Published online May 15, 2025. doi: 10.4239/wjd.v16.i5.95431
Revised: January 13, 2025
Accepted: March 24, 2025
Published online: May 15, 2025
Processing time: 380 Days and 0.3 Hours
Type 2 diabetes mellitus is characterized by pancreatic β-cell dysfunction and insulin resistance. Studies have suggested that β-cell dedifferentiation is one of the pathogeneses of β-cell dysfunction, but the detailed mechanism is still unclear. Most studies of β-cell dedifferentiation rely on rodent models and human pa
To investigate the molecular mechanism of β-cell dedifferentiation. Hence, an in vitro model of β-cell dedifferentiation induced by palmitic acid and high glucose was established using the INS-1 832/13 cell line.
The study was further analyzed using RNA-sequencing, transmission electron microscopy, quantitative real-time polymerase chain reaction and Western blot.
Results showed that the treatment of palmitic acid and high glucose significantly up-regulated β-cell forbidden genes and endocrine precursor cell marker genes, and down-regulated the expression of β-cell specific markers. Data showed that dedifferentiated INS-1 cells up-regulated the expression of endoplasmic reticulum (ER) stress-related genes. Moreover, the results also showed that forkhead box O1 (Foxo1) inhibition potentiated genetic changes in β-cell dedifferentiation induced by palmitic acid and high glucose.
ER stress is sufficient to trigger β-cell dedifferentiation and is necessary for palmitic acid and high glucose-induced β-cell dedifferentiation. Foxo1 inhibition can further enhance these phenomena.
Core Tip: β-cell dedifferentiation is one cause of β cell dysfunction, but the underlying mechanisms remain unclear. In this study, we established an in vitro model of β-cell dedifferentiation induced by high-glucose and palmitic acid and showed that the inhibition of forkhead box O1 can further enhance these phenomena. We found that high glucose and palmitic acid treatments significantly downregulated the expression of β-cell-specific markers and upregulated the expression of β-cell-disabling genes and endocrine precursor cell marker genes by RNA-sequencing, and that endoplasmic reticulum stress is the key to β-cell dedifferentiation. This study will be beneficial for investigating the mechanism of β-cell dysfunction caused by dedifferentiation.
- Citation: Wang LK, Kong CC, Yu TY, Sun HS, Yang L, Sun Y, Li MY, Wang W. Endoplasmic reticulum stress and forkhead box protein O1 inhibition mediate palmitic acid and high glucose-induced β-cell dedifferentiation. World J Diabetes 2025; 16(5): 95431
- URL: https://www.wjgnet.com/1948-9358/full/v16/i5/95431.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i5.95431
The rapid growth of type 2 diabetes (T2D) worldwide is one of the most serious health issues today. According to the International Diabetes Federation, there are 463 million people with diabetes in the world, more than 90% of whom are T2D patients[1]. T2D is a complicated multifactorial disorder, characterized by peripheral insulin resistance and pancreatic β-cell loss or β-cell dysfunction[2]. It has been reported that β-cell dysfunction can be caused by β-cell death, β-cell senescence, β-cell transdifferentiation and/or β-cell dedifferentiation[3].
Accumulating evidence reveals that a primary mechanism for T2D is β-cell dedifferentiation[4], a process in which terminally differentiated β cells degenerate to an immature or precursor-like state under stress. Although there is ambiguity about dedifferentiation, the following characteristics have reached consensus: (1) Downregulation of β-cell-enriched genes, including β-cell specific transcription factors, glucose metabolism genes, insulin synthesis and secretion genes; (2) Upregulation of forbidden genes with low expression in mature β cells; and (3) Upregulation of endocrine precursor cell marker genes[5,6]. These changes in β cells lead to the loss of their identity, which impairs their ability to synthesize and secrete insulin in response to elevated blood glucose levels[7]. These observations of β-cell dedifferentiation have been confirmed in diabetic animal models or diabetic patients[8-11].
Both genetic and environmental factors contribute to β-cell dedifferentiation. In mice, the Kir6.2 gain-of-function mutant results in ectopic expression, causing β cells to dedifferentiate into neurogenin 3 (Ngn3)-positive and insulin-negative states and lose their mature identity, leading to neonatal diabetes[12]. Likewise, there is a loss of another key component of the Kadenosine triphosphate-channel Abcc8 in mice β cells, an increase of β-cell dedifferentiation marker aldehyde dehydrogenase 1 family member A3 (Aldh1a3), and an alteration of β-cell identity[13]. These data suggest that β-cell membrane depolarization of glucose-stimulated insulin secretion is one of the important mechanisms for β-cell dedifferentiation.
Inflammation can also induce β-cell dedifferentiation. For example, tumor necrosis factor-α, interleukin-6 and interleukin-8 facilitate β-cell dedifferentiation in cultured mouse and human islets[14]. Similar results were found in patients with nondiabetic chronic pancreatitis, increasing the proportion of β-cell dedifferentiation[10]. These data suggest that inflammation-induced β-cell dedifferentiation is an environmental factor for diabetes progress. Moreover, metabolic stress is another factor for β-cell dedifferentiation. Both db/db mice and obese mice induced by a high-fat diet showed β-cell dedifferentiation[9,11]. Loss of the β-cell specific gene forkhead box O1 (Foxo1) in mice led to β-cell dedifferentiation under aging and multiparity, indicating decreased insulin expression and increased expression of the islet progenitor markers myelocytomatosis viral oncogene homolog 1, homeobox transcription factor, octamer-binding transcription factor 4 (Oct4) and Ngn3[15]. However, β-cell dedifferentiation is not always detrimental to the diabetes process. A recent study showed that β-cell specific deletion of the unfolded protein response sensor inosital-requiring enzyme-1 before insulitis caused transient β-cell dedifferentiation and prevented autoimmune destruction in NOD mice[16]. More importantly, dedifferentiated β cells can be induced to redifferentiate into functional β cells[12,17], which provides information for the development of antidiabetic strategies targeting β-cell dedifferentiation[17-20].
Although β cell dedifferentiation is considered an important mechanism of β-cell dysfunction in T2D, most studies rely on in vivo mouse models or isolated mouse/human islets and cadaveric pancreatic tissues. Moreover, the mechanism linking β-cell dedifferentiation to cellular stress and genetic factors is still unclear. Hence, the development of in vitro systems helps to implicate the underlying mechanism of β-cell dedifferentiation. A recent study modeled human β-cell dedifferentiation by using fibroblast growth factor-2 to stimulate the human β-cell line EndoC-βH1[21].
In this study, we investigated the molecular mechanism of β-cell dedifferentiation by inducing the rat β-cell line INS-1 to dedifferentiate by stimulation with palmitic acid (PA) and high glucose. We found that Foxo1 inhibition potentiated the dedifferentiated state of INS-1 cells. Additionally, we performed RNA-sequencing (RNA-seq) to analyze the genes and pathways in dedifferentiated INS-1 cells. Data further suggested that PA and high glucose induced INS-1 cell dedifferentiation through endoplasmic reticulum (ER) stress, and Foxo1 inhibition can enhance this process.
PA (Sigma Aldrich P9767) was diluted in water at 65 °C to a concentration of 100 mmol/L and finally diluted in serum-free Roswell Park Memorial Institute (RPMI) 1640 containing 10% bovine serum albumin (BSA) (Sigma Aldrich A730) and 25 mmol/L glucose to a concentration of 300 μM. The rat insulinoma INS-1 cell line (INS-1 832/13) was cultured in RPMI 1640 medium (11 mmol/L glucose) supplemented with 10% fetal bovine serum, 100 μg/mL streptomycin, 50 μM β-mercaptoethanol, 25 mmol/L 4-(2-hydroxyethyl)piperazine-1-ethane-sulfonate, 100 U/mL penicillin, and 2 mmol/L L-glutamine at 5% carbon dioxide and 37 °C. Cells were divided into the control group, glucose (Glu) + PA (25 mmol/L glucose + 300 μmol/L PA), and the treatment group, Glu + PA + AS (25 mmol/L glucose + 300 μmol/L PA + 1 μM AS1842856). The cells were treated for 36 hours before further experiments.
Treated cells were washed three times with cold phosphate-buffered saline (PBS) before adding TRIZOL reagent (Thermo Fisher Scientific, Waltham, MA, United States) to extract total RNA. Absorbance at 260 nm and 280 nm was measured using the nanodrop1000 spectrophotometer (SpectraMax 190, Molecular Devices, United States), to estimate total RNA concentration. The integrity of ribosomal RNA at 18S and 28S was assessed on a 1% agarose gel. Three repeated biological tests were conducted for each treatment group. The complementary DNA (cDNA) was reverse transcribed by M-MLV (Promega, Madison, WI, United States), followed by quantitative polymerase chain reaction (qPCR) using SYBR green qPCR mix kit (Thermo Fisher Scientific, Carlsbad, CA, United States). Forty cycles of amplification (95 °C denaturation for 15 seconds, 60 °C annealing and extension for 1 minute) were performed after an initial 95 °C for 2 minutes. The 2-ΔΔCt method was used to quantify the relative level of mRNA expression for each target gene. 18S was chosen as an internal reference gene.
INS-1 cells treated under different conditions were washed with PBS and collected quickly, then lysed in cold radioimmunoprecipitation assay lysis buffer (Jrdun, Shanghai, China) containing phosphatase and protease inhibitors. The supernatant was collected by centrifugation (12000 rpm, 10 minutes), and the protein concentration was determined using a BCA protein detection kit (American Thermal Science Company). The same amount of protein (50 μg) was isolated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to the polyvinylidene fluoride fixed membrane. Primary antibodies against Aldh1a3 (1:1000, Novus), pancreatic and duodenal homeobox 1 (Pdx1) (1:1000, Abcam), Foxo1 (1:1000, Proteintech), Oct4 (1:1000, Abcam), Tubulin (1:5000, Abcam), Caspase-3 (1:1000, Abcam), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1000, Proteintech), Akt (1:1000, Proteintech) and p-Akt (1:1500, Proteintech) were incubated at 4 °C overnight, followed by adding the goat anti-rabbit/mouse secondary antibody (1:5000, Yeasen), and then incubated at room temperature for 2 hours. The signal was detected by a chemiluminescence reagent. The protein density was quantified using Image J software. Tubulin or GAPDH was used to verify the equal loading of proteins. Three independent biological experiments were performed for each group.
Cells were seeded on coverslips and allowed to attach overnight. The treated cells were washed in PBS, fixed with 4% paraformaldehyde, followed by permeabilization and blocking in PBS containing 5% normal goat serum and 0.2% Triton X-100 for 2 hours at room temperature, and then incubated overnight with the primary antibody against Aldh1a3 (1:1000, Novus), Pdx1 (1:1000, Abcam) and Insulin (1:1000, Dako). After washing, cells were incubated with the following secondary antibodies: Alexa Fluor 488 anti rabbit (Thermo, China), Alexa Fluor 594 anti guinea pig (Thermo, China) or Alexa Fluor 647 anti rabbit (Thermo, China) at room temperature for 2 hours. 4’,6-Diamidino-2’-phenylindole (DAPI) staining of the nucleus was performed for 5 minutes. The slides were examined and imaged using a confocal microscope (Leica SP8). The confocal microscopy software package (Leica SP8) was used for quantitative analysis of Insulin, Aldh1a3 and Pdx1 in three different experiments. The fluorescence intensity of cells in three visual fields was calculated, as expressed by mean ± SEM, with P < 0.05 for the treatment group and P < 0.01 for the control group.
The isolated RNA was subsequently used for RNA-seq. The Beijing Genomics Institute (BGI, China) analyzed the total RNA of all samples. Three parallel replicates were set for each group. Magnetic beads were used to extract total RNA, followed by treatment using the RNA enrichment method. The BGISEQ-500 platform was then used to perform cDNA library construction and sequencing. The sequencing data was filtered with SOAPnuke (v1.5.2)[22] by deleting the reads with adaptor sequences, the reads with a low base ratio of over 20%, and the reads with an unknown base “N” of over 5%. After filtering, clean reads were mapped to the rat genome (Rattus norvegicus, Rnor_6.0) using HISAT2 (v2.0.4)[23]. Bowtie2 (v2.2.5) was used to align clean reads with rat coding genes[24], and RNA-Seq by expectation maximization (v1.2.12) was used to calculate the level of gene expression[25]. Analysis of partial data was conducted on the Dr. Tom network platform of BGI (http://report.bgi.com), including quantitative gene analysis and differentially expressed gene (DEG) analysis, according to gene expression, enrichment analyses of Gene Ontology (GO), and pathway significance enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG), clustering, etc. The adjusted P value ≤ 0.001 and genes with |log2 fold change| ≥ 1.00 were considered as differentially expressed genes (DEGs) with statistical significance. Deposited in the National Center for Biotechnology Information database, these sequence data are available through BioProject ID: PRJNA661135.
DEGs were used for network construction by the search tool for the retrieval of interacting genes/proteins (STRING) (https://string-db.org/). The required confidence (combined score) greater than 0.4 was used as the threshold for positive protein-protein interaction (PPI). Cytoscape software (version: 3.7.2) was used to visualize the PPI network. Subsequently, the top 20 genes were chosen as target hub genes through the Cytoscape plugin cytohubba based on the highest degree of connectivity.
INS-1 cells were seeded in the 12-well plate and cultured for 1 day in 5% CO2 and 37 °C. After cells were pretreated under different experimental conditions, the medium was removed and washed with Krebs-Ringer bicarbonate HEPES buffer (KRBH) twice. KRBH buffer containing 0.1% BSA was used to starve cells for 1 hour, while KRBH buffer containing 2.8 mmol/L and 16.7 mmol/L glucose was used to incubate cells for 2 hours, and the culture medium was collected. The supernatant was gathered by spinning at 1000 g at 4 °C for 5 minutes and put on ice, and then incubated according to the instructions of the rat insulin immunoassay kit (ImmunoDiagnostics, HK, China). Absorbance at 450 nm was detected. The insulin concentration of the samples was determined according to the standard curve.
Small clumps of cells were immediately fixed in 2.5% glutaraldehyde for at least 2 hours at 4 °C. After fixation, cells were washed with 0.1 M phosphate buffer containing dibasic sodium phosphate and sodium dihydrogen phosphate, then dehydrated and resin embedded. Images were taken using a Hitachi HT-7800 microscope. Morphology was used to identify β cells. Image J was used to measure the width of the ER lumen of four cells in each group, with at least 15 different positions of the same cells for measurement. Data are expressed as mean ± SEM, with P < 0.05 for the treatment group and P < 0.01 for the control group.
Statistics were analyzed with one-way analysis of variance followed by Bonferroni’s post hoc test or t-test (GraphPad Prism 8 software). Data were indicated as mean or mean ± SEM. P < 0.05 was considered significant.
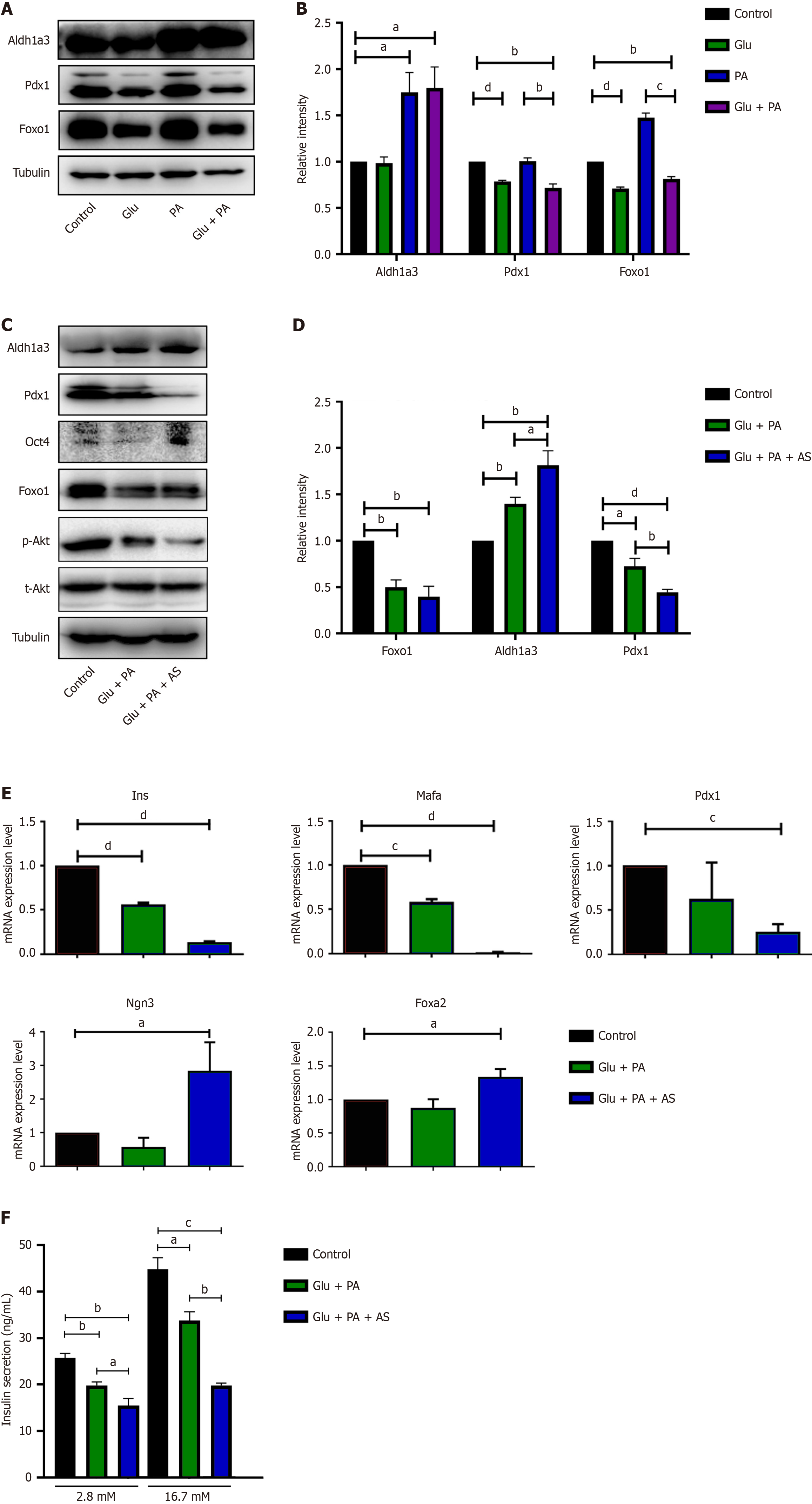
To simulate in vivo conditions, similar conditions were applied, including high glucose (25 mmol/L), PA (300 μmol/L), and high glucose plus PA (Glu + PA, Glu 25 mmol/L + PA 300 μmol/L), to stimulate INS-1 832/13 cells in vitro for 36 hours. As shown in Figure 1A and B, the β-cell dedifferentiation marker Aldh1a3 was significantly upregulated upon the stimulation of PA and Glu + PA, but not high glucose. Meanwhile, the two key transcription factors Pdx1 and Foxo1 were inhibited by Glu + PA stimulation (Figure 1A and B). These data indicated that INS-1 stimulation with metabolic stress of glucose and fatty acid led to β-cell dedifferentiation in vitro.
Foxo1 has been suggested to maintain β cell stability and prevent β-cell dedifferentiation during diabetes progression [26]. In addition, PA and high glucose induced β-cell dedifferentiation in INS-1 cells and suppressed Foxo1 protein levels (Figure 1A). Hence, we investigated the role of Foxo1 in the process of in vitro β-cell dedifferentiation. The Foxo1-specific inhibitor AS1842856 was selected to potently inhibit Foxo1 transactivation without affecting its transcription and protein levels[27,28]. INS-1 832/13 cells were treated with BSA (control), 25 mmol/L glucose plus 300 μmol/L PA (Glu + PA), or 25 mmol/L glucose plus 300 μmol/L PA plus 1 μmol/L AS1842856 (Glu + PA + AS) for 36 hours. These treatments failed to induce β-cell apoptosis, as cleaved caspase-3 protein was not detected and there were no significant cell fragments or floating cells in any of the three groups (Supplementary Figure 1 and data not shown). After treatment, we detected Akt and p-Akt levels to demonstrate the effect of AS1842856 on INS-1 cells. There was no significant difference in total Akt expression, but p-Akt expression decreased compared to the control group and Glu + PA + AS group (Supplementary Figure 1). Moreover, we measured several key markers for β-cell identity and β-cell dedifferentiation after treatment. As shown in Figure 1C and D, Aldh1a3 protein levels significantly increased in the Glu + PA + AS group compared to the Glu + PA group. Additionally, Pdx1 protein, a critical marker of differentiated β cells, significantly decreased. Quantitative real-time polymerase chain reaction (qRT-PCR) was also performed to analyze the mRNA levels of several genes. The β-cell maturation markers Ins, Mafa, and Pdx1 were all downregulated in the Glu + PA group, and the expression levels of these genes were much lower in the Glu + PA + AS group (Figure 1E). Although the progenitor cell markers Foxa2 and Ngn3 were not changed in the Glu + PA group, they were significantly downregulated in the Glu + PA + AS group (Figure 1E).
Because dedifferentiated β cells often lose their specific identity and function, we measured their insulin secretion under different conditions. The results showed that after treatment, insulin secretion stimulated by 2.8 mmol/L and 16.7 mmol/L glucose significantly decreased in the Glu + PA group and further decreased after Foxo1 inhibition in the Glu + PA + AS group (Figure 1F). These data suggest that insulin secretion was impaired in dedifferentiated β cells.
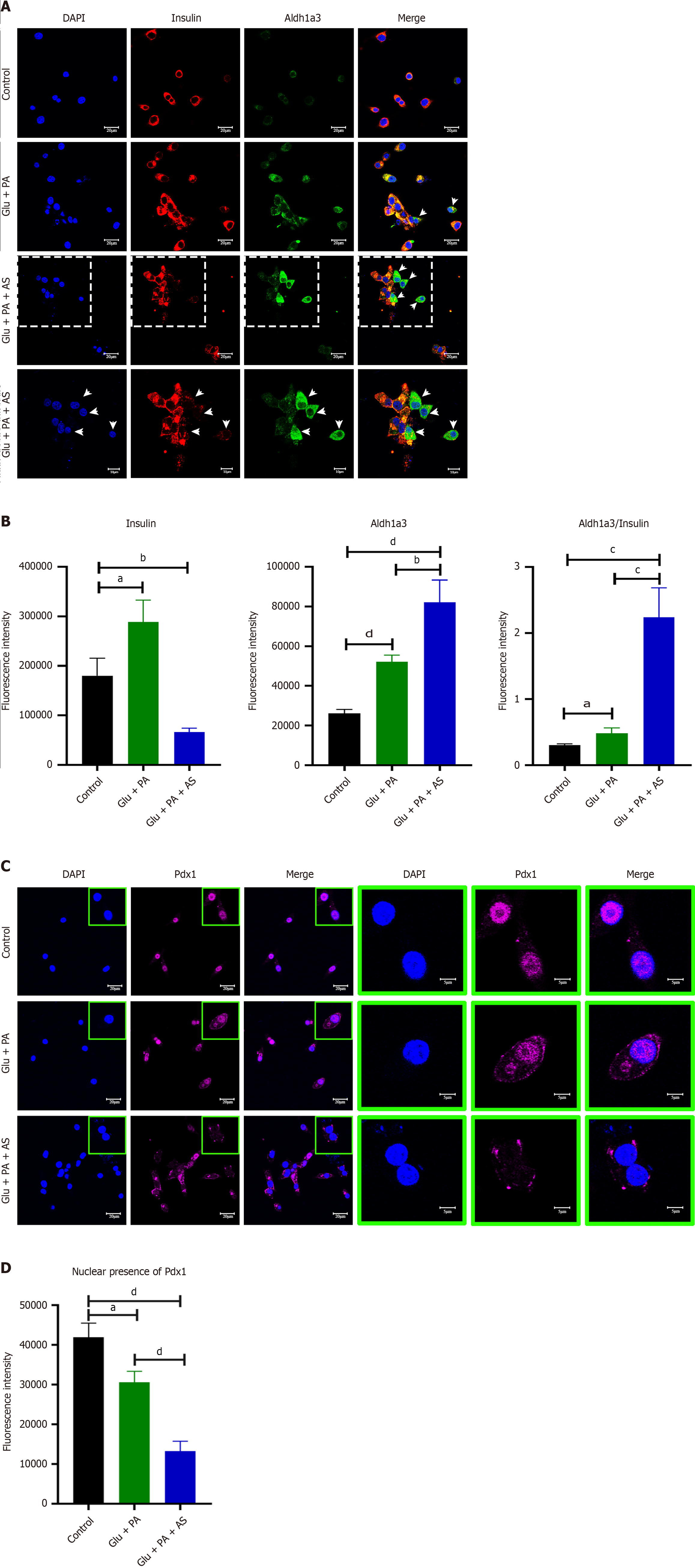
To further confirm the results obtained from Western blot and qRT-PCR, immunofluorescence staining was performed for insulin, Aldh1a3 and Pdx1. The insulin signal was dramatically reduced in the Glu + PA + AS group, but it was increased in the Glu + PA group (Figure 2A and B). Both the percentage of Aldh1a3-positive cells and fluorescence increased in the Glu + PA group, and this trend was more obvious in the Glu + PA + AS group (Figure 2A and B). Interestingly, an increasing number of cells with cytoplasmic Pdx1 was observed in the Glu + PA group and the Glu + PA + AS group (Figure 2C and D). Taken together, these data suggest that PA and high glucose stimulation induced β-cell dedifferentiation and Foxo1 inhibition potentiated the process.
RNA-seq analysis confirmed PA and high glucose-induced β-cell dedifferentiation
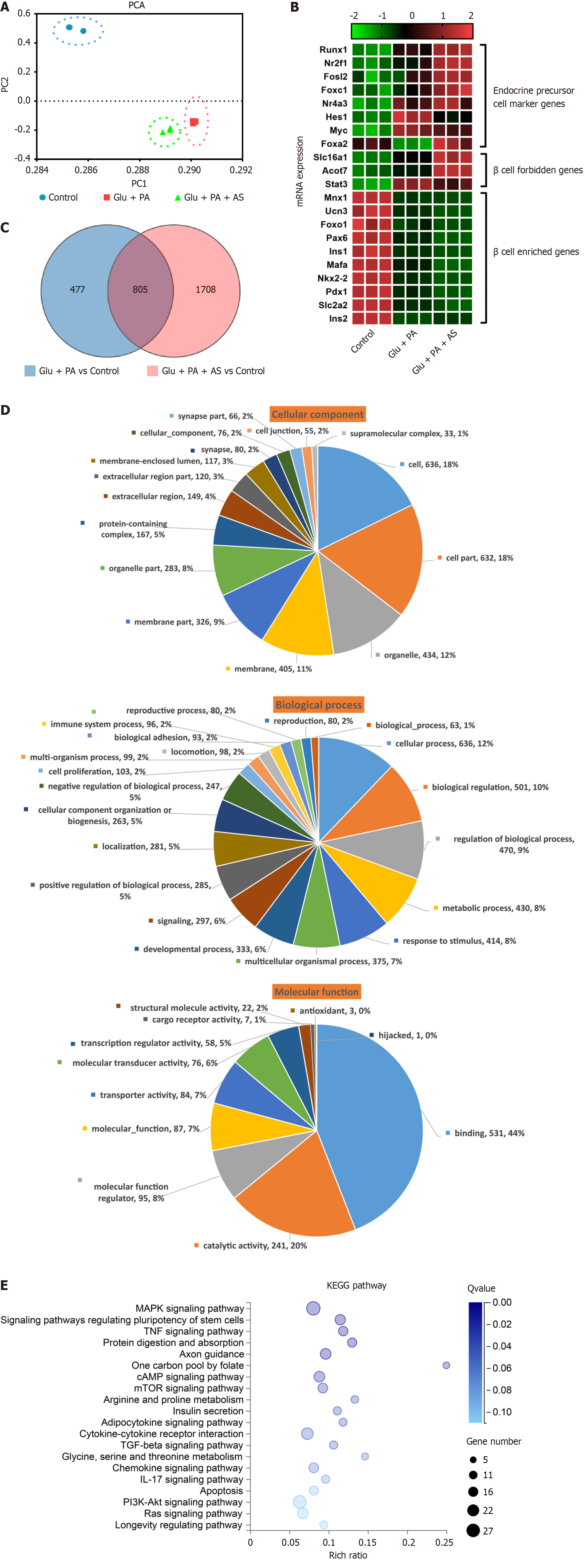
To further investigate the underlying molecular mechanism of β-cell dedifferentiation induced by Glu + PA and Glu + PA + AS, we performed RNA-seq to analyze transcriptional profiles. A total of nine libraries were constructed from three groups (control, Glu + PA, Glu + PA + AS), each with three biological replicates. After sequencing and low-quality data filtration, a total of 202.63 million clean reads were generated with an average of 22.51 million reads for individual samples. The percentages of total mapping and unique mapping were 94.86%-95.43% and 80.67%-83.62%, respectively (Table 1). The results of principal component analysis indicated distinct clustering of datasets by treatment group (Figure 3A). Because β-cell dedifferentiation is tightly controlled by many genes[5], we analyzed genes related to β-cell dedifferentiation, including endocrine precursor cell marker genes, β-cell forbidden genes and β-cell enriched genes. Both Glu + PA and Glu + PA + AS groups upregulated endocrine precursor cell marker genes (Myc, Hes1, Runx1, Nr2f1, Nr4a3, Fosl2 and Foxo1) and β-cell forbidden genes (Slc16a1, Stat3 and Acot7), while β-cell enriched genes (Mnx1, Ucn3, Foxo1, Pax6, Ins1, Ins2, Mafa, Nkx2.2, Pdx1 and Slc2a2) were downregulated (Figure 3B). Interestingly, most genes in the Glu + PA + AS group changed more dramatically than those in the Glu + PA group (Figure 3B). To further evaluate the transcriptome sequencing results, we performed qRT-PCR to measure genes in the categories of β cell identity, developmental process, metabolism, development, and cell differentiation. The qRT-PCR results of these genes showed an upregulation or downregulation consistent with the data obtained from RNA-seq, indicating that the DEGs obtained by transcriptome sequencing were reliable (Supplementary Figure 2). Taken together, these data confirm that PA and high glucose stimulation induced INS-1 β cell dedifferentiation.
| Sample | Total raw reads (M) | Total clean reads (M) | Total clean bases (Gb) | Clean reads Q20 (%) | Clean reads Q30 (%) | Clean reads ratio (%) |
| Control_1 | 23.62 | 22.29 | 1.11 | 98.99 | 95.71 | 94.37 |
| Control_2 | 23.62 | 22.47 | 1.12 | 99.04 | 96.06 | 95.12 |
| Control_3 | 25.78 | 23.71 | 1.19 | 98.94 | 95.78 | 91.98 |
| Glu + PA_1 | 23.62 | 22.38 | 1.12 | 98.96 | 95.59 | 94.76 |
| Glu + PA_2 | 23.63 | 22.36 | 1.12 | 98.95 | 95.9 | 94.64 |
| Glu + PA_3 | 23.64 | 22.33 | 1.12 | 98.87 | 95.56 | 94.45 |
| Glu + PA + AS_1 | 23.64 | 22.37 | 1.12 | 98.9 | 95.38 | 94.65 |
| Glu + PA + AS_2 | 23.64 | 22.36 | 1.12 | 98.96 | 95.61 | 94.61 |
| Glu + PA + AS_3 | 23.62 | 22.36 | 1.12 | 98.89 | 95.27 | 94.67 |
DEGs were then analyzed. We identified 1282 DEGs in the Glu + PA group (664 upregulated and 618 downregulated) and 2513 DEGs in the Glu + PA + AS group (938 upregulated and 1575 downregulated) (Supplementary Figure 3). Compared with the control group, Venn diagram analysis further revealed that 805 DEGs were shared by Glu + PA and Glu + PA + AS groups (Figure 3C), followed by GO and KEGG analyses. Based on GO terms, the shared 805 DEGs were annotated and classified into three categories: Cellular component, biological process, and molecular function. In the “cellular component” category, both “cell” (18%) and “cell part” (18%) were the top two categories, followed by “organelle” (12%). The “regulation of biological process” (9%), “biological regulation” (10%), and “cellular process” (12%) were the major sub-classes of the “biological process” category. The “catalytic activity” (20%) and “binding” (44%) were the two main sub-classes of the “molecular function” category (Figure 3D).
KEGG pathway enrichment analysis was performed on the 805 shared DEGs, and a total of 55 significant pathways were enriched (P < 0.05). The top 20 enriched pathways are shown in Table 2. The cytokine-cytokine receptor interaction, phosphatidylinositol-3-kinase (PI3K)-Akt signaling pathway and mitogen-activated protein kinase (MAPK) signaling pathway were enriched with 21, 27, and 28 genes, respectively (Figure 3E and Table 2). Moreover, several metabolic-related pathways, such as the cyclic adenosine monophosphate (cAMP) signaling pathway, mammalian target of rapamycin signaling pathway, insulin secretion pathway and Ras signaling pathway, were also enriched with more than 10 genes (Figure 3E and Table 2). These data suggest that PA and high glucose stimulation resulted in metabolic remodeling in β cells, which may in turn induce β-cell dedifferentiation.
| Pathway terms | Categories | Genes |
| MAPK signaling pathway | Signal transduction | Relb, Ddit3, Fgfr3, Tgfb2, Dnah12, Csf1, Ntrk2, Bdnf, Rasgrf1, Fgf21, Tnf, Atf4, Efna1, Ins2, Jund, Cacnb1, Jun, Myc, Ntrk1, Fgf22, LOC108348082, Prkcg, LOC108348108, Cacna2d4, Fgfr4, Pla2g4e, Igf1r |
| Signaling pathways regulating pluripotency of stem cells | Cellular community-eukaryotes | Esrrb, Fgfr3, Acvr1c, Bmpr1b, Pik3cb, Wnt10b, Id2, Wnt7a, Wnt4, Inhbe, Wnt9a, Myc, Onecut1, Id1, Fgfr4, Fzd6, Igf1r |
| TNF signaling pathway | Signal transduction | Cxcl1, Ccl20, Pik3cb, Csf1, Creb5, Cxcl10, Nfkbia, Tnf, Atf4, Tnfaip3, Cx3cl1, Jun, Icam1, LOC108348082 |
| Protein digestion and absorption | Digestive system | Slc3a2, Col6a6, Col5a3, Mep1a, Col12a1, Kcnk5, Col3a1, Slc6a19, Col5a2, Col27a1, Dpp10, Slc16a10, Slc1a5 |
| Axon guidance | Development | Robo2, Sema3c, Bmpr1b, Epha1, Pik3cb, Unc5d, Epha7, Epha4, Ephb4, Ntn1, Ntn4, Efna1, Wnt4, Ablim2, Dcc, Ptch1, Sema6a, Unc5b |
| One carbon pool by folate | Metabolism of cofactors and vitamins | Mtr, Aldh1L2, LOC100910688, Mthfd1 L, Mthfd2 |
| cAMP signaling pathway | Signal transduction | Cngb3, Pld1, Pde3a, Pik3cb, Bdnf, Adcy1, Adcyap1r1, Creb5, Atp2b4, Tiam1, Nfkbia, Jun, Ppara, Ptch1, Cngb1, Gipr, Npy, Gpr119 |
| mTOR signaling pathway | Signal transduction | Slc3a2, Ddit4, Rnf152, Pik3cb, Wnt10b, Eif4e2, Tnf, Wnt7a, Wnt4, Ins2, Wnt9a, Prkcg, Sgk1, Fzd6, Igf1r |
| Arginine and proline metabolism | Amino acid metabolism | Gatm, Pycr1, Ckmt1, Arg1, Aldh4a1, Azin2, Maob, Odc1 |
| Insulin secretion | Endocrine system | Cckar, Adcy1, Kcnj11, Adcyap1r1, Creb5, Atf4, Pdx1, Ins2, Prkcg, Gpr119 |
| Adipocytokine signaling pathway | Endocrine system | Ppargc1a, Prkcq, G6pc, Nfkbia, Tnf, Ppara, Irs3, Npy, Rxrg |
| Cytokine-cytokine receptor interaction | Signaling molecules and interaction | Cxcl1, Ltb, Ccl20, Tnfrsf21, Tgfb2, Acvr1c, Bmpr1b, Cntfr, Csf1, Cxcl11, Cxcl10, Tnf, Thpo, Cx3cl1, Inhbe, Cxcl16, Il9r, Il12rb1, Il2rb, Il17re, Ackr3 |
| TGF-beta signaling pathway | Signal transduction | Tgfb2, Acvr1c, Bmpr1b, Tgif1, Id2, Tnf, Inhbe, Myc, Id1, Chrd |
| Glycine, serine and threonine metabolism | Amino acid metabolism | Gatm, Agxt2, Psat1, Sds, Maob, Psph |
| Chemokine signaling pathway | Immune system | Cxcl1, Grk5, Ccl20, Pik3cb, Adcy1, Cxcl11, Tiam1, Cxcl10, Nfkbia, Shc4, Cx3cl1, Lyn, Cxcl16, Elmo1, Gng8 |
| IL-17 signaling pathway | Immune system | Cxcl1, Mapk15, Ccl20, Cxcl10, Nfkbia, Tnf, Tnfaip3, Jund, Jun, Il17re |
| Apoptosis | Cell growth and death | Ddit3, Pik3cb, Ctsz, Nfkbia, Tnf, Atf4, Ern1, Jun, Ctss, Endog, Ntrk1, Hrk, Casp6 |
| PI3K-Akt signaling pathway | Signal transduction | Fgfr3, Ddit4, Col6a6, Vwf, Pik3cb, Csf1, Ntrk2, Bdnf, Creb5, Fgf21, G6pc, Eif4e2, Atf4, Efna1, Ins2, Spp1, Myc, Ntrk1, Fgf22, Gng8, Il2rb, Fgfr4, Sgk1, Ppp2r5b, Igf1r |
| Ras signaling pathway | Signal transduction | Pld1, Fgfr3, Pik3cb, Csf1, Ntrk2, Bdnf, Rasgrf1, Fgf21, Tiam1, Shc4, Efna1, Ins2, Ntrk1, Fgf22, Gng8, Prkcg, Fgfr4, Pla2g4e, Igf1r |
| Longevity regulating pathway | Aging | Ppargc1a, Pik3cb, Adcy1, Creb5, Eif4e2, Atf4, Ins2, Irs3, Igf1r |
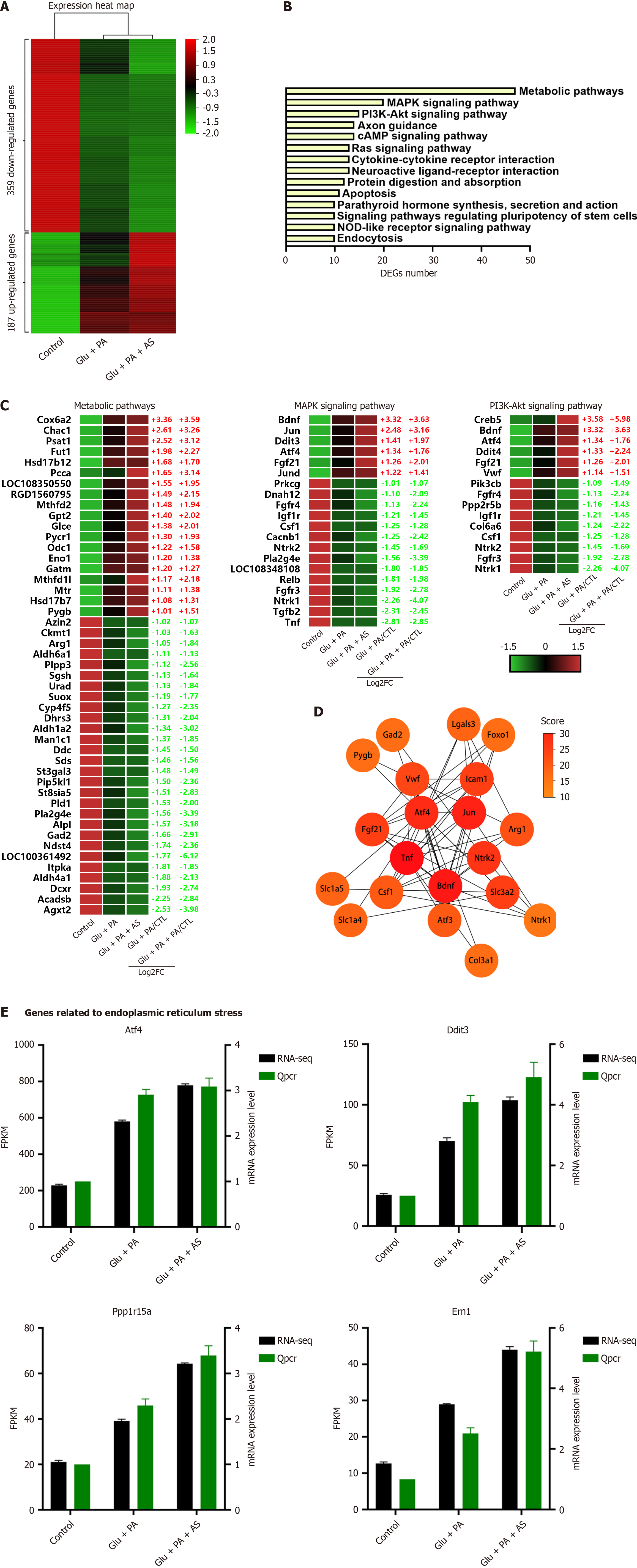
Because Foxo1 inhibition enhanced high glucose and PA-induced β-cell dedifferentiation (Figure 1), we characterized the genes associated with this phenomenon. After screening for genes with a consistent trend from DEGs, we found that the expression of 359 DEGs was gradually downregulated and 187 DEGs were gradually upregulated (Figure 4A). KEGG pathway analysis showed that metabolic pathway, MAPK pathway and PI3K-Akt pathway were the top three enriched pathways (Figure 4B). For the metabolic pathway, 47 genes were altered with 19 genes upregulated and 28 genes downregulated (Figure 4C). These genes primarily belonged to pathways of carbohydrate metabolism, lipid metabolism, cofactor and vitamin metabolism, and amino acid metabolism, such as Cox6a2, Gpt2, Sds, Gad2 and Agxt2. Moreover, 20 genes belonged to the MAPK signaling pathway and 15 genes belonged to the PI3K-Akt signaling pathway. Interestingly, many DEGs were shared by the two pathways, including growth factor (GF), receptor tyrosine kinases (RTKs), and cAMP-response element binding protein. These genes belonging to GF (Bdnf, Fgf21, Csf1) and RTKs (Fgfr3, Fgfr4, Ntrk1, Ntrk2, Igf1r) were all significantly downregulated (Figure 4C). These data indicate that β-cell dedifferentiation reinforced by Foxo1 deficiency under PA and high glucose was related to the PI3K-Akt signaling pathway, MAPK signaling pathway, and metabolic pathway.
The STRING database was used for PPI network analysis to explore the functional connectivity of 546 DEGs. STRING mapped Foxo1 and 546 DEGs to the PPI network with 356 nodes and 788 edges (Supplementary Figure 4), and then the network from STRING was further calculated by the Cytoscape software. The hub genes obtained from the PPI network by the cytoHubba plugin and Degree algorithm are shown in Supplementary Figure 3. The top 20 genes were Tnf, Bdnf, Jun, Atf4, Ntrk2, Slc3a2, Icam1, Fgf21, Vwf, Arg1, Atf3, Csf1, Lgals3, Slc1a5, Slc1a4, Col3a1, Foxo1, Gad2, Pygb and Ntrk1 (Figure 4D), indicating that these genes were the core genes in β-cell dedifferentiation enhanced by Foxo1 inhibition. In addition, Atf4 ranked in the top five among these 20 genes and its expression level was the most abundant among these genes. Hence, the Atf4 pathway and ER stress pathway were investigated. Both RNA-seq and qRT-PCR results indicated that ER stress-related genes (Atf4, Ddit3, Ppp1r15a and Ern1) were dramatically increased (Figure 4E). Additionally, these genes were dramatically increased under the Foxo1 inhibition condition (Figure 4E). These data suggested that ER stress was associated with β-cell dedifferentiation induced by PA and high glucose.
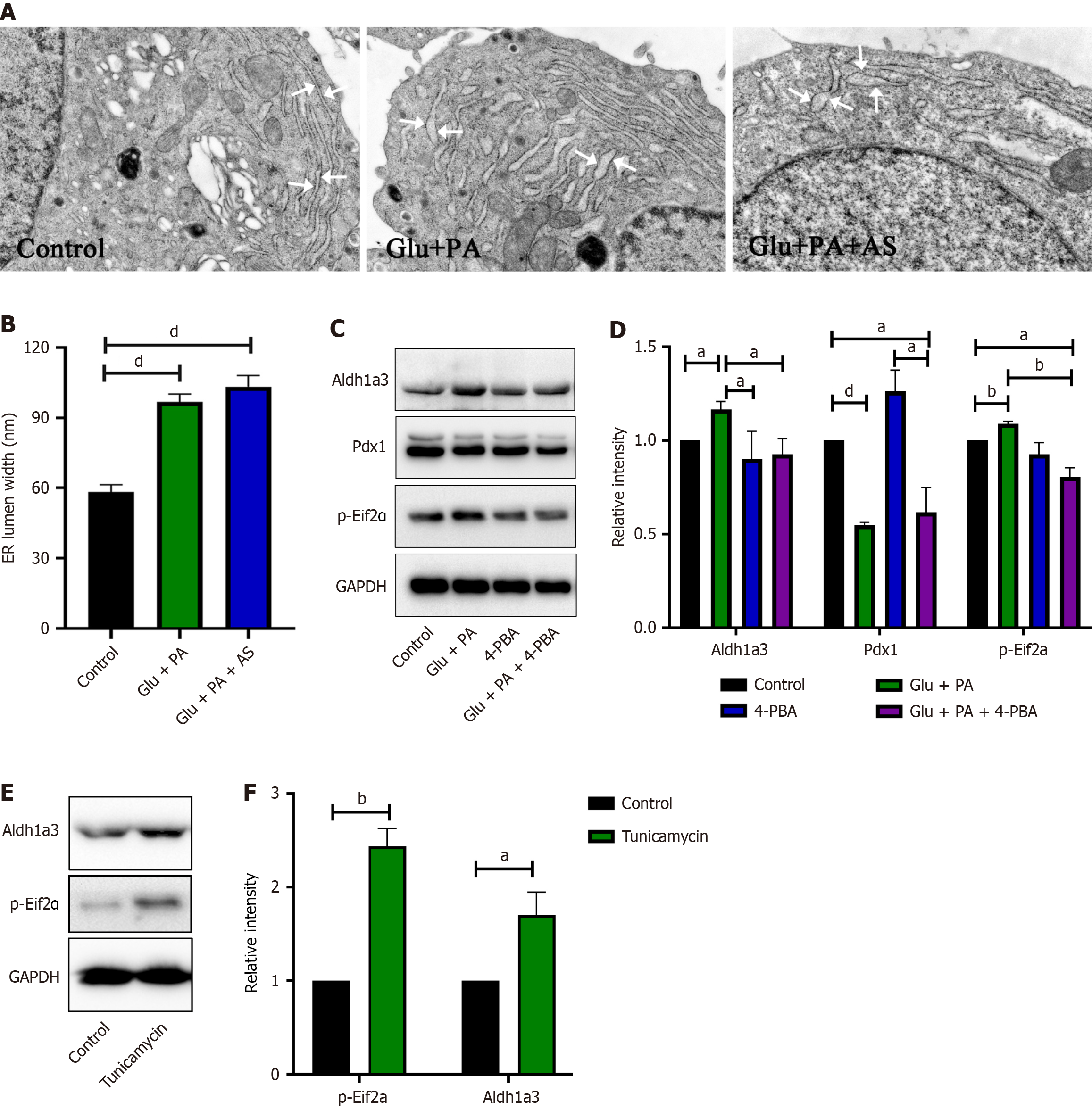
To further explore the association between ER stress and β-cell dedifferentiation induced by PA and high glucose, we performed transmission electron microscopy and analyzed cellular ultrastructure after different treatments. As shown in Figure 5A and B, the ER lumen width of the control group was 58.30 nm ± 22.24 nm. In comparison, the ER lumen width of Glu + PA and Glu + PA + AS groups significantly increased to 96.79 nm ± 29.21 nm and 103.21 nm ± 33.47 nm, respectively. Moreover, INS-1 cells treated with Glu + PA increased the ER stress-related gene p-Eif2α level (Figure 5C and D). However, the dedifferentiation marker Aldh1a3 level was significantly reduced when INS-1 cells were incubated with the ER stress reducer chemical chaperone 4-phenyl butyric acid (4-PBA) (Figure 5C and D). These data revealed that PA and high glucose induced ER stress in β cells, while relieving ER stress reduced β-cell dedifferentiation induced by PA and high glucose. Furthermore, after treatment of INS-1 cells with the ER stress inducer tunicamycin, Aldh1a3 protein levels significantly increased (Figure 5E and F). Taken together, these data suggested that ER stress is sufficient to induce β-cell dedifferentiation and is necessary for β-cell dedifferentiation induced by PA and high glucose.
β-cell dedifferentiation plays an important role in the pathological process of diabetes[9-11,29,30]. However, the molecular mechanism of β-cell dedifferentiation remains unclear. The development of a β-cell dedifferentiation model in vitro is beneficial to elucidate the detailed mechanism. This study showed that stimulation of INS-1β cells with PA and high glucose caused a loss of β-cell identity due to dedifferentiation.
Previous studies have revealed that the overnutrition state of mice induced β-cell dysfunction due to β-cell dedifferentiation, such as db/db mice and high-fat diet mice[9,11]. Therefore, the overnutrition state with high glucose plus PA (Glu + PA, Glu 25 mmol/L plus PA 300 μmol/L) was simulated using the INS-1 β cell line. PA and high glucose treatment downregulated the expression of β-cell functional genes, and upregulated the expression of β-cell dedifferentiation marker Aldh1a3 (Figure 1), forbidden genes in mature β cells, and endocrine precursor marker genes (Figure 3B). These changed characteristics were highly consistent with β-cell dedifferentiation and observations from in vivo studies. Moreover, the RNA-seq data revealed that high glucose and PA stimulation caused changes in many metabolism-related pathways, suggesting metabolic remodeling in the induction of β cell dedifferentiation (Figure 4).
Foxo1, which belongs to the forkhead transcription factor family, is deemed to be a nutrient sensing factor and an important metabolic regulator[31,32]. Moreover, Foxo1 is thought to maintain β cell stability and prevent them from dedifferentiation during the diabetes progression[15,26]. Knockout mice by β-cell specific Foxo1 genes showed elevated blood glucose under stress conditions, such as aging and multiparity, accompanied by β-cell dedifferentiation[15]. Also, a substantial reduction of Foxo1 was observed in islets of insulin resistant GIRKO (Insrflox/flox; GLUT4-cre) mice, severely hyperglycemic db/db mice, and high-fat diet mice, as well as patients with diabetes mellitus and chronic pancreatitis[4,26,29,33,34]. Hence, these data suggested that downregulation of Foxo1 is one of the key steps for β-cell dedifferentiation. In this study, PA and high glucose stimulation significantly decreased Foxo1 protein levels (Figure 1A). More impor
To further explore the regulatory network of Foxo1 during the β-cell dedifferentiation, RNA-seq was used for INS-1 cells treated with PA plus high glucose (Glu + PA) and PA plus high glucose plus Foxo1 inhibitor (Glu + PA + AS). The analysis of these RNA-seq data found that the metabolic pathway, MAPK pathway and PI3K-Akt pathway are the top three pathways regulated by Foxo1 in dedifferentiated INS-1 cells (Figure 4B). Next, the protein interaction network between Foxo1 and enriched genes was analyzed. Tnf, Bdnf, Jun, Atf4 and Ntrk2 were the top five genes of the hub genes for analysis, suggesting that these genes play a key role in the β-cell dedifferentiation reinforced by Foxo1 inhibition. In addition, Atf4 plays an important role in the response to ER stress[35]. Moreover, ER stress is highly associated with β-cell dedifferentiation[5]. Therefore, the gene expression related to ER stress was detected, showing that several ER stress genes were upregulated in dedifferentiated INS-1 cells (Figure 4E), which suggests that ER stress is involved in the Foxo1-regulated β-cell dedifferentiation.
To investigate the association between ER stress and β-cell dedifferentiation induced by PA and high glucose, ER ultrastructure was analyzed under different conditions. The results found that the ER lumen of Glu + PA and Glu + PA + AS treated cells were dramatically dilated (Figure 5A and B), suggesting that Glu + PA and Glu + PA + AS induced ER stress in INS-1 β cells. Moreover, the presence of 4-PBA, an ER stress reducer, suppressed the increase of Aldh1a3 induced by Glu + PA (Figure 5C). Meanwhile, the presence of tunicamycin, an ER stress inducer, is sufficient to induce INS-1 β cell dedifferentiation (Figure 5E and F). All these data revealed that Glu + PA induced β-cell dedifferentiation through ER stress.
In summary, an in vitro model of β-cell dedifferentiation by stimulation of INS-1 cells with high glucose plus PA was established. Through Western blot, immunostaining, qRT-PCR and RNA-seq, we found that the dedifferentiated INS-1 increased expression of the β-cell dedifferentiation marker Aldh1a3, downregulated β-cell specific transcription factors, upregulated the β-cell forbidden genes and endocrine precursor cell marker genes. Based on RNA-seq analysis, PA and high glucose induced the metabolic remodeling of INS-1 cells during dedifferentiation. Moreover, these results showed that Foxo1 inhibition enhanced INS-1 dedifferentiation, and β-cell dedifferentiation reinforced by Foxo1 inhibition was associated with metabolic pathway, MAPK pathway and PI3K-Akt pathway. Meanwhile, the data showed that the expression of several ER stress-related genes was upregulated in dedifferentiated INS-1 cells, further indicating that Glu + PA induced β-cell dedifferentiation through ER stress.
We thank members of the Li lab and Wang Lab for constructive discussions.
| 1. | Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, Colagiuri S, Guariguata L, Motala AA, Ogurtsova K, Shaw JE, Bright D, Williams R; IDF Diabetes Atlas Committee. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2019;157:107843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5345] [Cited by in RCA: 5908] [Article Influence: 984.7] [Reference Citation Analysis (8)] |
| 2. | DeFronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ, Hu FB, Kahn CR, Raz I, Shulman GI, Simonson DC, Testa MA, Weiss R. Type 2 diabetes mellitus. Nat Rev Dis Primers. 2015;1:15019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 865] [Cited by in RCA: 1319] [Article Influence: 131.9] [Reference Citation Analysis (0)] |
| 3. | Eizirik DL, Pasquali L, Cnop M. Pancreatic β-cells in type 1 and type 2 diabetes mellitus: different pathways to failure. Nat Rev Endocrinol. 2020;16:349-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 514] [Article Influence: 102.8] [Reference Citation Analysis (0)] |
| 4. | Moin ASM, Butler AE. Alterations in Beta Cell Identity in Type 1 and Type 2 Diabetes. Curr Diab Rep. 2019;19:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 5. | Bensellam M, Jonas JC, Laybutt DR. Mechanisms of β-cell dedifferentiation in diabetes: recent findings and future research directions. J Endocrinol. 2018;236:R109-R143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 169] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 6. | Zhang J, Liu F. The De-, Re-, and trans-differentiation of β-cells: Regulation and function. Semin Cell Dev Biol. 2020;103:68-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 7. | Rutter GA, Pullen TJ, Hodson DJ, Martinez-Sanchez A. Pancreatic β-cell identity, glucose sensing and the control of insulin secretion. Biochem J. 2015;466:203-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 259] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 8. | Lu TT, Heyne S, Dror E, Casas E, Leonhardt L, Boenke T, Yang CH, Sagar, Arrigoni L, Dalgaard K, Teperino R, Enders L, Selvaraj M, Ruf M, Raja SJ, Xie H, Boenisch U, Orkin SH, Lynn FC, Hoffman BG, Grün D, Vavouri T, Lempradl AM, Pospisilik JA. The Polycomb-Dependent Epigenome Controls β Cell Dysfunction, Dedifferentiation, and Diabetes. Cell Metab. 2018;27:1294-1308.e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 9. | Neelankal John A, Ram R, Jiang FX. RNA-Seq Analysis of Islets to Characterise the Dedifferentiation in Type 2 Diabetes Model Mice db/db. Endocr Pathol. 2018;29:207-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 10. | Sun J, Ni Q, Xie J, Xu M, Zhang J, Kuang J, Wang Y, Ning G, Wang Q. β-Cell Dedifferentiation in Patients With T2D With Adequate Glucose Control and Nondiabetic Chronic Pancreatitis. J Clin Endocrinol Metab. 2019;104:83-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 11. | Tersey SA, Levasseur EM, Syed F, Farb TB, Orr KS, Nelson JB, Shaw JL, Bokvist K, Mather KJ, Mirmira RG. Episodic β-cell death and dedifferentiation during diet-induced obesity and dysglycemia in male mice. FASEB J. 2018;32:fj201800150RR. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 12. | Wang Z, York NW, Nichols CG, Remedi MS. Pancreatic β cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab. 2014;19:872-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 325] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 13. | Stancill JS, Cartailler JP, Clayton HW, O'Connor JT, Dickerson MT, Dadi PK, Osipovich AB, Jacobson DA, Magnuson MA. Chronic β-Cell Depolarization Impairs β-Cell Identity by Disrupting a Network of Ca(2+)-Regulated Genes. Diabetes. 2017;66:2175-2187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Nordmann TM, Dror E, Schulze F, Traub S, Berishvili E, Barbieux C, Böni-Schnetzler M, Donath MY. The Role of Inflammation in β-cell Dedifferentiation. Sci Rep. 2017;7:6285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 15. | Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150:1223-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 939] [Cited by in RCA: 1114] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 16. | Lee H, Lee YS, Harenda Q, Pietrzak S, Oktay HZ, Schreiber S, Liao Y, Sonthalia S, Ciecko AE, Chen YG, Keles S, Sridharan R, Engin F. Beta Cell Dedifferentiation Induced by IRE1α Deletion Prevents Type 1 Diabetes. Cell Metab. 2020;31:822-836.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 17. | Lenz A, Toren-Haritan G, Efrat S. Redifferentiation of adult human β cells expanded in vitro by inhibition of the WNT pathway. PLoS One. 2014;9:e112914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Aloy-Reverté C, Moreno-Amador JL, Nacher M, Montanya E, Semino CE. Use of RGD-Functionalized Sandwich Cultures to Promote Redifferentiation of Human Pancreatic Beta Cells After In Vitro Expansion. Tissue Eng Part A. 2018;24:394-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Russ HA, Sintov E, Anker-Kitai L, Friedman O, Lenz A, Toren G, Farhy C, Pasmanik-Chor M, Oron-Karni V, Ravassard P, Efrat S. Insulin-producing cells generated from dedifferentiated human pancreatic beta cells expanded in vitro. PLoS One. 2011;6:e25566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 20. | Toren-Haritan G, Efrat S. TGFβ Pathway Inhibition Redifferentiates Human Pancreatic Islet β Cells Expanded In Vitro. PLoS One. 2015;10:e0139168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Diedisheim M, Oshima M, Albagli O, Huldt CW, Ahlstedt I, Clausen M, Menon S, Aivazidis A, Andreasson AC, Haynes WG, Marchetti P, Marselli L, Armanet M, Chimienti F, Scharfmann R. Modeling human pancreatic beta cell dedifferentiation. Mol Metab. 2018;10:74-86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 22. | Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2015] [Cited by in RCA: 2326] [Article Influence: 136.8] [Reference Citation Analysis (0)] |
| 23. | Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9910] [Cited by in RCA: 14951] [Article Influence: 1495.1] [Reference Citation Analysis (0)] |
| 24. | Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39162] [Cited by in RCA: 36964] [Article Influence: 2843.4] [Reference Citation Analysis (0)] |
| 25. | Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10832] [Cited by in RCA: 14462] [Article Influence: 1033.0] [Reference Citation Analysis (0)] |
| 26. | Kim-Muller JY, Kim YJ, Fan J, Zhao S, Banks AS, Prentki M, Accili D. FoxO1 Deacetylation Decreases Fatty Acid Oxidation in β-Cells and Sustains Insulin Secretion in Diabetes. J Biol Chem. 2016;291:10162-10172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 27. | Diep CH, Charles NJ, Gilks CB, Kalloger SE, Argenta PA, Lange CA. Progesterone receptors induce FOXO1-dependent senescence in ovarian cancer cells. Cell Cycle. 2013;12:1433-1449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 28. | Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol. 2010;78:961-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 218] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 29. | Cinti F, Bouchi R, Kim-Muller JY, Ohmura Y, Sandoval PR, Masini M, Marselli L, Suleiman M, Ratner LE, Marchetti P, Accili D. Evidence of β-Cell Dedifferentiation in Human Type 2 Diabetes. J Clin Endocrinol Metab. 2016;101:1044-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 426] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 30. | Téllez N, Vilaseca M, Martí Y, Pla A, Montanya E. β-Cell dedifferentiation, reduced duct cell plasticity, and impaired β-cell mass regeneration in middle-aged rats. Am J Physiol Endocrinol Metab. 2016;311:E554-E563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Kim-Muller JY, Zhao S, Srivastava S, Mugabo Y, Noh HL, Kim YR, Madiraju SR, Ferrante AW, Skolnik EY, Prentki M, Accili D. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metab. 2014;20:593-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 32. | Li Y, Ma Z, Jiang S, Hu W, Li T, Di S, Wang D, Yang Y. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog Lipid Res. 2017;66:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 33. | Ishida E, Kim-Muller JY, Accili D. Pair Feeding, but Not Insulin, Phloridzin, or Rosiglitazone Treatment, Curtails Markers of β-Cell Dedifferentiation in db/db Mice. Diabetes. 2017;66:2092-2101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 34. | Xiao X, Fischbach S, Zhang T, Chen C, Sheng Q, Zimmerman R, Patnaik S, Fusco J, Ming Y, Guo P, Shiota C, Prasadan K, Gangopadhyay N, Husain SZ, Dong H, Gittes GK. SMAD3/Stat3 Signaling Mediates β-Cell Epithelial-Mesenchymal Transition in Chronic Pancreatitis-Related Diabetes. Diabetes. 2017;66:2646-2658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 35. | Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. Surviving Stress: Modulation of ATF4-Mediated Stress Responses in Normal and Malignant Cells. Trends Endocrinol Metab. 2017;28:794-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 433] [Article Influence: 54.1] [Reference Citation Analysis (0)] |