JE. Cell surface syndecan-1 on distinct cell types differs in fine structure and ligand binding of its heparan sulfate chains. J Biol Chem. 1994;269:18881-18890. [PubMed] |
| 20. | Iba K, Albrechtsen R, Gilpin B, Fröhlich C, Loechel F, Zolkiewska A, Ishiguro K, Kojima T, Liu W, Langford JK. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. J Cell Biol. 2000;149:1143-1156. [PubMed] |
| 21. | Underhill GH, Kolli KP, Kansas GS. Complexity within the plasma cell compartment of mice deficient in both E- and P-selectin: implications for plasma cell differentiation. Blood. 2003;102:4076-4083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 22. | Teng YH, Aquino RS, Park PW. Molecular functions of syndecan-1 in disease. Matrix Biol. 2012;31:3-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 282] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 23. | De Luca M, Klimentidis YC, Casazza K, Chambers MM, Cho R, Harbison ST, Jumbo-Lucioni P, Zhang S, Leips J, Fernandez JR. A conserved role for syndecan family members in the regulation of whole-body energy metabolism. PLoS One. 2010;5:e11286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 24. | Kasza I, Suh Y, Wollny D, Clark RJ, Roopra A, Colman RJ, MacDougald OA, Shedd TA, Nelson DW, Yen MI. Syndecan-1 is required to maintain intradermal fat and prevent cold stress. PLoS Genet. 2014;10:e1004514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 25. | Nikolova V, Koo CY, Ibrahim SA, Wang Z, Spillmann D, Dreier R, Kelsch R, Fischgräbe J, Smollich M, Rossi LH. Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis. 2009;30:397-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 155] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 26. | Lee JG, Moon H, Park C, Shin SH, Kang K, Kim TJ. Reversible expression of CD138 on mature follicular B cells is downregulated by IL-4. Immunol Lett. 2013;156:38-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 27. | Dhodapkar MV, Sanderson RD. Syndecan-1 (CD 138) in myeloma and lymphoid malignancies: a multifunctional regulator of cell behavior within the tumor microenvironment. Leuk Lymphoma. 1999;34:35-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Zajonc DM, Cantu C, Mattner J, Zhou D, Savage PB, Bendelac A, Wilson IA, Teyton L. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6:810-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 256] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 29. | Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 30. | Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, Wong CH, Schneider E, Dy M, Leite-de-Moraes MC. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med. 2007;204:995-1001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 475] [Cited by in RCA: 483] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 31. | Godfrey DI, Hammond KJ, Poulton LD, Smyth MJ, Baxter AG. NKT cells: facts, functions and fallacies. Immunol Today. 2000;21:573-583. [PubMed] |
| 32. | Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12:845-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 364] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 33. | Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 494] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 34. | Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 161] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 35. | Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, Iwamura C, Nakajima H, Nakayama T, Taniguchi M. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J Exp Med. 2008;205:2727-2733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 193] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 36. | Georgiev H, Ravens I, Benarafa C, Förster R, Bernhardt G. Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat Commun. 2016;7:13116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 37. | Georgiev H, Ravens I, Shibuya A, Förster R, Bernhardt G. CD155/CD226-interaction impacts on the generation of innate CD8(+) thymocytes by regulating iNKT-cell differentiation. Eur J Immunol. 2016;46:993-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 38. | Lee YJ, Starrett GJ, Lee ST, Yang R, Henzler CM, Jameson SC, Hogquist KA. Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst γδ T, Innate Lymphoid, and Th Cells. J Immunol. 2016;197:1460-1470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 39. | Doisne JM, Soulard V, Bécourt C, Amniai L, Henrot P, Havenar-Daughton C, Blanchet C, Zitvogel L, Ryffel B, Cavaillon JM. Cutting edge: crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1.1- invariant NKT cells to bacteria. J Immunol. 2011;186:662-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 127] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 40. | Pichavant M, Goya S, Meyer EH, Johnston RA, Kim HY, Matangkasombut P, Zhu M, Iwakura Y, Savage PB, DeKruyff RH. Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J Exp Med. 2008;205:385-393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 241] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 41. | Simoni Y, Gautron AS, Beaudoin L, Bui LC, Michel ML, Coumoul X, Eberl G, Leite-de-Moraes M, Lehuen A. NOD mice contain an elevated frequency of iNKT17 cells that exacerbate diabetes. Eur J Immunol. 2011;41:3574-3585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Li S, Joseph C, Becourt C, Klibi J, Luce S, Dubois-Laforgue D, Larger E, Boitard C, Benlagha K. Potential role of IL-17-producing iNKT cells in type 1 diabetes. PLoS One. 2014;9:e96151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3754] [Cited by in RCA: 3794] [Article Influence: 237.1] [Reference Citation Analysis (0)] |
| 44. | Ahmed M, Gaffen SL. IL-17 inhibits adipogenesis in part via C/EBPα, PPARγ and Krüppel-like factors. Cytokine. 2013;61:898-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 45. | Zúñiga LA, Shen WJ, Joyce-Shaikh B, Pyatnova EA, Richards AG, Thom C, Andrade SM, Cua DJ, Kraemer FB, Butcher EC. IL-17 regulates adipogenesis, glucose homeostasis, and obesity. J Immunol. 2010;185:6947-6959. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 314] [Cited by in RCA: 298] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 46. | Molet S, Hamid Q, Davoine F, Nutku E, Taha R, Pagé N, Olivenstein R, Elias J, Chakir J. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J Allergy Clin Immunol. 2001;108:430-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 704] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 47. | Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1059] [Cited by in RCA: 1074] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 48. | Tard C, Rouxel O, Lehuen A. Regulatory role of natural killer T cells in diabetes. Biomed J. 2015;38:484-495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |