Published online Aug 10, 2016. doi: 10.4239/wjd.v7.i15.302
Peer-review started: March 31, 2016
First decision: May 17, 2016
Revised: May 30, 2016
Accepted: June 27, 2016
Article in press: June 29, 2016
Published online: August 10, 2016
Processing time: 133 Days and 8.9 Hours
Type 2 diabetes mellitus (T2DM) is a silent progressive polygenic metabolic disorder resulting from ineffective insulin cascading in the body. World-wide, about 415 million people are suffering from T2DM with a projected rise to 642 million in 2040. T2DM is treated with several classes of oral antidiabetic drugs (OADs) viz. biguanides, sulfonylureas, thiazolidinediones, meglitinides, etc. Treatment strategies for T2DM are to minimize long-term micro and macro vascular complications by achieving an optimized glycemic control. Genetic variations in the human genome not only disclose the risk of T2DM development but also predict the personalized response to drug therapy. Inter-individual variability in response to OADs is due to polymorphisms in genes encoding drug receptors, transporters, and metabolizing enzymes for example, genetic variants in solute carrier transporters (SLC22A1, SLC22A2, SLC22A3, SLC47A1 and SLC47A2) are actively involved in glycemic/HbA1c management of metformin. In addition, CYP gene encoding Cytochrome P450 enzymes also play a crucial role with respect to metabolism of drugs. Pharmacogenetic studies provide insights on the relationship between individual genetic variants and variable therapeutic outcomes of various OADs. Clinical utility of pharmacogenetic study is to predict the therapeutic dose of various OADs on individual basis. Pharmacogenetics therefore, is a step towards personalized medicine which will greatly improve the efficacy of diabetes treatment.
Core tip: Type 2 diabetes mellitus (T2DM) is a highly prevalent metabolic disorder, characterized by chronic hyperglycemia. It results from an interaction of environmental as well as genetic factors. Several genes have been identified associated with disease development and therapeutic outcomes. Inter-individual variations in the human genome affect both, risk of T2DM development and personalized response to identical drug therapies. Pharmacogenetic approaches focus on single nucleotide polymorphisms and their influence on individual drug response, efficacy and toxicity. In the present study, an effort has been made to review the genetic polymorphisms in candidate genes associated with efficacy of oral antidiabetic drugs.
- Citation: Singh S, Usman K, Banerjee M. Pharmacogenetic studies update in type 2 diabetes mellitus. World J Diabetes 2016; 7(15): 302-315
- URL: https://www.wjgnet.com/1948-9358/full/v7/i15/302.htm
- DOI: https://dx.doi.org/10.4239/wjd.v7.i15.302
Type 2 diabetes mellitus (T2DM) has been considered as a major health problem for both developed as well as developing countries. The global burden of diabetes is presently 415 million affected people, expected to rise to 642 million in 2040 and about 193 million people still undiagnosed. The Indian estimate is also alarming which shows 69.2 million people affected with T2DM in 2015 which will rise to 123.5 million in 2040[1].
Diabetes is traditionally known as a “silent disease” manifesting no symptoms until it progresses to severe damage of target organs. Diabetes has been classified under various categories depending upon their age of onset and severity[2]. The most prevalent adult-onset diabetes is T2DM characterized by hyperglycemia caused by defects in both insulin secretion and insulin signaling cascade. T2DM is a potential contributor to considerable morbidity in the form of metabolic complications viz. heart disease, stroke, neuropathy, kidney disease, vision disorder, peripheral vascular disease, ulcerations and amputations, infection, digestive diseases, oral complications and depression. T2DM is a multifactorial disease with high genetic variability in which certain candidate genes interfere with management of glycemic control in the body. Polymorphisms in the candidate genes may affect the susceptibility or risk of disease development and progression[3-7].
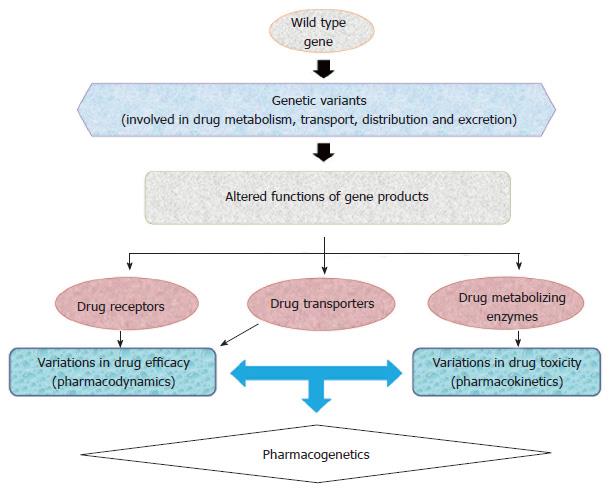
Pharmacogenomics establishes the use of an individual’s genetic information to guide treatment therapy and has become an important tool in achieving “personalized medicine”. The discoveries of novel genetic polymorphisms in drug transporters, and metabolizing enzymes have given an insight into the biological phenomena of drug efficacy and toxicity (Figure 1). Pharmacologically, several classes of drugs are currently being prescribed to treat T2DM patients, including biguanides, sulfonylureas, meglitinides, thiazolidinediones (TZDs), α-Glucosidase inhibitors, dipeptidyl peptidase-4 (DPP-4) inhibitors, glucagon-like peptide-1 (GLP-1) agonist, sodium-glucose co-transporter-2 inhibitors, insulin and its analogues[8-10]. Clinically, it is often observed that T2DM patients who receive identical antidiabetic regimens often exhibit significant variation in glycemic control, glycated haemoglobin (HbA1c) level, drug efficacy, tolerability and incidence of adverse effects[11]. Inter-individual differences can be attributed to polymorphisms of certain candidate genes involved in drug absorption, transportation, distribution, metabolism and signaling cascade of oral antidiabetic drugs (OADs)[11].
The term “pharmacogenetics” was coined by Vogel et al[12] which explains the differential response of individuals to identical medication. Clinical observations of inherited inter-individual differences during treatment were documented for the first time in 1950s[13-15] giving rise to a new field, i.e., pharmacogenetics and later pharmacogenomics. Pharmacogenomics is being used for genome-wide approaches to recognize the inherited inter-individual differences in response to drugs. Pharmacogenetics reveals that single nucleotide variations in genes (encoding drug receptor, transporters and metabolizing enzymes) are related to the efficacy and toxicity of drugs[16-18], for example CYP2D6, CYP2C8 and CYP2C9 are marked CYP enzymes that are actively involved in metabolism of various therapeutic agents[19].
The inter-individual differences are contributed by numerous factors, i.e., physical inactivity, race/ethnic diversity, hypertension, age, gender, etc[20]. During past decades, pharmacogenetic study was restricted to observations of familial response to a particular drug. However, genome-wide association studies, candidate gene approach and linkage analysis have transformed the area of pharmacogenetics/pharmacogenomics. These studies have elucidated the role of genetic variations for a particular drug and their doses on a personalized basis.
Treatment strategy for T2DM is mainly based on efficacy of OADs assessed by level of fasting/postprandial plasma glucose and/or HbA1c[10].
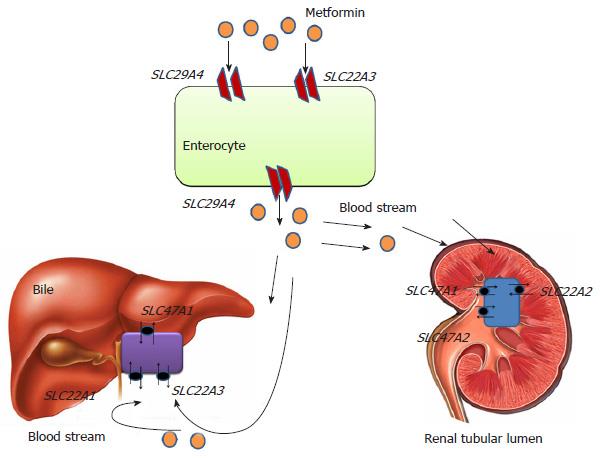
Metformin (N’,N’-dimethylbiguanide) is prescribed as a first-line medication for newly diagnosed T2DM patients[21]. Antihyperglycemic effects of metformin includes down regulation of hepatic gluconeogenesis, improvement in insulin sensitivity and significant reduction in insulin resistance[22]. The precise mechanism(s) of metformin action are still not fully elucidated. At physiological pH metformin serves as an organic cation being transported across the membrane by different isoforms of organic cation transporters (OCTs) viz. OCT1 expressed in hepatocytes, OCT2 in basolateral membrane of kidney. Metformin is transported from intestinal lumen into the epithelial cells via OCT3 and plasma membrane monoamine transporter. Uptake of metfomin from blood into hepatocytes is mediated by OCT1 and OCT3 (Figure 2). Metformin interferes with mitochondrial respiratory chain complex 1 by increasing AMP/ATP ratio, which promotes the activation of AMP kinase[23,24]. Metformin-induced AMP kinase activation leads to transcriptional inhibition of hepatic gluconeogenesis[25]. Metformin is not metabolized and excreted-out through urine via active renal tubular secretion. Metformin excretion in bile and urine is also facilitated by various isoforms of Multidrug and Toxin Extrusion transporters (MATE1 and MATE2)[26,27]. Therapeutic response of metformin differs inter-individually due to genetic polymorphisms. Single nucleotide polymorphisms (SNPs) in the genes encoding metformin transporters viz. OCT1, OCT2, MATE1, MATE2, etc., leads to significant association with the different degrees of efficacy and toxicity (Figure 2).
Solute carrier family 22 member 1 (SLC22A1) gene encodes the OCT1 which is expressed in hepatocytes and mediates the electrogenic transport of drugs[28]. OCT1 helps in transport of metformin into the liver (hepatocytes) and subsequent activity. It has been hypothesized that highly polymorphic SLC22A1 gene will influence the therapeutic success rate of metformin. In a South Indian study[29], it was reported that rs622342 variant of SLC22A1 gene was significantly associated with efficacy of metformin. They found that T2DM patients with rs622342 “AA” homozygotes had 5.6 times increased possibility of responding to metformin treatment. A recent pharmacogenetic study performed in a Chinese population demonstrated that T2DM patients with “AA” genotype of SLC22A1 rs594709 might have maximum plasma glucose lowering effect from metformin monotherapy[30]. Shu et al[31] studied the effect of loss of function polymorphism in SLC22A1 gene variants, i.e., rs12208357 (R61C), rs34130495 (G401S), rs72552763 (420del), rs34059508 (G465R). They concluded the study as these variants were significantly associated with lower efficacy of metformin in glucose tolerance test. However, in a subsequent GoDARTs study, two common SLC22A1 variants, R61C (rs12208357) and 420del (rs72552763) were reported to have no association with impaired initial response to metformin, or metformin monotherapy failure[32].
Solute carrier family 22 member 2 (SLC22A2) gene encodes the OCT2. OCT2 is a drug transporter and expressed in renal tubular cells thought to be responsible for their elimination[33,34]. Loss of function mutation in SLC22A2 gene has been significantly correlated with metformin disposition. In several studies, SLC22A2 gene has been reported as highly polymorphic in nature[34-37]. Zolk et al[38] found that SLC22A2 variant 808G > T (270Ala > Ser) significantly transforms the uptake of drugs. In healthy subjects, rs316019 (A270S) variant appeared responsible for decreased renal clearance of Metformin[30] while in a contradictory study a significant correlation of rs316019 was reported with increased metformin renal clearance[39]. Song et al[40] investigated the influence of rs201919874 (T199I) and rs145450955 (T201M) to the disposition of metformin in healthy individuals and reported that both were significantly associated with increased metformin plasma concentration and reduced renal clearance. A recent randomized cohort study performed in T2DM patients with one year follow-up demonstrated that efficacy of metformin was also influenced by SLC22A2 variant, rs316019 (808G > T)[41].
Solute carrier family 22 member 3 (SLC22A3) gene encodes for OCT3 which is expressed in liver, kidney and placenta. In public SNP database (http://www.ncbi.nlm.nih.gov/SNP/) five non-synonymous variants (ssj0008476, rs8187717, rs8187725, rs12212246, rs9365165) of human SLC22A3 gene were reported[42]. However, compared to OCT1 and OCT2, very few studies have reported about OCT3 variants and metformin therapeutics. In a pharmacologic study, Chen et al[43] studied the role of OCT3 variants ssj0008476 (T44M), rs8187725 (T400I) and V423F were found to be significantly associated with altered response to metformin action.
Solute carrier family 47 member 1 (SLC47A1) gene encodes the multidrug toxin extrusion receptor 1 expressed on apical domain of proximal and distal renal tubular cells and serves as an electro neutral organic cation/H+ exchanger. Since genetic polymorphisms in SLC47A1 associated with altered transport/excretion function might have great influence on metformin disposition, it is important to identify them in various ethnic populations and correlate in terms of therapeutic response. An intronic variant rs2289669 (G > A) in SLC47A1 was demonstrated to reduce HbA1c level significantly in metformin users[44]. While in a DPP (Diabetes Preventation Programme) study, SLC47A1 variant rs8065082 (C > T) was reported for lower diabetes incidence in individuals treated with metformin[45]. In a recent case control study the polymorphic effect of rs594709 in SLC22A1 and rs2289669 in SLC47A1 was evaluated in T2DM cases and no significant association was reported. The study concluded that carriers of allele “A” of rs594709 showed better efficacy for metformin[30]. In Chinese population, the SLC47A1 variant rs2289669 (G > A) appeared to promote metformin efficacy by delaying its excretion mechanism[46].
Solute carrier family 47 member 2 (SLC47A2) encodes for multidrug toxin extrusion receptor 2 (MATE2), expressed in apical membrane proximal tubule cells. It facilitates the disposition of metformin from renal tubular cells into urine. Choi et al[47] characterized variants of SLC47A2 to recognize their association with metformin. The study showed that homozygous individuals for rs12943590 (130G > A) of MATE2-K is significantly associated with poor plasma glucose control of metformin assessed by relative differences in HbA1C level.
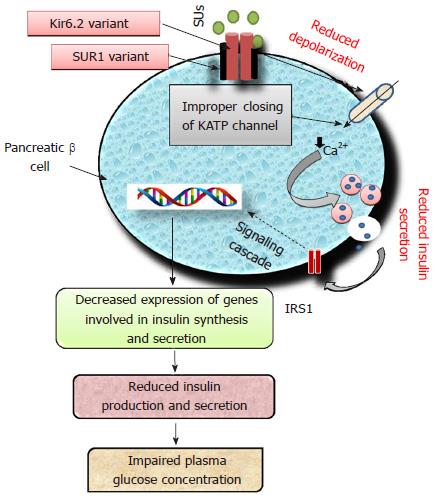
Sulfonylureas (SUs), insulin secretagogues are one of the most common classes of OADs being prescribed either alone or in combination since 1960s[8,48]. The second generation drugs viz. glimepiride, glibenclamide (glyburide), gliclazide and glipizide are most common representatives belonging to the group of SUs. The first-generation drugs viz. tolbutamide and chlorpropamide are no longer prescribed[10]. All SUs stimulate insulin secretion by binding to sulfonylurea receptor 1 (SUR1), a protein having 1581-amino acids. This interaction depolarizes the cell membrane of pancreatic beta cells by closing ATP-sensitive potassium (KATP) channels. Subsequent effect of depolarization leads to Ca2+ influx which trigger an enhanced insulin secretion from beta cells in glucose-independent manner[49]. KATP channel is a heterooctameric protein complex constructed by four inward-rectifier K+ channel, i.e., Kir6.2 (forming pore of KATP channel) coupled with SUR1, surrounding the pore[49]. In neonates, inactivating mutations in genes encoding Kir6.2 (KCNJ11) and SUR1 (ABCC8) are responsible for T2DM, while activating mutations lead to hypoglycemia[50]. Polymorphisms in the genes ABCC8, KCNJ11, CYP2C9, TCF7L2, NOS1AP (nitric oxide synthase 1 adaptor protein) have been reported for altered response to SUs[51,52]. Impairment of Kir6.2 (KCNJ11) and SUR1 (ABCC8) will lead to improper signaling cascade of insulin as shown in Figure 3.
ATP-sensitive potassium channel (KATP) is a transmembrane protein of pancreatic β-cells encoded by potassium inwardly-rectifying channel and subfamily J, member 11 (KCNJ11). Two hundred and nineteen SNPs have been reported for the KCNJ11 gene located on chromosome 11p15.1. Polymorphisms in KCNJ11 have been reported for development of diabetes because of its key role in insulin secretion[53]. Only 6 SNPs viz. rs5210, rs5215, rs5218, rs5219, rs886288, rs2285676 have been reported to be associated with diabetes[54]. A study found that in T2DM patients the rs5210 variant located at 3’ UTR of KCNJ11 improves the clinical efficacy of gliclazide[52]. The most widely studied KCNJ11 gene variant rs5219 (E23K) was significantly associated with the onset of T2DM in Asian Indian and Chinese populations[55,56]. However, studies performed on Caucasian individuals demonstrated for no significant differences in glycated hemoglobin[57,58]. Some studies have reported that diabetic patients having KCNJ11 gene variants respond better to pharmacotherapy with SUs as compared to insulin[59-61].
ATP-binding cassette, subfamily C member 8 (ABCC8) located at 11p15.1, encodes for SUR1 which modulate the activity of KATP channel[62]. Variants of ABCC8 gene rs1799854 (C/T) and rs1801261 have been studied extensively and are reported for inconsistent association with T2DM[63-70]. ABCC8 variant rs1799854 has been reported for significant association with sulfonylurea efficacy in terms of HbA1c level[57]. In one study, the genetic variants of ABCC8 were reported for significant reduction in HbA1c concentration[71]. Activating mutation in the genes encoding SUR1 (ABCC8) and Kir6.2 (KCNJ11) may lead to altered signaling cascade of insulin secretagogues resulting in therapeutic failure of SUs. The Arg972 variant of insulin receptor substrate 1 is reported for an enhanced risk of secondary failure to SUs in T2DM patients[72]. A study carried out in Chinese T2DM patients with two months follow-up, demonstrated that Ser1369Ala variant of ABCC8 is significantly associated with therapeutic success of gliclazide[73]. Carriers and non-carriers of SUR1-437A/T variant did not differ in insulin response stimulated by tolbutamide during OGT test[74].
SUs viz. tolbutamide, glimperide, glipizide and glibenclamide are metabolized to active metabolites in the liver mainly by cytochrome P450 2C9 (CYP2C9)[75] which are ultimately excreted by the kidney[76]. It has been reported that CYP2C9 variants were significantly associated with efficacy of SUs in diabetic patients[77]. Two variants of CYP2C9 gene, i.e., rs1057910 (CYP2C9*3) and rs1799853 (CYP2C9*2) have been significantly associated with missense amino acid polymorphisms resulting in decreased metabolism of SUs in healthy volunteers[74]. While in T2DM patients treated with SUs, CYP2C9*3 variant was reported with an enhanced risk of severe hypoglycemia[78,79]. Certain T2DM patients with CYP2C9 gene variants Ile359Leu and Arg144Cys were reported for 30%-80% reduction in renal clearance of glibenclamide suggesting lower doses of this antidiabetic drug to decrease the risk of hypoglycemia[51,75,77,80,81].
Transcription factor 7-like 2 (TCF7L2) is encoded by TCF7L2 gene which is actively involved in proliferation and differentiation of cells. It is required for secretion of glucose stimulated insulin from pancreatic β-cells. TCF7L2 is a key transcription factor, which regulates glucose metabolism in insulin dependent manner. It serves as a chief regulator in coordinating the proinsulin synthesis and its processing to produce mature insulin[82]. Hence, nucleotide variation in TCF7L2 gene may lead to alteration in insulin secretion[31] resulting reduced insulin secretion will lead to hyperglycemia. TCF7L2 gene is expressed in developing and mature pancreatic beta cells[83] and secretion of insulin is decreased in individuals having risk alleles[84-86]. Miyake et al[87] studied association of TCF7L2 variants with susceptibility to T2DM in 4087 Japanese patients. They found that rs7903146, rs12255372 and rs11196205 were significantly associated with T2DM while rs290487 and rs11196218 were reported for no association. Polymorphisms in TCF7L2 gene has been reported for strong association with T2DM affecting therapeutic response to SUs[88]. TCF7L2 SNPs were reported to influence the risk of T2DM[89]. Polymorphisms in TCF7L2, rs12255372 and rs7903146 were reported for decreased response to sulfonylurea efficacy[88]. The SNPs rs12255372T and rs7903146T were represented to be significantly associated with enhanced expression of TCF7L2 gene in beta cells, altering insulin release and predisposing individuals to T2DM[90,91].
TZDs are insulin sensitizers, they promote uptake of glucose by tissue and skeletal muscles, down regulate glucose output from liver[92]. Rosiglitazone and pioglitazone are medical representatives of TZD group. The exact molecular mechanism of TZDs is far from clear. However, data indicates that TZDs primarily bind with the peroxisome proliferator-activated receptor γ (PPARγ) in adipose tissue and affect their metabolism. On binding with PPARγ, TZDs stimulate adipocytes differentiation[93] and decrease plasma glucose level in T2DM patients[94,95]. Several studies have reported that TZDs improve both glucose homeostasis and insulin cascading in T2DM cases[96-98], hence may prevent the progression from altered plasma glucose tolerance to T2DM development[99]. Numerous potential mechanisms are reported by which TZDs improve molecular action of insulin in both liver[98,100] and skeletal muscles[97,101]. These include reduced content of intra-hepatocellular and intra-myocellular triglycerides[98,102] and altered body composition[97,103]. It also decreases synthesis and/or action of proinflammatory cytokines[104,105]. TZDs upregulate expression of genes in adipocytes resulting in increased level of adiponectin in plasma circulation[106-108], with positive effects on insulin sensitivity[109] and reduced hyperglycemia. Some previous studies reported that use of rosiglitazone (a TZD drug) as compared to pioglitazone could cause severe side effects, the risk of myocardial infarction and also lead to death due to cardiovascular dysfunction[110,111]. Several gene variants have been identified for significant association with therapeutic outcome of TZDs. Adiponectin, resistin, leptin, TNF-α and PPARγ are commonly called adipocytokines which are key regulators of insulin resistance[112].
PPARγ
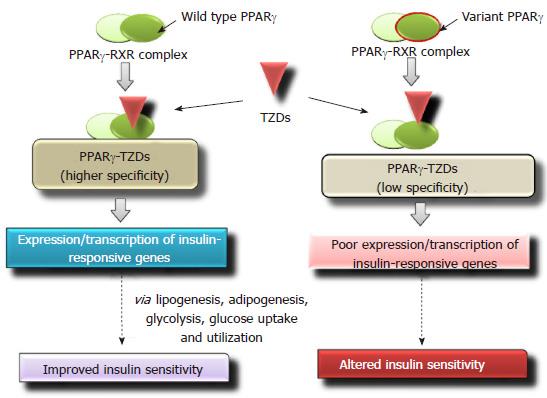
PPARγ belonging to the nuclear receptor family regulates metabolism of carbohydrates, regulates lipid homeostasis and adipocyte differentiation[113]. It is also a key mediator of insulin signaling[114]. In humans, TZDs bind to PPARγ with high specificity. At physiological pH PPARγ forms a dimer with retinoid X receptor (RXR). Binding of TZDs to the PPARγ-RXR complex stimulates a conformational change[115] which subsequently leads to the binding of the above heterodimer complex to the PPARγ response elements (PPRE) in the target genes[116]. It results in improved insulin sensitivity via glycolysis, lipogenesis, adipogenesis and increased glucose uptake and utilization[117]. Single nucleotide variations in PPARγ gene may affect the binding affinity with TZDs and its therapeutic efficacy (Figure 4). In PPARγ gene, loss-of-function mutations are significantly associated with insulin resistance and T2DM[118]. Multiple studies have reported that missense polymorphism Pro12Ala (CCA-to-GCA) in PPARγ gene is associated with decreased risk of T2DM development[119-121] and improved insulin sensitivity[122,123]. A pilot study performed on South Indian population, evaluated the effect of Pro12Ala variants on therapeutic success to pioglitazone, and reported a significant association with glycemic control[124]. T2DM cases with Pro12Ala variant of PPARγ gene, showed significant glycemic control [fasting plasma glucose (FPG) and HbA1C level] for rosiglitazone treatment as compared with carriers having wild-type genotype[125]. Zhang et al[126] demonstrated that in Chinese patients, amino acid variants Thr394Thr and Gly482Ser of peroxisome proliferator-activated receptor gamma coactivator 1-alpha were also significantly associated with efficacy of rosiglitazone.
Variants of adiponectin (ADIPOQ) gene have been reported for changes in FPG and level of HbA1c after 12 wk of rosiglitazone treatment. A study carried out by Liu et al[127] in T2DM Chinese patients demonstrated that sequence variation in leptin and TNF alpha gene interferes with therapeutic response to rosiglitazone. Nucleotide variants rs2241766 (45T/G) and rs266729 (-11377C/G) of ADIPOQ gene[128], rs1800629 (-308 G/A) of TNF-α and rs7799039 (-2548G/A) of leptin gene[127] were found to affect the rosiglitazone therapeutics and reverse insulin resistance in Chinese patients. In a pilot study, it was found that single nucleotide polymorphism at -420 (G/G) in resistin gene may serve as an independent predictor for down regulation of insulin resistance and hyperglycemia associated with pioglitazone therapeutics[129].
Metabolism of rosiglitazone is mainly metabolized by CYP2C8 and CYP2C9[78] while biotransformation of pioglitazone is mainly metabolized by CYP2C8 and CYP3A4[130]. Nucleotide polymorphisms in CYP2C8 gene were significantly associated with impaired clearance of rosiglitazone. Polymorphisms in CYP2C8*3 encoding for a reduced functioning of CYP2C8 enzyme, was reported for altered drug clearance[131]. Hence genetic variants of CYP2C8 may contribute to the degree of TZD therapeutics.
Meglitinide, insulin secretagogues act by inhibiting KATP channel leading to promote insulin secretion. Molecular mechanism of both sulphonylureas and meglitinide are similar. Sulphonylureas and meglitinide inhibit the activity of KATP channel by binding at two different sites of the SUR1 subunit[132]. Meglitinides have shorter duration of action and more rapid onset as compared with SUs. Repaglinide (a benzoic acid derivative) and nateglinide (a derivative of d-phenylalanine) belonging to meglitinide stimulate early secretion of insulin. Due to their short action, a potential adverse effect of meglitinide is to induce hypoglycemia[133]. Repaglinide is 100% metabolized in liver and hence excreted mainly via bile. Genetic polymorphisms associated with response to meglitinide were mapped in SLCO1B1, CYP2C8, CYP3A4, TCF7L2, SLC30A8, IGF2BP2, KCNJ11, KCNQ1, UCP2, NAMPT, MDR1, PAX4 and NeuroD1[78,134-139]. Out of these SLCO1B1 is reported to facilitate the hepatic uptake of a drug repaglinide[140].
Solute Carrier Organic anion transporter family member 1B1 (SLCO1B1) gene, mainly expressed in basolateral membrane (hepatocytes) encodes for organic anion-transporting polypeptide 1B1 (OATP1B1). Genetic polymorphisms in SLCO1B1 have been reported to exert significant influence on repaglinide pharmacokinetics with reduced exposure after administration of a single dose of repaglinide[141]. Genetic variant of SLCOB1 gene (521T > C) markedly affected the pharmacokinetics of nateglinide[134]. Cellular uptake of various drugs is regulated by OATP1B1. Several studies have demonstrated the pivotal role of SLCOB1 gene variants in pharmacokinetics of meglitinides[134,142-144]. Nateglinide is catabolized by CYP2C9. A study performed in Chinese male volunteers has demonstrated that genetic variants of SLCOB1 (521T > C) and CYP2C9 (CYP2C9*3) could affect the nateglinide efficacy[139].
CYP2C8 and CYP3A4, both are actively engaged in metabolism of repaglinide. Clinical studies demonstrate that individuals with CYP2C8*3 variant have greater clearance of OADs as compared to wild-type genotype[78]. A Chinese population treated with repaglinide and genotyped for KCNQ1 variants rs2237892 (C/T) and rs2237895 (C/A) were found to be associated with therapeutic efficacy of repaglinide[137]. Single nucleotide polymorphisms in SLC30A8 viz. Arg325Trp (rs13266634) and Arg325Gln (rs16889462) have been reported to be significantly associated with T2DM development and repaglinide efficacy[135]. KCNJ11 SNP rs5219 (Lys23Glu) has been found to be associated with poor regulation of fasting/postprandial glucose and HbA1c levels in T2DM patients with “GA” or “AA” genotype in contrast with “GG”. T2DM patients having “TT” genotype of TCF7L2 gene rs290487 (C/T) demonstrated better efficacy for repaglinide treatment with respect to triglyceride, LDL and fasting insulin as compared to patients with “CC” or “CT” genotype[107].
DPP-4 is involved in the degradation of two incretin hormones viz. GLP-1 and gastric inhibitory polypeptide. These hormones bring about a glucose dependent stimulation of insulin release. These hormones are also responsible for reduction in circulating plasma glucose levels by interrupting glucagon secretion and subsequently improve beta cell sensitization by glucose[145]. DPP-4 inhibitors inhibit function of DPP-4 enzyme, thus reducing glucagon secretion. Sitagliptin, vildagliptin and saxagliptin are medical representatives of DPP4 inhibitors. Sitagliptin was the first DPP-4 inhibitor approved by Food and Drug Administration (FDA) in 2006[146]. Metabolism of saxagliptin (a DPP-4 inhibitor) is catalyzed by CYP3A4/A5 while sitagliptin is metabolized by CYP3A4 with minor contribution of CYP2C8[147]. Zimdahl et al[148] investigated the effect of TCF7L2 variants for therapeutic efficacy of linagliptin, a DPP-4 inhibitor. Linagliptin was found to significantly improve glucose homeostasis in both cases with and without TCF7L2 risk alleles for diabetes. Effects of genetic polymorphisms associated with DPP-4 inhibitors remain to be investigated.
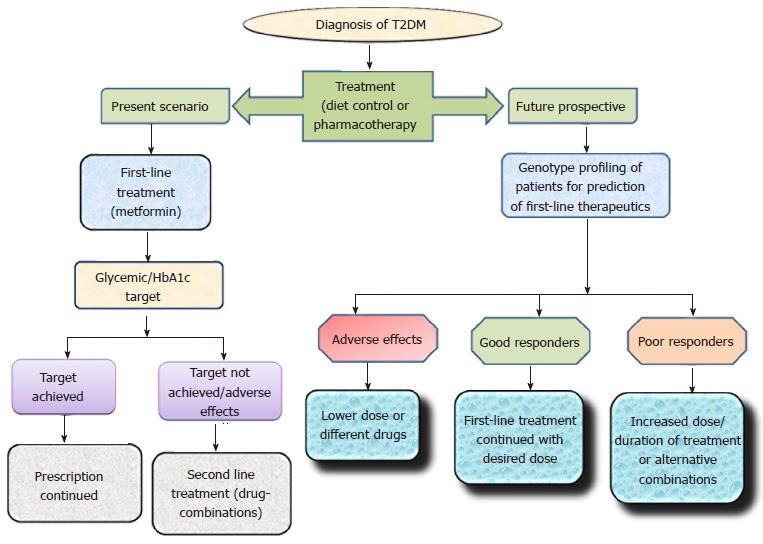
About 70 genetic loci have been identified to be associated with T2DM[149]. Pharmacogenetics, an expanding area of research provides a platform to understand and improve pharmacological treatment. Over the last decade, the number of available antidiabetic drugs has considerably increased. However, clinical treatment of T2DM patients has become more complex due to different degrees of therapeutic outcomes. Personalized differences during OADs therapeutics have been linked with numerous variants related to drug-transporters, drug-targets, drug catabolizing enzymes and T2DM risk genes (Table 1). Although inter-individual differences in respect to efficacy and toxicity of OADs are significantly associated with genetic makeup, it is clear that different degrees of response to antidiabetics cannot be predicted by studying the genetic differences alone. The role of genetic variations with respect to therapeutic outcomes must be further tested via clinical trials thus leading to a personalized pharmacotherapy. The present scenario and future prospect of Pharmacogenetic studies has been elaborated in Figure 5.
| Class | Common medical representatives | Mechanism of action | Candidate genes involved in pharmacotherapy | Ref. |
| Biguanide | Metformin | AMP-kinase activation | SLC22A1, SLC22A2, SLC22A3, SLC47A1, SLC47A2 | [28-39] |
| Sulfonylureas | Gliburide, gliclazide, Glimepiride, glipizide | Inhibition of KATP channel on plasma membrane of β-cells | KCNJ11, ABCC8, CYP2C9, TCF7L2 | [8,10,48-91] |
| Thiazolidinediones | Pioglitazone, rosiglitazone | Activates PPAR-γ | PPAR-γ, ADIPOQ, TNF-α, LEP, CYP2C8 | [92-131] |
| Meglitinides | Nateglinide, repaglinide | Inhibition of KATP channel on Plasma Membrane of β-cells | SLCOB1, CYP2C8, KCNQ1, SLC30A8, KCNJ11, TCF7L2 | [78,106,132-144] |
| DPP-4 inhibitors | Alogliptin, linagliptin, saxagliptin, sitagliptin, vildagliptin | Inhibits DPP-4, Affect GLP-1 receptor pathway | Possibly TCF7L2 | [145-148] |
| α-glucosidase inhibitors | Acarbose, miglitol, voglibose | Inhibits intestinal α-glucosidase | Yet to identify? | [10] |
| SGLT-2 inhibitors | Canagliflozin, dapagliflozin, empagliflozin | Inhibits SGLT2 transporters in kidney | Yet to identify? | [10] |
| GLP-1 agonist | Exenatide, liraglutide | Activate GLP-1 receptor | Yet to identify? | [10] |
The authors would like to thank Indian Council of Medical Research, New Delhi, India and Centre of Excellence, Higher Education, Government of Uttar Pradesh, Lucknow, India for funding diabetes research. SS would like to acknowledge BSR National Fellowship, University Grants Commission, New Delhi, India.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country of origin: India
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Baskin Y, Demonacos C S- Editor: Ji FF L- Editor: A E- Editor: Wu HL
| 1. | IDF. Diabetes Atlas, 7th ed. Brussels, Belgium: International Diabetes Federation 2015; 1-114. |
| 2. | Saxena M, Banerjee M. Diabetes: History, prevalence, insulin action and associated genes. J Applied Bioscience. 2008;34:139-151. |
| 3. | Saxena M, Srivastava N, Banerjee M. Genetic association of adiponectin gene polymorphisms (+45T/G and +10211T/G) with type 2 diabetes in North Indians. Diabetes Metab Syndr. 2012;6:65-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Gautam S, Pirabu L, Agrawal CG, Banerjee M. CD36 gene variants and their association with type 2 diabetes in an Indian population. Diabetes Technol Ther. 2013;15:680-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Saxena M, Srivastava N, Banerjee M. Association of IL-6, TNF-α and IL-10 gene polymorphisms with type 2 diabetes mellitus. Mol Biol Rep. 2013;40:6271-6279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 6. | Vats P, Chandra H, Banerjee M. Glutathione S-transferase and catalase gene polymorphism with type 2 diabetes mellitus. Dieses Mol Med. 2013;1:46-53. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 7. | Vats P, Sagar N, Singh TP, Banerjee M. Association of Superoxide dismutases (SOD1 and SOD2) and Glutathione peroxidase 1 (GPx1) gene polymorphisms with type 2 diabetes mellitus. Free Radic Res. 2015;49:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 8. | Inzucchi SE, Bergenstal RM, Buse JB, Diamant M, Ferrannini E, Nauck M, Peters AL, Tsapas A, Wender R, Matthews DR. Management of hyperglycemia in type 2 diabetes: a patient-centered approach: position statement of the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetes Care. 2012;35:1364-1379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2563] [Cited by in RCA: 2609] [Article Influence: 200.7] [Reference Citation Analysis (4)] |
| 9. | Topić E. The Role of Pharmacogenetics in the Treatment of Diabetes Mellitus/uloga farmakogenetike u lečnju dijabetes melitusa. J Med Biochem. 2014;33:58-70. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | American Diabetes Association. Standards of medical care in diabetes--2006. Diabetes Care. 2006;29 Suppl 1:S4-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 182] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 11. | Mannino GC, Sesti G. Individualized therapy for type 2 diabetes: clinical implications of pharmacogenetic data. Mol Diagn Ther. 2012;16:285-302. [PubMed] |
| 12. | Vogel F. Moderne probleme der Humangenetik. Ergeb Inn Med Kinderheild. 1959;12:52-125. [DOI] [Full Text] |
| 13. | Hughes HB, Biehl JP, Jones AP, Schmidt LH. Metabolism of isoniazid in man as related to the occurrence of peripheral neuritis. Am Rev Tuberc. 1954;70:266-273. [PubMed] |
| 14. | Alving AS, Carson PE, Flanagan CL, Ickes CE. Enzymatic deficiency in primaquine-sensitive erythrocytes. Science. 1956;124:484-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 468] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 15. | Evans DA, Manley KA, Mckusick VA. Genetic control of isoniazid metabolism in man. Br Med J. 1960;2:485-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 508] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 16. | Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1730] [Cited by in RCA: 1568] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 17. | Evans WE, Johnson JA. Pharmacogenomics: the inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet. 2001;2:9-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 249] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 18. | McLeod HL, Evans WE. Pharmacogenomics: unlocking the human genome for better drug therapy. Annu Rev Pharmacol Toxicol. 2001;41:101-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 186] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 19. | Rettie AE, Jones JP. Clinical and toxicological relevance of CYP2C9: drug-drug interactions and pharmacogenetics. Annu Rev Pharmacol Toxicol. 2005;45:477-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 180] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 20. | American Diabetes Association. Standards of medical care in diabetes--2013. Diabetes Care. 2013;36 Suppl 1:S11-S66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2371] [Cited by in RCA: 2483] [Article Influence: 206.9] [Reference Citation Analysis (0)] |
| 21. | American Diabetes Association. Standards of medical care in diabetes--2012. Diabetes Care. 2012;35 Suppl 1:S11-S63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 969] [Cited by in RCA: 1366] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 22. | Kirpichnikov D, McFarlane SI, Sowers JR. Metformin: an update. Ann Intern Med. 2002;137:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 724] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 23. | Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348 Pt 3:607-614. [PubMed] |
| 24. | Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3802] [Cited by in RCA: 4211] [Article Influence: 175.5] [Reference Citation Analysis (0)] |
| 25. | Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond). 2012;122:253-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1054] [Cited by in RCA: 1224] [Article Influence: 94.2] [Reference Citation Analysis (0)] |
| 26. | Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci USA. 2005;102:17923-17928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 433] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 27. | Masuda S, Terada T, Yonezawa A, Tanihara Y, Kishimoto K, Katsura T, Ogawa O, Inui K. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J Am Soc Nephrol. 2006;17:2127-2135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 289] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 28. | Jacobs C, Pearce B, Du Plessis M, Hoosain N, Benjeddou M. Genetic polymorphisms and haplotypes of the organic cation transporter 1 gene (SLC22A1) in the Xhosa population of South Africa. Genet Mol Biol. 2014;37:350-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Umamaheswaran G, Praveen RG, Damodaran SE, Das AK, Adithan C. Influence of SLC22A1 rs622342 genetic polymorphism on metformin response in South Indian type 2 diabetes mellitus patients. Clin Exp Med. 2015;15:511-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Xiao D, Guo Y, Li X, Yin JY, Zheng W, Qiu XW, Xiao L, Liu RR, Wang SY, Gong WJ. The Impacts of SLC22A1 rs594709 and SLC47A1 rs2289669 Polymorphisms on Metformin Therapeutic Efficacy in Chinese Type 2 Diabetes Patients. Int J Endocrinol. 2016;2016:4350712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 31. | Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, Sheardown SA, Yue L, Burchard EG, Brett CM. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther. 2008;83:273-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 392] [Cited by in RCA: 353] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 32. | Zhou K, Donnelly LA, Kimber CH, Donnan PT, Doney AS, Leese G, Hattersley AT, McCarthy MI, Morris AD, Palmer CN. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes. 2009;58:1434-1439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 132] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 33. | Motohashi H, Sakurai Y, Saito H, Masuda S, Urakami Y, Goto M, Fukatsu A, Ogawa O, Inui K. Gene expression levels and immunolocalization of organic ion transporters in the human kidney. J Am Soc Nephrol. 2002;13:866-874. [PubMed] |
| 34. | Wang ZJ, Yin OQ, Tomlinson B, Chow MS. OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet Genomics. 2008;18:637-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 197] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 35. | Leabman MK, Huang CC, Kawamoto M, Johns SJ, Stryke D, Ferrin TE, DeYoung J, Taylor T, Clark AG, Herskowitz I. Polymorphisms in a human kidney xenobiotic transporter, OCT2, exhibit altered function. Pharmacogenetics. 2002;12:395-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 142] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 36. | Ogasawara K, Terada T, Motohashi H, Asaka J, Aoki M, Katsura T, Kamba T, Ogawa O, Inui K. Analysis of regulatory polymorphisms in organic ion transporter genes (SLC22A) in the kidney. J Hum Genet. 2008;53:607-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 37. | Takane H, Shikata E, Otsubo K, Higuchi S, Ieiri I. Polymorphism in human organic cation transporters and metformin action. Pharmacogenomics. 2008;9:415-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 38. | Zolk O, Solbach TF, König J, Fromm MF. Functional characterization of the human organic cation transporter 2 variant p.270Ala& gt; Ser. Drug Metab Dispos. 2009;37:1312-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 39. | Chen Y, Li S, Brown C, Cheatham S, Castro RA, Leabman MK, Urban TJ, Chen L, Yee SW, Choi JH. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet Genomics. 2009;19:497-504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 172] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 40. | Song IS, Shin HJ, Shim EJ, Jung IS, Kim WY, Shon JH, Shin JG. Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin Pharmacol Ther. 2008;84:559-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 196] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 41. | Hou W, Zhang D, Lu W, Zheng T, Wan L, Li Q, Bao Y, Liu F, Jia W. Polymorphism of organic cation transporter 2 improves glucose-lowering effect of metformin via influencing its pharmacokinetics in Chinese type 2 diabetic patients. Mol Diagn Ther. 2015;19:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Sakata T, Anzai N, Kimura T, Miura D, Fukutomi T, Takeda M, Sakurai H, Endou H. Functional analysis of human organic cation transporter OCT3 (SLC22A3) polymorphisms. J Pharmacol Sci. 2010;113:263-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 43. | Chen L, Pawlikowski B, Schlessinger A, More SS, Stryke D, Johns SJ, Portman MA, Chen E, Ferrin TE, Sali A. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet Genomics. 2010;20:687-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 157] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 44. | Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes. 2009;58:745-749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 208] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 45. | Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, Hanson RL, Saxena R, Fowler S, Shuldiner AR, Knowler WC. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672-2681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 204] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 46. | He R, Zhang D, Lu W, Zheng T, Wan L, Liu F, Jia W. SLC47A1 gene rs2289669 G>A; A variants enhance the glucose-lowering effect of metformin via delaying its excretion in Chinese type 2 diabetes patients. Diabetes Res Clin Pract. 2015;109:57-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 47. | Choi JH, Yee SW, Ramirez AH, Morrissey KM, Jang GH, Joski PJ, Mefford JA, Hesselson SE, Schlessinger A, Jenkins G. A common 5’-UTR variant in MATE2-K is associated with poor response to metformin. Clin Pharmacol Ther. 2011;90:674-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 48. | Sola D, Rossi L, Schianca GP, Maffioli P, Bigliocca M, Mella R, Corlianò F, Fra GP, Bartoli E, Derosa G. Sulfonylureas and their use in clinical practice. Arch Med Sci. 2015;11:840-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 327] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 49. | Ashcroft FM, Rorsman P. Electrophysiology of the pancreatic beta-cell. Prog Biophys Mol Biol. 1989;54:87-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 785] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 50. | Flanagan SE, Clauin S, Bellanné-Chantelot C, de Lonlay P, Harries LW, Gloyn AL, Ellard S. Update of mutations in the genes encoding the pancreatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat. 2009;30:170-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 51. | Bozkurt O, de Boer A, Grobbee DE, Heerdink ER, Burger H, Klungel OH. Pharmacogenetics of glucose-lowering drug treatment: a systematic review. Mol Diagn Ther. 2007;11:291-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 52. | Xu H, Murray M, McLachlan AJ. Influence of genetic polymorphisms on the pharmacokinetics and pharmaco-dynamics of sulfonylurea drugs. Curr Drug Metab. 2009;10:643-658. [PubMed] |
| 53. | Bonfanti DH, Alcazar LP, Arakaki PA, Martins LT, Agustini BC, de Moraes Rego FG, Frigeri HR. ATP-dependent potassium channels and type 2 diabetes mellitus. Clin Biochem. 2015;48:476-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 54. | Haghvirdizadeh P, Mohamed Z, Abdullah NA, Haghvirdizadeh P, Haerian MS, Haerian BS. KCNJ11: Genetic Polymorphisms and Risk of Diabetes Mellitus. J Diabetes Res. 2015;2015:908152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 55. | Zhou D, Zhang D, Liu Y, Zhao T, Chen Z, Liu Z, Yu L, Zhang Z, Xu H, He L. The E23K variation in the KCNJ11 gene is associated with type 2 diabetes in Chinese and East Asian population. J Hum Genet. 2009;54:433-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 56. | Phani NM, Guddattu V, Bellampalli R, Seenappa V, Adhikari P, Nagri SK, D Souza SC, Mundyat GP, Satyamoorthy K, Rai PS. Population specific impact of genetic variants in KCNJ11 gene to type 2 diabetes: a case-control and meta-analysis study. PLoS One. 2014;9:e107021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 57. | Nikolac N, Simundic AM, Katalinic D, Topic E, Cipak A, Zjacic Rotkvic V. Metabolic control in type 2 diabetes is associated with sulfonylurea receptor-1 (SUR-1) but not with KCNJ11 polymorphisms. Arch Med Res. 2009;40:387-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | Klen J, Dolžan V, Janež A. CYP2C9, KCNJ11 and ABCC8 polymorphisms and the response to sulphonylurea treatment in type 2 diabetes patients. Eur J Clin Pharmacol. 2014;70:421-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 59. | Pearson ER, Flechtner I, Njølstad PR, Malecki MT, Flanagan SE, Larkin B, Ashcroft FM, Klimes I, Codner E, Iotova V. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med. 2006;355:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 708] [Cited by in RCA: 671] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 60. | Siklar Z, Ellard S, Okulu E, Berberoğlu M, Young E, Savaş Erdeve S, Mungan IA, Hacihamdioğlu B, Erdeve O, Arsan S. Transient neonatal diabetes with two novel mutations in the KCNJ11 gene and response to sulfonylurea treatment in a preterm infant. J Pediatr Endocrinol Metab. 2011;24:1077-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 61. | Dupont J, Pereira C, Medeira A, Duarte R, Ellard S, Sampaio L. Permanent neonatal diabetes mellitus due to KCNJ11 mutation in a Portuguese family: transition from insulin to oral sulfonylureas. J Pediatr Endocrinol Metab. 2012;25:367-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 62. | Patch AM, Flanagan SE, Boustred C, Hattersley AT, Ellard S. Mutations in the ABCC8 gene encoding the SUR1 subunit of the KATP channel cause transient neonatal diabetes, permanent neonatal diabetes or permanent diabetes diagnosed outside the neonatal period. Diabetes Obes Metab. 2007;9 Suppl 2:28-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 63. | Inoue H, Ferrer J, Welling CM, Elbein SC, Hoffman M, Mayorga R, Warren-Perry M, Zhang Y, Millns H, Turner R. Sequence variants in the sulfonylurea receptor (SUR) gene are associated with NIDDM in Caucasians. Diabetes. 1996;45:825-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 64. | Hani EH, Clément K, Velho G, Vionnet N, Hager J, Philippi A, Dina C, Inoue H, Permutt MA, Basdevant A. Genetic studies of the sulfonylurea receptor gene locus in NIDDM and in morbid obesity among French Caucasians. Diabetes. 1997;46:688-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 65. | Hart LM, de Knijff P, Dekker JM, Stolk RP, Nijpels G, van der Does FE, Ruige JB, Grobbee DE, Heine RJ, Maassen JA. Variants in the sulphonylurea receptor gene: association of the exon 16-3t variant with Type II diabetes mellitus in Dutch Caucasians. Diabetologia. 1999;42:617-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Hart LM, Dekker JM, van Haeften TW, Ruige JB, Stehouwer CD, Erkelens DW, Heine RJ, Maassen JA. Reduced second phase insulin secretion in carriers of a sulphonylurea receptor gene variant associating with Type II diabetes mellitus. Diabetologia. 2000;43:515-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 67. | Rissanen J, Markkanen A, Kärkkäinen P, Pihlajamäki J, Kekäläinen P, Mykkänen L, Kuusisto J, Karhapää P, Niskanen L, Laakso M. Sulfonylurea receptor 1 gene variants are associated with gestational diabetes and type 2 diabetes but not with altered secretion of insulin. Diabetes Care. 2000;23:70-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 68. | Weisnagel SJ, Rankinen T, Nadeau A, Rao DC, Chagnon YC, Pérusse L, Bouchard C. Decreased fasting and oral glucose stimulated C-peptide in nondiabetic subjects with sequence variants in the sulfonylurea receptor 1 gene. Diabetes. 2001;50:697-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 69. | Reis AF, Hani EH, Beressi N, Robert JJ, Bresson JL, Froguel P, Velho G. Allelic variation in exon 18 of the sulfonylurea receptor 1 (SUR1) gene, insulin secretion and insulin sensitivity in nondiabetic relatives of type 2 diabetic subjects. Diabetes Metab. 2002;28:209-215. [PubMed] |
| 70. | Venkatesan R, Bodhini D, Narayani N, Mohan V. Association study of the ABCC8 gene variants with type 2 diabetes in south Indians. Indian J Hum Genet. 2014;20:37-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 71. | Zhang H, Liu X, Kuang H, Yi R, Xing H. Association of sulfonylurea receptor 1 genotype with therapeutic response to gliclazide in type 2 diabetes. Diabetes Res Clin Pract. 2007;77:58-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 72. | Sesti G, Marini MA, Cardellini M, Sciacqua A, Frontoni S, Andreozzi F, Irace C, Lauro D, Gnasso A, Federici M. The Arg972 variant in insulin receptor substrate-1 is associated with an increased risk of secondary failure to sulfonylurea in patients with type 2 diabetes. Diabetes Care. 2004;27:1394-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 73. | Feng Y, Mao G, Ren X, Xing H, Tang G, Li Q, Li X, Sun L, Yang J, Ma W. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese type 2 diabetic patients. Diabetes Care. 2008;31:1939-1944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 122] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 74. | Hansen T, Ambye L, Grarup N, Hansen L, Echwald SM, Ferrer J, Pedersen O. Genetic variability of the SUR1 promoter in relation to beta-cell function and Type II diabetes mellitus. Diabetologia. 2001;44:1330-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 75. | Holstein A, Plaschke A, Ptak M, Egberts EH, El-Din J, Brockmöller J, Kirchheiner J. Association between CYP2C9 slow metabolizer genotypes and severe hypoglycaemia on medication with sulphonylurea hypoglycaemic agents. Br J Clin Pharmacol. 2005;60:103-106. [PubMed] |
| 76. | Kirchheiner J, Bauer S, Meineke I, Rohde W, Prang V, Meisel C, Roots I, Brockmöller J. Impact of CYP2C9 and CYP2C19 polymorphisms on tolbutamide kinetics and the insulin and glucose response in healthy volunteers. Pharmacogenetics. 2002;12:101-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 128] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 77. | Becker ML, Visser LE, Trienekens PH, Hofman A, van Schaik RH, Stricker BH. Cytochrome P450 2C9 *2 and *3 polymorphisms and the dose and effect of sulfonylurea in type II diabetes mellitus. Clin Pharmacol Ther. 2008;83:288-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 98] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 78. | Kirchheiner J, Roots I, Goldammer M, Rosenkranz B, Brockmöller J. Effect of genetic polymorphisms in cytochrome p450 (CYP) 2C9 and CYP2C8 on the pharmacokinetics of oral antidiabetic drugs: clinical relevance. Clin Pharmacokinet. 2005;44:1209-1225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 140] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 79. | Ragia G, Petridis I, Tavridou A, Christakidis D, Manolopoulos VG. Presence of CYP2C9*3 allele increases risk for hypoglycemia in Type 2 diabetic patients treated with sulfonylureas. Pharmacogenomics. 2009;10:1781-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 80. | Rydberg T, Jönsson A, Røder M, Melander A. Hypoglycemic activity of glyburide (glibenclamide) metabolites in humans. Diabetes Care. 1994;17:1026-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 81. | Distefano JK, Watanabe RM. Pharmacogenetics of Anti-Diabetes Drugs. Pharmaceuticals (Basel). 2010;3:2610-2646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 82. | Zhou Y, Park SY, Su J, Bailey K, Ottosson-Laakso E, Shcherbina L, Oskolkov N, Zhang E, Thevenin T, Fadista J. TCF7L2 is a master regulator of insulin production and processing. Hum Mol Genet. 2014;23:6419-6431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 142] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 83. | Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, Balkau B, Charpentier G, Pattou F, Stetsyuk V. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55:2903-2908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 235] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 84. | Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med. 2006;355:241-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 645] [Cited by in RCA: 621] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 85. | Melzer D, Murray A, Hurst AJ, Weedon MN, Bandinelli S, Corsi AM, Ferrucci L, Paolisso G, Guralnik JM, Frayling TM. Effects of the diabetes linked TCF7L2 polymorphism in a representative older population. BMC Med. 2006;4:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 86. | Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjögren M, Florez JC, Almgren P, Isomaa B, Orho-Melander M. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890-2895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 288] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 87. | Miyake K, Horikawa Y, Hara K, Yasuda K, Osawa H, Furuta H, Hirota Y, Yamagata K, Hinokio Y, Oka Y. Association of TCF7L2 polymorphisms with susceptibility to type 2 diabetes in 4,087 Japanese subjects. J Hum Genet. 2008;53:174-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 88. | Pearson ER, Donnelly LA, Kimber C, Whitley A, Doney AS, McCarthy MI, Hattersley AT, Morris AD, Palmer CN. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56:2178-2182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 220] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 89. | Brunetti A, Chiefari E, Foti D. Recent advances in the molecular genetics of type 2 diabetes mellitus. World J Diabetes. 2014;5:128-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 80] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (1)] |
| 90. | Hattersley AT. Prime suspect: the TCF7L2 gene and type 2 diabetes risk. J Clin Invest. 2007;117:2077-2079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 91. | Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, Sjögren M, Ling C, Eriksson KF, Lethagen AL. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155-2163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 571] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 92. | Chen L, Yang G. PPARs Integrate the Mammalian Clock and Energy Metabolism. PPAR Res. 2014;2014:653017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 166] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 93. | Spiegelman BM. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1335] [Cited by in RCA: 1300] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 94. | Aronoff S, Rosenblatt S, Braithwaite S, Egan JW, Mathisen AL, Schneider RL. Pioglitazone hydrochloride monotherapy improves glycemic control in the treatment of patients with type 2 diabetes: a 6-month randomized placebo-controlled dose-response study. The Pioglitazone 001 Study Group. Diabetes Care. 2000;23:1605-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 449] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 95. | Scherbaum WA, Göke B. Metabolic efficacy and safety of once-daily pioglitazone monotherapy in patients with type 2 diabetes: a double-blind, placebo-controlled study. Horm Metab Res. 2002;34:589-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 96. | Petersen KF, Krssak M, Inzucchi S, Cline GW, Dufour S, Shulman GI. Mechanism of troglitazone action in type 2 diabetes. Diabetes. 2000;49:827-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 99] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 97. | Miyazaki Y, Mahankali A, Matsuda M, Mahankali S, Hardies J, Cusi K, Mandarino LJ, DeFronzo RA. Effect of pioglitazone on abdominal fat distribution and insulin sensitivity in type 2 diabetic patients. J Clin Endocrinol Metab. 2002;87:2784-2791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 439] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 98. | Bajaj M, Suraamornkul S, Pratipanawatr T, Hardies LJ, Pratipanawatr W, Glass L, Cersosimo E, Miyazaki Y, DeFronzo RA. Pioglitazone reduces hepatic fat content and augments splanchnic glucose uptake in patients with type 2 diabetes. Diabetes. 2003;52:1364-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 220] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 99. | Buchanan TA, Xiang AH, Peters RK, Kjos SL, Marroquin A, Goico J, Ochoa C, Tan S, Berkowitz K, Hodis HN. Preservation of pancreatic beta-cell function and prevention of type 2 diabetes by pharmacological treatment of insulin resistance in high-risk hispanic women. Diabetes. 2002;51:2796-2803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1023] [Cited by in RCA: 908] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 100. | Ye JM, Frangioudakis G, Iglesias MA, Furler SM, Ellis B, Dzamko N, Cooney GJ, Kraegen EW. Prior thiazolidinedione treatment preserves insulin sensitivity in normal rats during acute fatty acid elevation: role of the liver. Endocrinology. 2002;143:4527-4535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 101. | Maggs DG, Buchanan TA, Burant CF, Cline G, Gumbiner B, Hsueh WA, Inzucchi S, Kelley D, Nolan J, Olefsky JM. Metabolic effects of troglitazone monotherapy in type 2 diabetes mellitus. A randomized, double-blind, placebo-controlled trial. Ann Intern Med. 1998;128:176-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 202] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 102. | Mayerson AB, Hundal RS, Dufour S, Lebon V, Befroy D, Cline GW, Enocksson S, Inzucchi SE, Shulman GI, Petersen KF. The effects of rosiglitazone on insulin sensitivity, lipolysis, and hepatic and skeletal muscle triglyceride content in patients with type 2 diabetes. Diabetes. 2002;51:797-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 499] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 103. | Nakamura T, Funahashi T, Yamashita S, Nishida M, Nishida Y, Takahashi M, Hotta K, Kuriyama H, Kihara S, Ohuchi N. Thiazolidinedione derivative improves fat distribution and multiple risk factors in subjects with visceral fat accumulation--double-blind placebo-controlled trial. Diabetes Res Clin Pract. 2001;54:181-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 104. | Sigrist S, Bedoucha M, Boelsterli UA. Down-regulation by troglitazone of hepatic tumor necrosis factor-alpha and interleukin-6 mRNA expression in a murine model of non-insulin-dependent diabetes. Biochem Pharmacol. 2000;60:67-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 105. | Alleva DG, Johnson EB, Lio FM, Boehme SA, Conlon PJ, Crowe PD. Regulation of murine macrophage proinflammatory and anti-inflammatory cytokines by ligands for peroxisome proliferator-activated receptor-gamma: counter-regulatory activity by IFN-gamma. J Leukoc Biol. 2002;71:677-685. [PubMed] |
| 106. | Albrektsen T, Frederiksen KS, Holmes WE, Boel E, Taylor K, Fleckner J. Novel genes regulated by the insulin sensitizer rosiglitazone during adipocyte differentiation. Diabetes. 2002;51:1042-1051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 107. | Yu M, Xu XJ, Yin JY, Wu J, Chen X, Gong ZC, Ren HY, Huang Q, Sheng FF, Zhou HH. KCNJ11 Lys23Glu and TCF7L2 rs290487(C/T) polymorphisms affect therapeutic efficacy of repaglinide in Chinese patients with type 2 diabetes. Clin Pharmacol Ther. 2010;87:330-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 108. | Iwaki M, Matsuda M, Maeda N, Funahashi T, Matsuzawa Y, Makishima M, Shimomura I. Induction of adiponectin, a fat-derived antidiabetic and antiatherogenic factor, by nuclear receptors. Diabetes. 2003;52:1655-1663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 577] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 109. | Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002;8:731-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1564] [Cited by in RCA: 1532] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 110. | Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N Engl J Med. 2007;356:2457-2471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3497] [Cited by in RCA: 3343] [Article Influence: 185.7] [Reference Citation Analysis (0)] |
| 111. | Chen X, Yang L, Zhai SD. Risk of cardiovascular disease and all-cause mortality among diabetic patients prescribed rosiglitazone or pioglitazone: a meta-analysis of retrospective cohort studies. Chin Med J (Engl). 2012;125:4301-4306. [PubMed] |
| 112. | Semiz S, Dujic T, Causevic A. Pharmacogenetics and personalized treatment of type 2 diabetes. Biochem Med (Zagreb). 2013;23:154-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 113. | Mudaliar S, Henry RR. New oral therapies for type 2 diabetes mellitus: The glitazones or insulin sensitizers. Annu Rev Med. 2001;52:239-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 156] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 114. | Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 1569] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 115. | Berger J, Bailey P, Biswas C, Cullinan CA, Doebber TW, Hayes NS, Saperstein R, Smith RG, Leibowitz MD. Thiazolidinediones produce a conformational change in peroxisomal proliferator-activated receptor-gamma: binding and activation correlate with antidiabetic actions in db/db mice. Endocrinology. 1996;137:4189-4195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 213] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 116. | Schoonjans K, Martin G, Staels B, Auwerx J. Peroxisome proliferator-activated receptors, orphans with ligands and functions. Curr Opin Lipidol. 1997;8:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 368] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 117. | Hauner H. The mode of action of thiazolidinediones. Diabetes Metab Res Rev. 2002;18 Suppl 2:S10-S15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 273] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 118. | Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880-883. [PubMed] |
| 119. | Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1044] [Cited by in RCA: 1038] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 120. | Azab MM, Abdel-Azeez HA, Zanaty MF, El Alawi SM. Peroxisome proliferator activated receptor γ2 gene Pro12Ala gene polymorphism in type 2 diabetes and its relationship with diabetic nephropathy. Clin Lab. 2014;60:743-749. [PubMed] |
| 121. | Dubinina IA, Chistiakov DA, Eremina IA, Brovkin AN, Zilberman LI, Nikitin AG, Kuraeva TL, Nosikov VV, Peterkova VA, Dedov II. Studying progression from glucose intolerance to type 2 diabetes in obese children. Diabetes Metab Syndr. 2014;8:133-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 122. | Koch M, Rett K, Maerker E, Volk A, Haist K, Deninger M, Renn W, Häring HU. The PPARgamma2 amino acid polymorphism Pro 12 Ala is prevalent in offspring of Type II diabetic patients and is associated to increased insulin sensitivity in a subgroup of obese subjects. Diabetologia. 1999;42:758-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 123. | Stumvoll M, Wahl HG, Löblein K, Becker R, Machicao F, Jacob S, Häring H. Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-gamma2 gene is associated with increased antilipolytic insulin sensitivity. Diabetes. 2001;50:876-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 124. | Priya SS, Sankaran R, Ramalingam S, Sairam T, Somasundaram LS. Genotype Phenotype Correlation of Genetic Polymorphism of PPAR Gamma Gene and Therapeutic Response to Pioglitazone in Type 2 Diabetes Mellitus- A Pilot Study. J Clin Diagn Res. 2016;10:FC11-FC14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 125. | Kang ES, Park SY, Kim HJ, Kim CS, Ahn CW, Cha BS, Lim SK, Nam CM, Lee HC. Effects of Pro12Ala polymorphism of peroxisome proliferator-activated receptor gamma2 gene on rosiglitazone response in type 2 diabetes. Clin Pharmacol Ther. 2005;78:202-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 126. | Zhang KH, Huang Q, Dai XP, Yin JY, Zhang W, Zhou G, Zhou HH, Liu ZQ. Effects of the peroxisome proliferator activated receptor-γ coactivator-1α (PGC-1α) Thr394Thr and Gly482Ser polymorphisms on rosiglitazone response in Chinese patients with type 2 diabetes mellitus. J Clin Pharmacol. 2010;50:1022-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 127. | Liu HL, Lin YG, Wu J, Sun H, Gong ZC, Hu PC, Yin JY, Zhang W, Wang D, Zhou HH. Impact of genetic polymorphisms of leptin and TNF-alpha on rosiglitazone response in Chinese patients with type 2 diabetes. Eur J Clin Pharmacol. 2008;64:663-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 128. | Sun H, Gong ZC, Yin JY, Liu HL, Liu YZ, Guo ZW, Zhou HH, Wu J, Liu ZQ. The association of adiponectin allele 45T/G and -11377C/G polymorphisms with Type 2 diabetes and rosiglitazone response in Chinese patients. Br J Clin Pharmacol. 2008;65:917-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 129. | Makino H, Shimizu I, Murao S, Kondo S, Tabara Y, Fujiyama M, Fujii Y, Takada Y, Nakai K, Izumi K. A pilot study suggests that the G/G genotype of resistin single nucleotide polymorphism at -420 may be an independent predictor of a reduction in fasting plasma glucose and insulin resistance by pioglitazone in type 2 diabetes. Endocr J. 2009;56:1049-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 130. | Jaakkola T, Laitila J, Neuvonen PJ, Backman JT. Pioglitazone is metabolised by CYP2C8 and CYP3A4 in vitro: potential for interactions with CYP2C8 inhibitors. Basic Clin Pharmacol Toxicol. 2006;99:44-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 131. | Kirchheiner J, Thomas S, Bauer S, Tomalik-Scharte D, Hering U, Doroshyenko O, Jetter A, Stehle S, Tsahuridu M, Meineke I. Pharmacokinetics and pharmacodynamics of rosiglitazone in relation to CYP2C8 genotype. Clin Pharmacol Ther. 2006;80:657-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 95] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 132. | Yan FF, Casey J, Shyng SL. Sulfonylureas correct trafficking defects of disease-causing ATP-sensitive potassium channels by binding to the channel complex. J Biol Chem. 2006;281:33403-33413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 133. | Huang C, Florez JC. Pharmacogenetics in type 2 diabetes: potential implications for clinical practice. Genome Med. 2011;3:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 134. | Zhang W, He YJ, Han CT, Liu ZQ, Li Q, Fan L, Tan ZR, Zhang WX, Yu BN, Wang D. Effect of SLCO1B1 genetic polymorphism on the pharmacokinetics of nateglinide. Br J Clin Pharmacol. 2006;62:567-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 135. | Huang Q, Yin JY, Dai XP, Wu J, Chen X, Deng CS, Yu M, Gong ZC, Zhou HH, Liu ZQ. Association analysis of SLC30A8 rs13266634 and rs16889462 polymorphisms with type 2 diabetes mellitus and repaglinide response in Chinese patients. Eur J Clin Pharmacol. 2010;66:1207-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 136. | Sheng FF, Dai XP, Qu J, Lei GH, Lu HB, Wu J, Xu XJ, Pei Q, Dong M, Liu YZ. NAMPT -3186C/T polymorphism affects repaglinide response in Chinese patients with Type 2 diabetes mellitus. Clin Exp Pharmacol Physiol. 2011;38:550-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 137. | Dai XP, Huang Q, Yin JY, Guo Y, Gong ZC, Lei MX, Jiang TJ, Zhou HH, Liu ZQ. KCNQ1 gene polymorphisms are associated with the therapeutic efficacy of repaglinide in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol. 2012;39:462-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 138. | Xiang Q, Cui YM, Zhao X, Yan L, Zhou Y. The Influence of MDR1 G2677T/a genetic polymorphisms on the pharmacokinetics of repaglinide in healthy Chinese volunteers. Pharmacology. 2012;89:105-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 139. | Cheng Y, Wang G, Zhang W, Fan L, Chen Y, Zhou HH. Effect of CYP2C9 and SLCO1B1 polymorphisms on the pharmacokinetics and pharmacodynamics of nateglinide in healthy Chinese male volunteers. Eur J Clin Pharmacol. 2013;69:407-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 140. | Takane H. Genetic polymorphisms of SLCO1B1 for drug pharmacokinetics and its clinical implications. Yakugaku Zasshi. 2011;131:1589-1594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 141. | He J, Qiu Z, Li N, Yu Y, Lu Y, Han D, Li T, Zhao D, Sun W, Fang F. Effects of SLCO1B1 polymorphisms on the pharmacokinetics and pharmacodynamics of repaglinide in healthy Chinese volunteers. Eur J Clin Pharmacol. 2011;67:701-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 142. | Kalliokoski A, Backman JT, Neuvonen PJ, Niemi M. Effects of the SLCO1B1*1B haplotype on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. Pharmacogenet Genomics. 2008;18:937-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 143. | Kalliokoski A, Neuvonen M, Neuvonen PJ, Niemi M. Different effects of SLCO1B1 polymorphism on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. J Clin Pharmacol. 2008;48:311-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 144. | Kalliokoski A, Neuvonen M, Neuvonen PJ, Niemi M. The effect of SLCO1B1 polymorphism on repaglinide pharmacokinetics persists over a wide dose range. Br J Clin Pharmacol. 2008;66:818-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 145. | Holstein A, Seeringer A, Kovacs P. Therapy with oral antidiabetic drugs: applied pharmacogenetics. Br J Diabetes Vascular Disease. 2011;11:10-16. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 146. | Dicker D. DPP-4 inhibitors: impact on glycemic control and cardiovascular risk factors. Diabetes Care. 2011;34 Suppl 2:S276-S278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 147. | Vincent SH, Reed JR, Bergman AJ, Elmore CS, Zhu B, Xu S, Ebel D, Larson P, Zeng W, Chen L. Metabolism and excretion of the dipeptidyl peptidase 4 inhibitor [14C]sitagliptin in humans. Drug Metab Dispos. 2007;35:533-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 114] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 148. | Zimdahl H, Ittrich C, Graefe-Mody U, Boehm BO, Mark M, Woerle HJ, Dugi KA. Influence of TCF7L2 gene variants on the therapeutic response to the dipeptidylpeptidase-4 inhibitor linagliptin. Diabetologia. 2014;57:1869-1875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 149. | Sun X, Yu W, Hu C. Genetics of type 2 diabetes: insights into the pathogenesis and its clinical application. Biomed Res Int. 2014;2014:926713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |