Published online Jul 15, 2023. doi: 10.4239/wjd.v14.i7.958
Peer-review started: December 23, 2022
First decision: April 11, 2023
Revised: May 1, 2023
Accepted: May 22, 2023
Article in press: May 22, 2023
Published online: July 15, 2023
Processing time: 201 Days and 21.5 Hours
Diabetes mellitus (DM) is a group of metabolic disorders defined by hyperglycemia induced by insulin resistance, inadequate insulin secretion, or excessive glucagon secretion. In 2021, the global prevalence of diabetes is anticipated to be 10.7% (537 million people). Noncoding RNAs (ncRNAs) appear to have an important role in the initiation and progression of DM, according to a growing body of research. The two major groups of ncRNAs implicated in diabetic disorders are miRNAs and long noncoding RNAs. miRNAs are single-stranded, short (17–25 nucleotides), ncRNAs that influence gene expression at the post-transcriptional level. Because DM has reached epidemic proportions worldwide, it appears that novel diagnostic and therapeutic strategies are required to identify and treat complications associated with these diseases efficiently. miRNAs are gaining attention as biomarkers for DM diagnosis and potential treatment due to their function in maintaining physiological homeostasis via gene expression regulation. In this review, we address the issue of the gradually expanding global prevalence of DM by presenting a complete and up-to-date synopsis of various regulatory miRNAs involved in these disorders. We hope this review will spark discussion about ncRNAs as prognostic biomarkers and therapeutic tools for DM. We examine and synthesize recent research that used novel, high-throughput technologies to uncover ncRNAs involved in DM, necessitating a systematic approach to examining and summarizing their roles and possible diagnostic and therapeutic uses.
Core tip: Diabetes mellitus is a chronic endocrinopathy characterized by disrupted glucose, lipid, and amino acid metabolism and has reached pandemic proportions. A vast body of evidence demonstrates that miRNAs play a key role in diabetic pathophysiology. Here, we explore numerous regulatory miRNAs involved in DM and discuss their potential diagnostic and therapeutic applications.
- Citation: Macvanin MT, Gluvic Z, Bajic V, Isenovic ER. Novel insights regarding the role of noncoding RNAs in diabetes. World J Diabetes 2023; 14(7): 958-976
- URL: https://www.wjgnet.com/1948-9358/full/v14/i7/958.htm
- DOI: https://dx.doi.org/10.4239/wjd.v14.i7.958
Diabetes mellitus (DM) is a chronic endocrinopathy caused by genetic and environmental factors that lead to disruption of carbohydrate, lipid and amino acid metabolism. If DM is left out of control, the delayed effects of such a metabolic derangement promote systemic, mainly vascular consequences[1-3]. The prevalence of DM is increasing rapidly and has reached pandemic proportions[4,5]. According to studies and International Diabetes Federation (IDF), the global prevalence of type 2 DM (T2DM) was 9.3% in 2019, expected to rise to 10.9% by 2045, and affects ~629 million people[6,7].
The pathophysiology of DM is based on hyperglycemia induced by either insulin resistance (IR), insulin deficiency, or both[4]. Ineffective glucose utilization favors the activation of alternative glucose metabolic pathways (i.e. polyol, protein kinase C, and hexosamine), contributing to mitochondrial dysfunction, reactive oxygen species (ROS) generation, and the development of cellular and tissue hypoxia[8]. DM can cause endothelial cell destruction and low-grade systemic inflammation, leading to vascular problems, including diabetic foot ulcers, peripheral neuropathy, nephropathy, maculopathy, and retinopathy[9-11]. In DM patients, vascular issues contribute to a two to four-fold increase in myocardial infarction, stroke, and overall mortality[12-14].
There are at least five types of DM, with type 1 DM (T1DM) and T2DM being the most prevalent in clinical settings[2,15]. T1DM represents 10% of all cases and is one of the most frequent chronic diseases of childhood, with a worldwide incidence that is increasing by 3% annually[16-18]. Multiple mechanisms, including autoimmunity, genetic susceptibility, and epigenetic modulation, have been implicated in T1DM pathogenesis[19]. The presence of autoantibodies against β-cell antigens, such as insulin, decarboxylase, tyrosine phosphatases-2 and -2b, and glutamic acid, have been detected in T1DM patients[19,20]. More than 50 mutations that describe disease susceptibility were discovered at 50 different genetic loci, with HLA class II mutations being the most common. In addition, environmental factors such as nutritional habits, viruses, and other epigenetic factors directly affect the expression of insulin genes and genes responsible for autoimmune responses[21].
T2DM is more common in older or obese patients and is distinguished by cells and tissues that are resistant to high insulin levels and even high blood sugar levels. T2DM accounts for most (90%–95%) cases of DM[2,7]. While genetic predisposition and obesity are the main risk factors for the development of T2DM, autoimmunity plays a pivotal role in the development of T1DM[1,22].
Basic scientific research continuously aims to identify improved biochemical markers of early or advanced endothelial injury. Besides C-reactive protein, homocysteine, nitric oxide, and inflammatory cytokines, miRNAs are promising tools for assessing vascular complications risk in DM patients. Due to accumulating evidence regarding their role in the onset and progression of DM, miRNAs are gaining substantial attention as novel diagnostic biomarkers for DM and potential therapeutic agents.
In this review, we address the problem of the global prevalence of DM by providing a systematic and up-to-date summary of various miRNAs involved in the pathogenesis and pathophysiology of DM. We also summarize the recent findings of numerous studies that used a novel high-throughput methodology to identify miRNAs involved in the pathogenesis of DM and their potential diagnostic and therapeutic applications.
The rapid advancement of high-throughput sequencing technologies, such as microarray, deep RNA sequencing, and next-generation sequencing, revealed novel insights into the structure and function of the human genome and transcriptome. An unexpected finding of such studies is that only ~2% of the human genome is transcribed into protein-coding mRNA[23,24], while the remainder of the genome is transcribed into noncoding RNAs (ncRNAs), including structural RNAs (rRNAs and tRNAs), and regulatory RNAs such as miRNAs and long noncoding RNAs (lncRNAs)[25]. miRNAs are small (17–25 nucleotides) single-stranded ncRNA molecules that regulate gene expression post-transcriptionally primarily by binding to complementary cis-elements in the 3'-untranslated region (UTR) of target mRNAs[26]. They may, however, bind anywhere along the mRNA transcript to inhibit translation and regulate mRNA stability and degradation[27,28]. miRNA interactions with coding areas and the 5′-UTR are thought to mute gene expression[29,30], whereas miRNA interactions with promoter regions are thought to activate transcription[31].
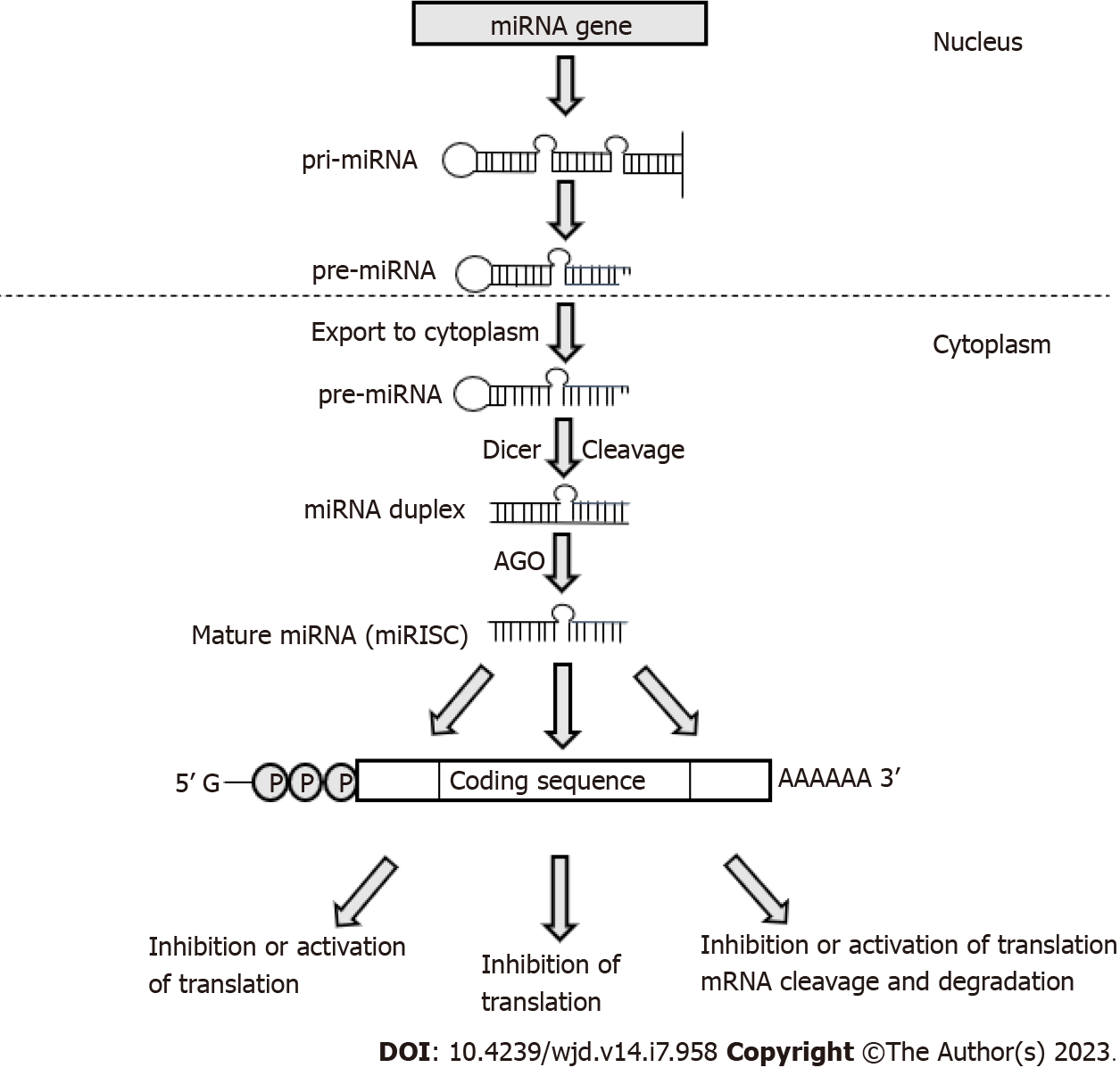
miRNAs are transcribed by RNA polymerase II as long precursor molecules, which are cleaved by the nuclear RNase III-type endoribonuclease DROSHA to approximately 70-nucleotide precursor miRNAs (pre-miRNAs)[32], before being transported into the cytoplasm and further processed by another RNase III enzyme, DICER, to generate mature double-stranded miRNAs[33]. The guide strand of mature miRNA associates with Argonaute (AGO) proteins and is incorporated into the minimal miRNA-induced silencing complex (miRISC) that interacts with complementary sites within the target mRNAs[33] (Figure 1). It has been estimated that around 60% of human transcripts contain potential miRNA-binding sites within their 3′-UTRs[34] and that a single miRNA can potentially bind to more than 100 target mRNAs, where several miRNAs may act synergistically to finely tune the expression of the same transcript[35-37].
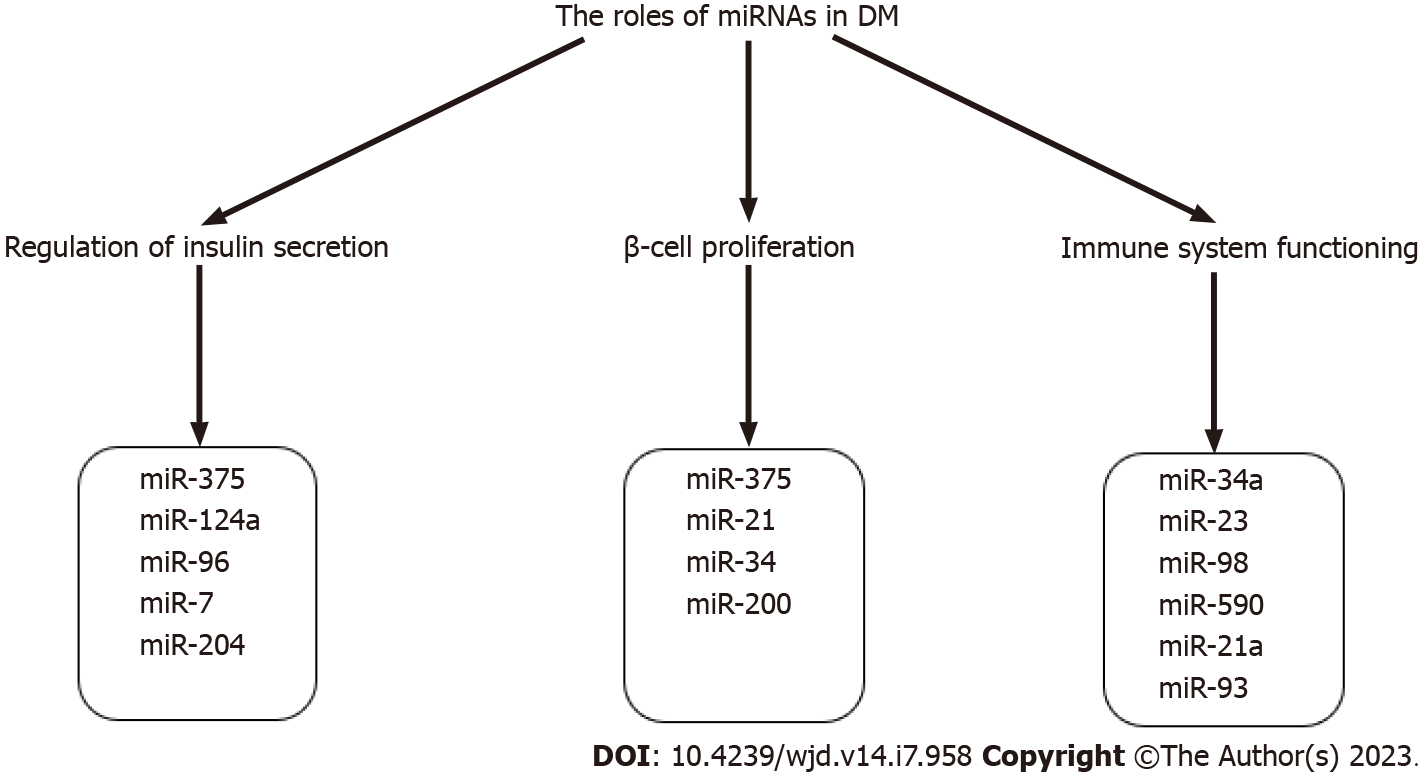
miRNAs regulate glucose homeostasis by controlling insulin production, secretion, and cell proliferation. Fine-tuned insulin secretion from pancreatic β-cells is required for blood glucose homeostasis; perturbations in this process can result in hyperglycemia and DM. The tissue-specific expression of miRNAs is an important aspect of their role in the pathophysiology of DM. For instance, miR-9, miR-375, miR-376 and miR-7 are highly expressed in the human pancreas, where they exhibit an important role in pancreatic islet function[38-40], participating in pancreas development and the regulation of islet mass, as well as β-cell proliferation and insulin secretion. Expanding knowledge of tissue-specific miRNA expression in animal models and human subjects is illustrated by several studies that provide valuable insights into connections between miRNAs and DM. For example, in streptozotocin-induced T1D mice, miRNA-microarray profiling revealed 64 upregulated and 72 downregulated pancreatic miRNAs, and subsequent qRT-PCR analysis validated the decreased expression of let-7, miR-148b-3p, miR-27a-3p, miR-7a-5p, miR-7b-5p, miR-26a-5p, and miR-26b-5p in diabetic mice[41]. Another study confirmed the downregulation of miR-26a-5p in pancreatic mouse tissues[42]. Regarding T2DM animal models, increased levels of miRNAs belonging to the miR-199 and miR-200 families, as well as miR-34a, miR-132, miR-146, let-7b, and miR-21, were observed in pancreatic islets of diabetic mice[43,44] whereas miR-30d, miR-184, miR-203, miR-210, miR-338–3p and miR-383 had significantly decreased levels[44-46]. miR-199a-5p and miR-184 were consistently dysregulated in several mouse models of obesity and/or IR. For instance, increased expression of miR-199a-5p was also reported in islets of diet-induced obese (DIO) mice[44,47], whereas decreased expression of miR-184 was confirmed in an independent study using the islets of mice on a high-fat diet[48]. In the nonobese spontaneous Goto-Kakizaki rat T2DM rat model, global evaluation of miRNA expression pattern in pancreatic islets identified 30 dysregulated miRNAs[49]. A study by Karolina et al[50] performed on pancreatic T2DM rat tissue showed a significant increase of miR-144, miR-150, miR-29a, miR-192, and miR-320a observed, while miR-146a, miR-30d, and miR-182 were highly downregulated[50].
Human studies show a cluster of highly expressed miRNAs expressed explicitly in human β-cells, such as miR-655, miR-656, miR-127, miR-136, miR-543, miR-369, miR-411, miR-432, miR-487, miR-495, miR-589, is significantly downregulated in islets from T2DM patients[51]. Other studies also found increased miR-124a and miR-187 expression in the islet tissue of T2DM patients[52,53]. However, miR-7a, and miR-184, implicated in the regulation of pancreatic β-cell function, showed a significantly decreased expression in human T2DM islets[47,48]. In T1DM patients, significant upregulation of miR-125a-5p compared to healthy controls was observed[54].
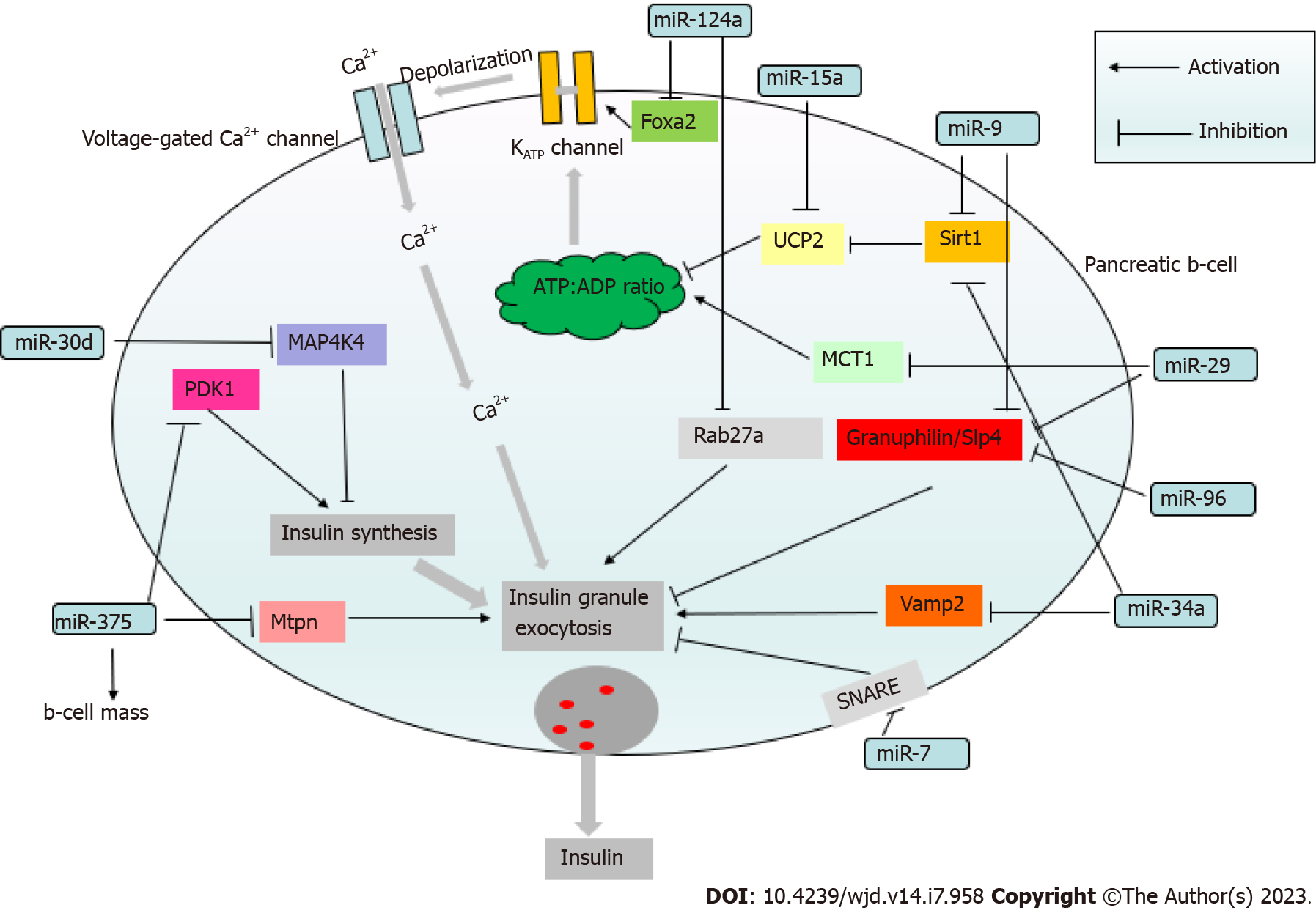
miR-375, which has an established role in regulating insulin secretion (Figure 2)[40], was the most highly expressed miRNA in human pancreatic islets[55]. The target of miR-375 is myotrophin (Mtpn) which is involved in the cytoskeletal remodeling by depolymerizing actin filaments[40] and mediating exocytosis by enabling the fusion of insulin vesicles on membranes of β-cells[40,56] (Figure 3). In addition, Mtpn was shown to upregulate the nuclear factor (NF)-κB), thus inducing the expression of proteins responsible for targeting insulin vesicles to the membrane[57,58].
Lovis et al[59] found that miR-124a and miR-96 could also operate as transcriptional regulators of proteins involved in insulin exocytosis and secretion (Figure 3)[59]. miR-124a increases the levels of Rab3A, SNAP25, and synapsin-1A while decreasing Noc2 and Rab27A levels (Figure 3). Rab27A is a GTPase that allows vesicles to be transported to the cell membrane. The direct binding of miR-124a to the 3'-UTR of Rab27A reduces Rab27A expression (Figure 3). miR-124a overexpression causes excessive insulin release at rest and decreases glucose-induced insulin secretion[59]. miR-96 has been shown to reduce glucose-induced insulin release by upregulating granuphilin, a negative regulator of insulin exocytosis, while suppressing Noc2 expression[59], a protein that is essential for the normal regulation of endocrine cell exocytosis[60]. miR-7 has also been found to inhibit glucose-induced insulin release in β-cells. It acts by directly regulating the expression of genes involved in the late stages of insulin granules fusion with the plasma membrane, including the soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex, which mediates membrane fusion of vesicles with their target cellular compartments[47].
Several miRNAs are implicated in the regulation of insulin production. miR-375 has been shown to inhibit 3'-phosphoinositide-dependent protein kinase (PDK)1, a crucial component of the PI3K cascade (Figure 3)[61]. Decreased PDK1 levels lead to the downregulation of insulin gene expression in response to glucose stimulation[61]. miR-204 has also been found as a negative regulator of insulin production, whose expression is influenced by thioredoxin-interacting protein (TXNIP), a redox state regulator in β-cells. TXNIP levels are increased in DM, resulting in elevated miR-204 expression that mediates increased degradation of the insulin transcription factor MAFA[62]. Another target of miR-204 is the glucagon-like peptide 1 receptor 3'-UTR, and this interaction also downregulates glucose-induced insulin secretion[63].
miR-375 and several other pancreatic miRNAs, including miR-21, miR-34, and miR-200, have been found to influence β-cell proliferation, survival, and apoptosis (Figure 2). For instance, decreased proliferation and viability are associated with a miR-375 knockdown in mice, resulting in a severely diabetic state[56]. MiR-21, whose expression is regulated by the NF-κB proinflammatory cytokine pathway, has been reported to regulate β-cell number[64,65]. miR-21 overexpression in vitro was associated with decreased β-cell number[64]. miR-34a knockdown increased β-cell quantity and mass, which could be explained by the role of miR-34 in targeting sirtuin (SIRT)1 and, as a result, causing p53-mediated apoptosis[64]. miR-200 also induces expression of proapoptotic genes in both T1DM and T2DM by suppressing Xiap, a caspase inhibitor, and Dnajc3, a β-cell heat shock protein, while activating the tumor suppressor protein Trp53[66].
The significance of miRNAs as global cellular regulators of gene expression is evident in their function in immune system balance and regulating immune cell differentiation, maturation, and activation, which is especially relevant in the context of T1DM pathogenesis (Figure 2). Various immune cells, such as CD4+, CD8+ T cells, natural killer (NK) cells, type B lymphocytes, dendritic cells, and chemokines and cytokines, are characteristic of T1DM-associated β-cell damage[67]. B lymphocytes play a protective and defensive role for β-cells preventing T1DM development. Mone et al[68] reported that miR-34a overexpression in diabetic mice leads to reduced activity of B lymphocytes via a negative expression of the Foxp1 gene[68] involved in B lymphopoiesis, resulting in a reduced ability of pancreatic islets to defend themselves against damage[68,69]. miRNA expression also regulates the production of specific T cells at the other end of the homeostatic chain. miR-590, miR-98, and miR-23 overexpression has also been demonstrated to influence the production of CD8+ T lymphocytes that target pancreatic islet antigens. The mechanisms of action operate by suppressing the expression of TRAIL and FAS ligands, implying that miRNA-mediated gene silencing may promote autoimmunity and the development of T1DM[70]. The three primary antibodies in T1DM occurring long before disease onset are autoantibodies against specific antigens of pancreatic islet cells IA, IA2B, and GAD (glutamic acid decarboxylase) and a cluster of 32 miRNAs participate in biosynthetic pathways that modify the expression of T1DM autoantibody sequences[71]. miRNA-21a and miR-93 are involved in the processes of inflammation and pathways leading to cell death in peripheral blood mononuclear cells (PBMCs) of T1DM patients[72], whereas miR-326, which targets significant immune system modulators such as the vitamin D receptor, was found to be overexpressed in T1DM patients' PBMC[73].
lncRNAs are common long (> 200 nucleotides) linear transcripts that regulate gene expression at the transcriptional and post-transcriptional levels, influencing mRNA stability, pre-mRNA splicing, and translation[74-77]. Mechanistically, lncRNAs can act as miRNA sponges, scaffolds for protein complexes, and decoys for regulatory factors[76]. The interaction of lncRNAs with transcription factors results in transcription regulation[75,78]. It has also been observed that some lncRNAs may interact with pre-mRNAs to influence splicing[79]. lncRNAs can potentially block protein interactions with target mRNAs or change protein catalytic activity, acting as decoys[74,76]. It was also discovered that lncRNAs binding to translating mRNAs change the target mRNA’s stability and translation[74].
lncRNAs regulate critical physiological processes such as cell proliferation, growth, differentiation, senescence, aging, and secretion[80,81]. They are also implicated in the pathogenesis of several diseases, including cardiovascular disease and DM. In humans, lncRNAs are primarily produced by RNA polymerase II or III[82] and are characterized as sense- or antisense-overlap, bidirectional, intronic, or intergenic lncRNAs based on their proximity to the next protein-coding gene[83]. The functions of lncRNAs are governed by their cellular location, with nuclear lncRNAs modulating transcription and splicing and cytoplasmic lncRNAs regulating post-transcriptional events such as mRNA stability, protein synthesis, and posttranscriptional alterations[76].
Various lncRNAs are expressed in a cell-type specific manner in pancreatic β-cells, such as GAS5 (growth arrest-specific transcript 5), PLUTO (PDX1 locus upstream transcript), TUG1 (taurine upregulated gene 1), MEG3 (maternally expressed gene 3), and βLINC (β-cell long intergenic ncRNAs). GAS5 is a lncRNA that regulates cell development and proliferation. GAS5 levels in diabetic patients’ serum are considerably lower than in healthy controls[84], and db/db mice[85], and GAS5 silencing in vitro is related to cell cycle arrest and decreased insulin production and secretion. PLUTO is an antisense transcript lncRNA upstream of the gene that codes for PDX1, a transcription factor involved in β-cell differentiation and pancreatic development. Both PLUTO and PDX1 are significantly downregulated in T2DM patients[86]. Reduced PLUTO expression is related to chromatin changes that limit the interaction of the PDX1 promoter with its enhancer, resulting in lower PDX1 expression[86], implying that PLUTO plays a role in the control of β-cell function. TUG1 and MEG3 are extensively expressed in the pancreas and are controlled by glucose levels[87,88]. TUG1 and MEG3 knockdown reduces insulin synthesis and secretion and promotes β-cell death[88], supporting their roles in β-cell development and insulin production control. βLINC1 is a highly conserved lncRNA linked to increased glucose intolerance and aberrant insulin secretion[89]. βLINC2 and βLINC3 are more abundant in pancreatic islets than other organs. βLINC2 expression levels correlate favorably with body weight, glycemia, and insulinemia, but βLINC3 expression correlates negatively with body mass index (BMI) and is considerably lower in T2DM patients compared to healthy controls[90].
Circular RNAs (circRNAs) are abundant, conserved tissue-specific covalently closed loop circular RNAs[91-95] produced by the direct ligation of 5′ and 3′ ends of linear RNAs as intermediates in RNA processing or generated by backsplicing where a downstream 5' splice donor attacks an upstream 3' splice acceptor site of pre-mRNA forming a covalently closed circRNA lacking the 5' and 3' ends[96,97]. circRNAs regulate gene expression by acting as miRNA sponges, modulating protein–protein interactions, binding to ribosomes, and interfering with translation or modifying transcription[98]. circRNAs are thought to play a role in the etiology of many diseases, including DM[99]. circRNA Cdr1as, for example, regulates insulin production and secretion by acting as a sponge for miR-7, reducing insulin secretion. Cdr1as contains about 60 miR-7 binding sites[93], and Cdr1as upregulation increases insulin secretion by inhibiting miR-7 activity[47]. miR-7 directly targets and inhibits the expression of paired box (Pax)6 and the myosin VIIA and Rab interacting protein (Myrip). Pax6 is a transcription factor that interacts with the promoters of the ins1 and ins2 genes to stimulate insulin production and secretion, whereas Myrip is involved in secretory granule transport and release. Cdr1as expression is downregulated in db/db mouse islets[100], but Cdr1as overexpression increases Pax 6 and Myrip expression, enhancing insulin transcription and secretion in pancreatic islets[101].
CircHIPK3 is abundantly expressed in pancreatic β-cells, and decreased circHIPK3 levels are associated with reduced proliferation of β-cells[100]. CircHIPK3 silencing decreases insulin mRNA levels and perturbs glucose-stimulated insulin secretion[100].
CircAFF1[101], another highly expressed circRNA in pancreatic islets, causes β-cell death in vitro, implying its role in β-cell growth and function[100].
Numerous investigations have shown that miRNAs act as endocrine signaling molecules, regulating insulin production and fat metabolism. Specific miRNAs directly regulated several insulin-signaling components, including insulin receptors (INSR) and several transcription factors. For example, miR-424-5p was found to target the 3′-UTR sequence of INSR mRNA, and its overexpression in human hepatocytes HepG2 cell line leads to decreased INSR levels and lipid accumulation[102]. Treatment of HepG2 cells with saturated fatty acids leads to increased miR-424-5 and a reduced expression of INSR[102]. Similarly, miR-15b[103], miR-195[104], and miR-96[105] bind to human INSR mRNA, and in the liver of mice on a high-fat diet or in HepG2 cells treated with saturated fatty acids, elevated expression of miR-96 and miR-195 was observed, accompanied by decreased INSR[104,105]. MiR-122, miR-144, and miR-146a were reported to indirectly modulate INSR by controlling the expression of protein tyrosine phosphatases that remove phosphate groups from tyrosine residues of the cytoplasmic domain of INSR and negatively affect insulin signaling[50,106,107]. In addition, INSR compartmentalization and signal transduction depends on the presence of caveolae, specialized microdomains in plasma membranes composed of caveolin proteins. It has been discovered that miR-107 and miR-103 bind to the 3′-UTR of caveolin-1 mRNA[108] and play a role in IR by increasing liver glucose production. AntagomiR-mediated silencing of miR-107 and miR-103 in adipocytes of DIO mice normalized glucose status[108].
miRNAs also target IR substrates (IRSs), altering insulin signaling and cholesterol and fatty acid metabolism. miR-96, miR-126, and miR-145 regulate IRS1 expression[109-111], whereas IRS2 is targeted by miR33a/b[112,113]. However, it has been reported that different miRNAs regulate IRS in various target tissues. IRS-1 is regulated in skeletal muscle by miR-29a, miR-29c, and miR-128a[104,114,115] and IRS2 by miR-135a[116]. IRS-1 mRNA contains a binding site for miR-29, which has been shown to activate lipid metabolism genes such as the peroxisome proliferator-activated receptor- coactivator-1 and 3-hydroxy-3-methylglutaryl-CoA synthase 2[117].
miR-26, which is significantly downregulated in the livers of overweight people and obese leptin-deficient mice, is another miRNA implicated in regulating insulin sensitivity, glucose and fat metabolism[118]. miR-26a levels correlate positively with BMI and are negatively related to homeostatic model assessment for IR (HOMA-IR). Furthermore, miR-26a overexpression prevented metabolic activity changes associated with obesity[118].
miRNAs regulate insulin-like growth factor (IGF)-1 and its receptor (IGF-1R) expression and secretion, as de-monstrated by Ling et al[119], who revealed that miR-320 modulates the insulin signaling pathways by influencing IGF-1 expression and insulin sensitivity in adipocytes[119]. This finding implies that specific miRNAs may bind to multiple targets to regulate glucose metabolism, which is supported by the discovery that miR-1 regulates IGF-1 and IGF-1R expression in cardiac and skeletal muscles[120], whereas let-7 may bind to the 3'-UTR regions of INSR, IRS-2, and IGF-1R[121]. The discovery that miR-143-3p, which regulates IGF-2R receptor expression, is significantly up-regulated in the serum of T2DM patients and multiple tissues of obese mice (including pancreas, skeletal muscle, and heart), contributing to IR associated with metabolic syndrome, lends support to the role of IGF signaling in metabolic diseases[122].
As previously mentioned, pancreatic β-cells are characterized by a high abundance of miR-375[40]. However, studies of miRNA distribution in other organs and tissues, such as liver and adipose tissue, suggest that miRNA expression profiles may serve as signatures of cell identity[123]. High-throughput omics studies have found a link between miRNA expression in different tissues, such as the pancreas, liver, and adipose tissue, and conditions like metabolic disease and obesity[124-126]. For instance, 221 out of 1736 genomic loci associated with obesity correspond to miRNAs[124]. The role of miRNAs in the pathophysiology of T2DM and associated metabolic disorders was investigated. Specific miRNAs such as miR-27a and miR-222 were upregulated in adipose tissue, whereas mir-122, miR-103, and miR-195 were enriched in the liver[127], with miR-122 accounting for nearly 70% of the total miRNA expressed in this tissue[128-130]. In addition, treating adipocytes with glucose results in increased expression of miR-27a, miR-29a, and miR-222 and downregulation of miR-10b in skeletal muscle. Other studies reported that muscle cells are enriched in miR-133a, miR-133b, miR-1, miR-486, miR-206, miR-208a, miR-208b, and miR-499[131-133].
Several studies show that specific miRNAs are differentially expressed in obese subjects’ white adipose tissue compared to nonobese controls[109-112], and visceral adipose tissue miRNAs are more important in metabolic dysregulation than subcutaneous tissue miRNAs[134,135]. A correlation has been found between miRNA expression in adipose tissue and metabolic parameters such as BMI, glycemia, leptinemia, and adipogenesis[136,137]. For instance, elevated miR-21 expression was observed in the white adipose tissue of obese humans, and it positively correlated with BMI[138]. Treatment with a miR-21 inhibitor [locked nucleic acid (LNA)-miR-21] resulted in decreased adipocyte size, significant weight loss, and inhibition of expression of transforming growth factor β-receptor 2 (TGFBR2) and phosphatase and tensin homolog (PTEN)[139].
Circulating miRNAs exert an additional level of the regulation of metabolic homeostasis, which can mediate communications between various types of cells. Extracellular miRNAs may be utilized for evaluating an individual’s metabolic condition because dysregulation of their expression is linked to various metabolic diseases, including T2DM, obesity, and cardiovascular diseases[140], and correlates with individual lifestyle characteristics such as exercise[141-144], dietary intake[145], and the composition of gut microbiota[146]. Adipocytes and adipose tissue macrophages (ATMs) can alter insulin-sensitive organs like the liver and muscles by releasing exosomal vesicles carrying miRNAs[147]. miRNA profiling of ATM exosomes revealed enhanced expression of miR-155, which targets the PPARG gene that encodes for peroxisome proliferator-activated receptor , which regulates glucose metabolism and fatty acid storage[148]. It has been established that adipose tissue is a major source of exosomal miRNAs that regulate gene expression in distant organs[149]. Several studies confirmed the link between adipose tissue and miRNA profiles, demonstrating that weight loss was associated with significant changes in circulating miRNA levels[150,151]. Thomou et al[149] observed that the liver could take up exosomal miR-99b from adipose tissue, resulting in a decreased expression of hepatic fibroblast growth factor 21 and, as a result, glucose intolerance[149].
Extracellular vesicles (EVs), specifically exosomes, play a vital role in interorgan communication by carrying lncRNAs and miRNAs that modulate metabolic pathways. EVs are tiny vesicles enclosed by a membrane, originate from endosomes, and are released by cells into the extracellular fluids depending on their cargo[152]. According to the Minimal Information for Studies of EVs 2018 recommendations, EVs are a component of the total secretome released by the cell, and no specific marker can distinguish EV subtypes and their subcellular origin[153]. Exosomes and microvesicles, two forms of EVs released by cells, are distinguished by their manner of synthesis rather than size. Cells undergo EV biogenesis, which includes inward invagination of the plasma membrane within the cytosol, forming early and late endosomes (LEs). These LEs join together to create multivesicular bodies, which invaginate to form intraluminal vesicles (ILVs)[154]. Exocytosis occurs when these ILVs fuse with the plasma membrane and release exosomes into the extracellular environment[155]. Exosomes are found in various bodily fluids and are released by various cell types, including lymphocytes and pancreatic islets[155]. The transfer of nucleic acids by exosomes is enhanced in inflammatory conditions, and miRNAs represent one of the main cargos transported by exosomes[156].
In DM, these molecules target specific tissues regulating their activity. Therefore, to understand the pathogenesis of T1DM and T2DM, it is crucial to investigate the communication between affected organs in response to elevated blood glucose levels. Exosomes act as messengers, linking the immune response to pancreatic damage and adipocyte stimulation, leading to IR in the liver and muscles. Exosomes containing lncRNAs and miRNAs also contribute to cellular communication by altering metabolic and insulin signals, impacting inflammatory processes in pancreatic cells. Exosome-carried miRNAs, particularly, hold great promise as biomarkers or in developing innovative therapeutics for diabetes and its consequences[157].
Islet insulitis is connected with the transfer of a specific group of miRNAs from lymphocytes to cells via exosomes, including miR-142-3p, miR-142-5p, and miR-155, leading to the selective death of insulin-secreting cells. Inactivation of these miRNAs protected cells against apoptosis caused by T cell exosomes in vitro and reduced T1DM development in NOD mice in vivo. As a result, it has been postulated that miRNA transfer mediated by exosomes released by lymphocytes causes β-cell death and may be one of the mechanisms contributing to the development of T1DM[158].
Katayama et al[159] used microarrays to evaluate the expression profile of exosomal miRNAs in healthy people and those with T2DM[159]. They discovered that miR-20b-5p in exosomes was abundant in people with T2DM. Further in vitro studies revealed that this miRNA targets the AKT interacting protein, which modifies AKT protein activity and decreases glycogen buildup in muscles and IR[159].
Downregulation of exosomal lncRNA-p3134 in diabetic mice reduced glucose-stimulated insulin production by lowering key regulators (Pdx-1, MafA, GLUT2 and Tcf7 L2) in β-cells. This shows that lncRNA-p3134 regulates insulin secretion and that its downregulation leads to diabetes pathogenesis[160].
Exosomal miRNAs (miR-20b-5p, miR-155, miR-450b-3p, miR-151-3p and miR-29b-3p) were elevated in diabetic mice and targeted skeletal muscles via insulin signaling regulatory proteins (PPAR, AKT, GLUT4 and FOXO). This contributes to DM pathogenesis by influencing insulin signaling and glucose absorption in skeletal muscles[161]. Other exosomal miRNAs (miR-122, 192, 27a/b, 155 and 29b-3p) were upregulated in diabetic models and targeted adipocytes via PPAR proteins. This impairs lipid metabolism and contributes to diabetes development[162].
Exosomal miRNAs (miR-142-3p and miR-142-5p) were also found to be enhanced in diabetic mice and target pancreatic cells via cytokines that are elevated. This contributes to DM pathogenesis by encouraging immune cell recruitment and β-cell death during autoimmune attacks[158]. Other exosomal miRNAs that target organs such as astrocytes, retinal tissue, and renal cells (miR-106, miR-146a, miR-222 and miR-486) have potential therapeutic roles in protecting pancreatic cells or treating diabetic complications[163].
Exosomal miRNAs and lncRNAs influence DM development in various ways, including regulating pancreatic inflammation and metabolic and insulin signaling in target organs. Despite mounting evidence, research on the involvement of exosomes harboring ncRNAs in diabetes is still in its early stages, but they have promise and significant roles in pathogenesis, diagnosis and treatment of DM[164,165].
Finally, chronic inflammation is a feature of metabolic disorders such as DM and obesity[166], and several miRNAs are associated with regulating the expression of inflammatory markers[167-169]. The overexpression of miR-132 in human adipose-derived stem cells results in increased production of interleukin (IL)-8 and monocyte chemoattractant protein (MCP)1[170], which was also found to be regulated by miR-126 and miR-193b[171]. Other studies have found a link between low miR-221 expression and high tumor necrosis factor (TNF)-α levels in human adipose tissue-derived mesenchymal stem cells from obese women[172]. miR-145 was found to boost TNF-α expression in adipocytes by activating the NF-κB signaling pathway[173]. The investigation of miRNA expression in leptin-deficient ob/ob mouse adipocytes and TNF-α-treated adipocytes in vitro revealed a similar expression pattern[174]. In adipocytes isolated from mice, TNF-α increased the expression of a set of miRNAs, including miR-130, miR-150, miR-146a, miR-146b, miR-221 and miR-222, while decreasing miR-143 and miR-103 levels[174-177]. In human preadipocytes, both TNF-α and leptin induce decreased expression of miR-221 and upregulation of miR-335[177,178].
Increasing evidence supports using extracellular circulatory miRNAs as biomarkers for various pathophysiological conditions[123,179]. Highly stable extracellular miRNAs that are protected from degradation due to their enclosure in exosomal vesicles[180] or association with AGO2 proteins[181,182] and high-density lipoproteins[183], are present in various biological fluids, such as serum[184], plasma[181], cerebrospinal fluid[185], saliva[180], urine and tears[186]. Although the concentration of miRNAs in body fluids is in the femtomolar range[187], highly sensitive assays such as qRT-PCR, microarrays, and RNA sequencing (RNA-seq) can detect them in small samples even after prolonged storage[186].
Several studies investigated the expression of miRNAs in the serum/plasma of diabetic patients, and several circulating miRNAs are consistently dysregulated in T1DM patients compared to controls. In T1DM, 11 circulating miRNAs implicated in immune system function, cell survival and proliferation, and insulin production were dysregulated[188]. This set comprised miR-100-5p, miR-21-5p, miR-150-5p, miR-24-3p, miR-146a-5p, miR-148a-3p, miR-181a-5p, miR-210-5p, miR-342-3p, miR-1275 and miR-375. In a study by Nielsen et al[189], global profiling of serum miRNAs expression in new-onset T1DM in children and age-matched healthy controls revealed a set of 12 upregulated miRNAs in T1DM patients, comprising miR-24, miR-152, miR-30a-5p, miR-200, miR-148a, miR-181a, miR-210, miR-27a, miR-29a, miR-26a, miR-27b and miR-25; some of which are implicated in β-cell function and glycemic control[189]. Erener et al[190] studied serum miRNA expression and signaling pathways involved in T1DM development[190]. They found six miRNA (miR-222-3p, miR-24-3p, miR-454-3p, miR-144-5p, miR-345-5p and miR-140-5p) that were specifically dysregulated in new-onset T1DM but not at later stages of DM, and were involved in Wnt, MAPK, and PI3K/Akt signaling pathways[190]. The plasma levels of miR-30d, miR29a, miR-21, miR-24, miR-34a, miR-148a, miR-126 and miR-146 were significantly upregulated in adult T1DM patients[191], and higher levels of miR-210, miR-21 and miR-181a in T1DM were also confirmed in other studies[192,193]. More than one independent study found upregulation of miR-21, miR-148a, miR-24, miR-210 and miR-181a-5p in T1DM, indicating their potential use as circulating biomarkers of T1DM.
Global profiling of circulating miRNAs in blood samples of T2DM patients revealed approximately 70 upregulated miRNAs and around 100 downregulated miRNAs in T2DM patients[50]. A subsequent meta-analysis suggested that plasma levels of miR-103, miR-29a, miR-107, miR34a, miR-142–3p, miR-132, miR-375 and miR-144 may potentially serve as biomarkers for T2DM[194]. miR-126a was the most significantly downregulated circulating miRNA in T2DM patients[195-199]. Comparison of plasma miR-126 levels with healthy controls, T2DM-susceptible, and T2DM patients demonstrated significant miRNA-126 downregulation in both T2DM-susceptible individuals and T2DM patients, suggesting a close association of miR-126 with the T2DM manifestation, thus making this miRNA a potential biomarker for the early identification of susceptibility to T2DM[199]. In addition, numerous studies reported an association of other serum miRNAs with T2DM[197,200-208]. For example, miRNA profiles comparing T2DM patients to a control group with normal glucose tolerance revealed significantly lower expression levels of miR-486, miR-96, miR-23a, miR-191, miR-186, miR192 and let-7[202] whereas increased levels of miR-9, miR34a, miR-27a, miR-15b, miR-29a, miR-124a, miR-30d, miR-192, miR-150, miR-375, miR-146b, miR-320a, miR-571, miR-486, miR-661, miR-1303 miR-770 and miR-892b were observed[203-206]. Whole blood evaluations demonstrated upregulation of miR-320, miR-144, miR-29a, miR-192 and miR-150, downregulation of miR-30d, miR-15a and miR-182[50,207], and decreased miR-103b expression in platelets of T2DM patients[208].
Regarding the utility of miRNAs as potential biomarkers of metabolic diseases, circulatory miRNAs miR-17-5p, miR-15a-5p, miR-221 and let-7g were reported as reliable predictive biomarkers of metabolic syndrome (MetS)[209]. These miRNAs have a well-documented role in regulating IR, β-cell apoptosis, and central obesity[210,211]. Circulating miR-192 and miR-194 have been suggested as prospective DM risk biomarkers[212], whereas plasma levels of miR-29a, miR-9, miR-28-3p, miR-30a-5p, miR-103, and miR-150 are reliable predictive biomarkers that distinguish between incident-T2DM and non-T2DM patients[145]. It should be emphasized that this group of miRNAs is linked to cell proliferation, insulin sensitivity, and secretion and that alterations in their levels can occur up to three years before T2DM development[123]. However, more controlled studies on large patient cohorts are required to fully establish the potential of many proposed miRNA-based biomarkers for T1DM, T2DM, and other metabolic diseases.
In recent years, bio-nanomedicine has turned its attention to EVs as a novel disease treatment approach. One of the most promising applications is the delivery of tolerogenic nanoparticles (TNPs) to combat autoimmune diseases like T1DM. EVs and nanoparticles (NPs), as opposed to traditional medicines, provide advantages such as tailored delivery, lower toxicity, and enhanced stability. TNPs can induce immunological tolerance in T1DM patients by regulating the immune response via various mechanisms[213]. In contrast, EVs can deliver cargo such as cytokines, growth factors, and miRNAs to recipient cells, influencing immune responses via a paracrine impact and during the development of the immunological synapse[214].
A recent study has focused on developing EVs to contain TNPs to treat T1DM. For example, immunomodulatory NPs containing antisense oligonucleotides to CD40, CD80 and CD86 have been utilized to prevent T1DM in mice by increasing Foxp3+ Treg cells[215]. Another study found that the coculture of islets and bone marrow stem cells enhanced islet-cell survival and functionality in mice, mediated by exosomes via a paracrine action[216]. Clinical studies[217] have shown that exosomes derived from mesenchymal stem cells can suppress immune targeting in allogeneic grafts. These findings imply that EVs have regenerative, antiapoptotic, immunomodulatory, and angiogenic activities, making them a prospective tool for restoring islet-cell function and treating autoimmune disorders[218].
Nanotechnology has gained prominence in diabetes research by leveraging nanomaterials, nanostructures, and NP design to obtain more exact information on DM diagnosis. NPs can be used to deliver RNA and proteins to identify and monitor illness progression[219]. A recent study aimed to identify critical miRNAs that are dysregulated in pancreatic islets during T1DM progression and to create a theranostic strategy to modulate their expression using an MRI-based nanodrug. Iron oxide NPs combined with miRNA-targeting oligonucleotides were used to treat a mouse model of T1DM[157,220].
RNA-based therapeutic approaches have several important advantages compared to other drugs. Theoretically, miRNAs can simultaneously target several mRNAs, and fine-tuning miRNAs expression may restore physiological homeostasis[221]. The mechanism of action of RNA molecules may be elucidated from various available bioinformatics tools, such as in silico analysis and RNA structure prediction. ncRNAs are not associated with the development of drug resistance[222]. Thus, miRNAs are intriguing pharmacological targets that can potentially treat various complex diseases such as DM, cardiovascular disease, and cancer at the molecular level[222-225]. In miRNA-based therapies, the ultimate goal is to restore specific miRNA functions to normal levels, achieved by either restoring the expression of downregulated miRNAs using miRNA mimics or inhibiting the activity of upregulated miRNAs with a miRNA inhibitor.
miRNA mimics are double-stranded RNAs with the same sequence as a specific endogenous miRNAs[226]. miRNA mimics have been used in vitro to stimulate the regeneration of insulin-producing cells from induced pluripotent stem cells[227-230]. The approach is based on discovering that different miRNA clusters control human iPSC reprogramming[231,232]. However, this approach is not used in vivo since it may be associated with unwanted side effects[233].
Antisense oligonucleotides (ASOs) containing a complementary sequence to the target miRNA are used as miRNA inhibitors. Since most miRNAs repress target gene expression, binding a miRNA inhibitor to the mature target miRNA typically activates target gene expression. Locked nucleic acid (LNA) anti-miRs and antagomiRs are in vivo efficient modified miRNA inhibitors[234,235]. Anti-miRs are ASOs fully or partially complementary to an endogenous miRNA and act by preventing its interaction with target genes. AntagomiRs are cholesterol-conjugated anti-miRs with improved intracellular delivery[236]. At present, anti-miRs are the most commonly used miRNA-based therapeutic tool[237,238]. LNA anti-miR-122 treatment has been tested in mice and nonhuman primates, showing reduced plasma cholesterol, improved liver steatosis, and no indication of hepatic toxicity[239,240]. Several other miRNA-targeting therapeutics are in different phases of preclinical and clinical studies. Regarding the potential treatment of T2DM and IR, ASOs were used to inhibit miR-103 and miR-107 in the liver and adipose tissue of obese mice resulting in improved insulin sensitivity and glucose homeostasis[108]. Modified GalNAc-conjugated oligonucleotides RG-125 (AZD4076) developed by Regulus Therapeutics, which targets miR-103/mir-107, was tested in phase I and IIa clinical trials to assess their effect on insulin sensitivity and liver fat content in patients with T2DM and nonalcoholic steatohepatitis[241]. Another anti-miR developed by Regulus Therapeutics for treating metabolic illnesses is 2'-fluoro/methoxyethyl modified, phosphorothioate backbone modified anti-miR-33, which has been proven to reduce atherosclerotic plaque in T2DM patients[242,243]. An LNA-modified ASO targeting miR-208A (MGN-9103) was developed by Viridian Therapeutics, and it demonstrated the potential to ameliorate insulin sensitivity and systemic glucose tolerance in MetS[244]. Several candidate miRNA molecules, such as mir-21 and miR-181a, were suggested as potential therapeutic targets for metabolic disease. miR-21 suppression with LNA-modified anti-miR-21 in adipose tissue of db/db mice led to a significant decrease in body weight[139], whereas oligonucleotide-mediated downregulation of miR-181 in DIO mice increased levels of SIRT1 and ameliorated insulin sensitivity and glucose homeostasis[245].
It is also worth noting that dietary substances like conjugated linoleic acid (CLA), polyphenols, and long-chain omega-3 polyunsaturated fatty acids (n-3 PUFAs) indirectly influence miRNA expression[246-249]. For instance, CLA treatment altered the expression of miRNAs related to adipocyte differentiation, lipid metabolism, and obesity, such as miR-143, miR-107, miR-222, miR-103 and miR-221[246]. Dietary polyphenols inhibited the increased expression of miR-103 and miR-107, which is associated with a high-fat diet[247], whereas n-3 PUFA consumption alters the expression of miRNAs involved in lipid metabolism and inflammation[249].
Gene editing based on clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 (CRISPR/Cas9) has recently emerged as a technique with broad applications in the research and treatment of various diseases. The CRISPR/Cas9 RNA-guided editing technology could be used for genome editing. It is composed of a Cas9 nuclease that binds to a conserved three-nucleotide sequence known as the proto-adjacent motif (PAM) and creates a double-stranded DNA break. CRISPR RNA (crRNA) is a guide for Cas9 and an adapter trans-activating RNA (tracrRNA). The crRNA and tracrRNA can be combined to create a single-guide RNA capable of directing Cas9 to any target within the PAM sequence[250-252]. The CRISPR/Cas9 platform has been used to target the expression of miRNAs implied in various pathophysiological conditions[253,254]. An innovative example of CRISPR/Cas9 use in the context of the development of antidiabetic therapeutics is a recent development of a therapeutic candidate VCTX210. In 2021, CRISPR Therapeutics and ViaCyt companies jointly developed CRISPR-edited stem cell therapy candidate VCTX210 for potentially treating T1DM and T2DM, which has been approved for a clinical trial in Canada. VCTX210 is a stem cell-derived therapy edited with CRISPR-Cas9 intended to replace the β-cells lost in diabetes. Nonetheless, the use of gene editing for therapeutic purposes in the future will necessitate precise delivery of CRISPR to specific target tissues as well as strict control of potential off-target effects[255].
Although most current animal and human studies focus on the detection rather than the functional analysis of various miRNAs associated with cell physiology and pathology, many highly specific miRNAs have recently been identified as potential prognostic markers and therapeutic tools in diabetes patients. MiRNAs are involved in the regulation of gene expression in glucose homeostasis, lipid metabolism, and immune system balance. Increasing evidence shows that miRNAs can be a biomarker to predict type 1 and type 2 diabetes. Future studies must extensively investigate the roles of identified miRNAs to find those with the most reliable prognostic and therapeutic potential. Developing highly precise miRNA-based therapies designed for clinical usage will improve predicting vascular risk and end-organ vascular damage. With further advancements in high-throughput methodologies, such as whole genome and transcriptome profiling, and the associated proteomic and metabolomic analyses, a more profound link between various miRNAs and the physiology and pathophysiology of glucose homeostasis and fat metabolism will be more firmly established.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: Serbia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Beg MMA, Kyrgyzstan; Wang Z, China; Wu QN, China S-Editor: Li L L-Editor: A P-Editor: Li L
| 1. | Nair M. Diabetes mellitus, part 1: physiology and complications. Br J Nurs. 2007;16:184-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 2. | Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol Rev. 2013;93:137-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1360] [Cited by in RCA: 1778] [Article Influence: 148.2] [Reference Citation Analysis (1)] |
| 3. | Katsarou A, Gudbjörnsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, Jacobsen LM, Schatz DA, Lernmark Å. Type 1 diabetes mellitus. Nat Rev Dis Primers. 2017;3:17016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 799] [Article Influence: 99.9] [Reference Citation Analysis (0)] |
| 4. | American Diabetes Association Professional Practice Committee. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2022. Diabetes Care. 2022;45:S17-S38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 1421] [Article Influence: 473.7] [Reference Citation Analysis (1)] |
| 5. | Forouhi NG, Wareham NJ. Epidemiology of diabetes. Medicine (Abingdon). 2014;42:698-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 264] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 6. | Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, Colagiuri S, Guariguata L, Motala AA, Ogurtsova K, Shaw JE, Bright D, Williams R; IDF Diabetes Atlas Committee. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract. 2019;157:107843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5345] [Cited by in RCA: 5942] [Article Influence: 990.3] [Reference Citation Analysis (8)] |
| 7. | Cho NH, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, Malanda B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3709] [Cited by in RCA: 4391] [Article Influence: 627.3] [Reference Citation Analysis (0)] |
| 8. | Kitada M, Zhang Z, Mima A, King GL. Molecular mechanisms of diabetic vascular complications. J Diabetes Investig. 2010;1:77-89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 9. | Stehouwer CDA. Microvascular Dysfunction and Hyperglycemia: A Vicious Cycle With Widespread Consequences. Diabetes. 2018;67:1729-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 198] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 10. | Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383:69-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1462] [Cited by in RCA: 1652] [Article Influence: 150.2] [Reference Citation Analysis (0)] |
| 11. | Gordin D, Groop PH. Aspects of Hyperglycemia Contribution to Arterial Stiffness and Cardiovascular Complications in Patients With Type 1 Diabetes. J Diabetes Sci Technol. 2016;10:1059-1064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Creager MA, Lüscher TF, Cosentino F, Beckman JA. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: Part I. Circulation. 2003;108:1527-1532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 940] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 13. | Rodríguez-Mañas L, López-Dóriga P, Petidier R, Neira M, Solís J, Pavón I, Peiró C, Sánchez-Ferrer CF. Effect of glycaemic control on the vascular nitric oxide system in patients with type 1 diabetes. J Hypertens. 2003;21:1137-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Rebnord EW, Strand E, Midttun Ø, Svingen GFT, Christensen MHE, Ueland PM, Mellgren G, Njølstad PR, Tell GS, Nygård OK, Pedersen ER. The kynurenine:tryptophan ratio as a predictor of incident type 2 diabetes mellitus in individuals with coronary artery disease. Diabetologia. 2017;60:1712-1721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 15. | Chen Q, Zhu L, Tang Y, Zhao Z, Yi T, Chen H. Preparation-related structural diversity and medical potential in the treatment of diabetes mellitus with ginseng pectins. Ann N Y Acad Sci. 2017;1401:75-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Mayer-Davis EJ, Kahkoska AR, Jefferies C, Dabelea D, Balde N, Gong CX, Aschner P, Craig ME. ISPAD Clinical Practice Consensus Guidelines 2018: Definition, epidemiology, and classification of diabetes in children and adolescents. Pediatr Diabetes. 2018;19 Suppl 27:7-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 377] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 17. | Ounissi-Benkalha H, Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol Med. 2008;14:268-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 18. | Atkinson MA. The pathogenesis and natural history of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 19. | Csorba TR, Lyon AW, Hollenberg MD. Autoimmunity and the pathogenesis of type 1 diabetes. Crit Rev Clin Lab Sci. 2010;47:51-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Gonzalez CD, Lee MS, Marchetti P, Pietropaolo M, Towns R, Vaccaro MI, Watada H, Wiley JW. The emerging role of autophagy in the pathophysiology of diabetes mellitus. Autophagy. 2011;7:2-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 245] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 21. | Esposito S, Toni G, Tascini G, Santi E, Berioli MG, Principi N. Environmental Factors Associated With Type 1 Diabetes. Front Endocrinol (Lausanne). 2019;10:592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Al-Goblan AS, Al-Alfi MA, Khan MZ. Mechanism linking diabetes mellitus and obesity. Diabetes Metab Syndr Obes. 2014;7:587-591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 459] [Cited by in RCA: 542] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 23. | Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PLoS Comput Biol. 2017;13:e1005457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 437] [Cited by in RCA: 554] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 24. | McGettigan PA. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17:4-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 193] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 25. | Hemberg M, Gray JM, Cloonan N, Kuersten S, Grimmond S, Greenberg ME, Kreiman G. Integrated genome analysis suggests that most conserved non-coding sequences are regulatory factor binding sites. Nucleic Acids Res. 2012;40:7858-7869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 26. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16090] [Article Influence: 1005.6] [Reference Citation Analysis (2)] |
| 27. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27850] [Article Influence: 1326.2] [Reference Citation Analysis (0)] |
| 28. | Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3232] [Cited by in RCA: 3113] [Article Influence: 207.5] [Reference Citation Analysis (0)] |
| 29. | Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879-14884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 468] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 30. | Zhou H, Rigoutsos I. MiR-103a-3p targets the 5' UTR of GPRC5A in pancreatic cells. RNA. 2014;20:1431-1439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 31. | Zhang Y, Fan M, Zhang X, Huang F, Wu K, Zhang J, Liu J, Huang Z, Luo H, Tao L, Zhang H. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA. 2014;20:1878-1889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 32. | Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016-3027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1468] [Cited by in RCA: 1576] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 33. | Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 450] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 34. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5833] [Cited by in RCA: 6543] [Article Influence: 384.9] [Reference Citation Analysis (0)] |
| 35. | Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2577] [Cited by in RCA: 2720] [Article Influence: 160.0] [Reference Citation Analysis (0)] |
| 36. | Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2851] [Cited by in RCA: 2957] [Article Influence: 164.3] [Reference Citation Analysis (0)] |
| 37. | Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1178] [Cited by in RCA: 1223] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 38. | Bolmeson C, Esguerra JL, Salehi A, Speidel D, Eliasson L, Cilio CM. Differences in islet-enriched miRNAs in healthy and glucose intolerant human subjects. Biochem Biophys Res Commun. 2011;404:16-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 39. | Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns. 2009;9:109-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 195] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 40. | Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1531] [Cited by in RCA: 1602] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 41. | Tian C, Ouyang X, Lv Q, Zhang Y, Xie W. Cross-talks between microRNAs and mRNAs in pancreatic tissues of streptozotocin-induced type 1 diabetic mice. Biomed Rep. 2015;3:333-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 42. | Ma H, Zhang S, Shi D, Mao Y, Cui J. MicroRNA-26a Promotes Regulatory T cells and Suppresses Autoimmune Diabetes in Mice. Inflammation. 2016;39:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 43. | Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C, Abderrahmani A, Regazzi R. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728-2736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 271] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 44. | Nesca V, Guay C, Jacovetti C, Menoud V, Peyot ML, Laybutt DR, Prentki M, Regazzi R. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia. 2013;56:2203-2212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 189] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 45. | Zhao X, Mohan R, Özcan S, Tang X. MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic β-cells. J Biol Chem. 2012;287:31155-31164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 46. | Jacovetti C, Abderrahmani A, Parnaud G, Jonas JC, Peyot ML, Cornu M, Laybutt R, Meugnier E, Rome S, Thorens B, Prentki M, Bosco D, Regazzi R. MicroRNAs contribute to compensatory β cell expansion during pregnancy and obesity. J Clin Invest. 2012;122:3541-3551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 47. | Latreille M, Hausser J, Stützer I, Zhang Q, Hastoy B, Gargani S, Kerr-Conte J, Pattou F, Zavolan M, Esguerra JL, Eliasson L, Rülicke T, Rorsman P, Stoffel M. MicroRNA-7a regulates pancreatic β cell function. J Clin Invest. 2014;124:2722-2735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 237] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 48. | Tattikota SG, Rathjen T, McAnulty SJ, Wessels HH, Akerman I, van de Bunt M, Hausser J, Esguerra JL, Musahl A, Pandey AK, You X, Chen W, Herrera PL, Johnson PR, O'Carroll D, Eliasson L, Zavolan M, Gloyn AL, Ferrer J, Shalom-Feuerstein R, Aberdam D, Poy MN. Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metab. 2014;19:122-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 49. | Esguerra JL, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One. 2011;6:e18613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 50. | Karolina DS, Armugam A, Tavintharan S, Wong MT, Lim SC, Sum CF, Jeyaseelan K. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011;6:e22839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 298] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 51. | Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ, Chen Y, Choi I, Vourekas A, Won KJ, Liu C, Vivek K, Naji A, Friedman JR, Kaestner KH. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19:135-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 272] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 52. | Sebastiani G, Po A, Miele E, Ventriglia G, Ceccarelli E, Bugliani M, Marselli L, Marchetti P, Gulino A, Ferretti E, Dotta F. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol. 2015;52:523-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 53. | Locke JM, da Silva Xavier G, Dawe HR, Rutter GA, Harries LW. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia. 2014;57:122-128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 54. | Sebastiani G, Ventriglia G, Stabilini A, Socci C, Morsiani C, Laurenzi A, Nigi L, Formichi C, Mfarrej B, Petrelli A, Fousteri G, Brusko TM, Dotta F, Battaglia M. Regulatory T-cells from pancreatic lymphnodes of patients with type-1 diabetes express increased levels of microRNA miR-125a-5p that limits CCR2 expression. Sci Rep. 2017;7:6897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 55. | Filios SR, Shalev A. β-Cell MicroRNAs: Small but Powerful. Diabetes. 2015;64:3631-3644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 56. | Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813-5818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 605] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 57. | Hammar EB, Irminger JC, Rickenbach K, Parnaud G, Ribaux P, Bosco D, Rouiller DG, Halban PA. Activation of NF-kappaB by extracellular matrix is involved in spreading and glucose-stimulated insulin secretion of pancreatic beta cells. J Biol Chem. 2005;280:30630-30637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 58. | Xia HQ, Pan Y, Peng J, Lu GX. Over-expression of miR375 reduces glucose-induced insulin secretion in Nit-1 cells. Mol Biol Rep. 2011;38:3061-3065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389:305-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 214] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 60. | Matsumoto M, Miki T, Shibasaki T, Kawaguchi M, Shinozaki H, Nio J, Saraya A, Koseki H, Miyazaki M, Iwanaga T, Seino S. Noc2 is essential in normal regulation of exocytosis in endocrine and exocrine cells. Proc Natl Acad Sci U S A. 2004;101:8313-8318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 61. | El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3'-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708-2717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 338] [Cited by in RCA: 355] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 62. | Xu G, Chen J, Jing G, Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med. 2013;19:1141-1146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 219] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 63. | Jo S, Chen J, Xu G, Grayson TB, Thielen LA, Shalev A. miR-204 Controls Glucagon-Like Peptide 1 Receptor Expression and Agonist Function. Diabetes. 2018;67:256-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 64. | Backe MB, Novotny GW, Christensen DP, Grunnet LG, Mandrup-Poulsen T. Altering β-cell number through stable alteration of miR-21 and miR-34a expression. Islets. 2014;6:e27754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 65. | Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, Regazzi R. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978-986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 262] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 66. | Belgardt BF, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, von Meyenn F, Villena FN, Herrmanns K, Bosco D, Kerr-Conte J, Pattou F, Rülicke T, Stoffel M. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21:619-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 226] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 67. | Burrack AL, Martinov T, Fife BT. T Cell-Mediated Beta Cell Destruction: Autoimmunity and Alloimmunity in the Context of Type 1 Diabetes. Front Endocrinol (Lausanne). 2017;8:343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 212] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 68. | Mone P, de Donato A, Varzideh F, Kansakar U, Jankauskas SS, Pansini A, Santulli G. Functional role of miR-34a in diabetes and frailty. Front Aging. 2022;3:949924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 69. | Fomison-Nurse I, Saw EEL, Gandhi S, Munasinghe PE, Van Hout I, Williams MJA, Galvin I, Bunton R, Davis P, Cameron V, Katare R. Diabetes induces the activation of pro-ageing miR-34a in the heart, but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death Differ. 2018;25:1336-1349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 70. | Margaritis K, Margioula-Siarkou G, Giza S, Kotanidou EP, Tsinopoulou VR, Christoforidis A, Galli-Tsinopoulou A. Micro-RNA Implications in Type-1 Diabetes Mellitus: A Review of Literature. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 71. | Taplin CE, Barker JM. Autoantibodies in type 1 diabetes. Autoimmunity. 2008;41:11-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 157] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 72. | Salas-Pérez F, Codner E, Valencia E, Pizarro C, Carrasco E, Pérez-Bravo F. MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology. 2013;218:733-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 73. | Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev. 2011;27:862-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 74. | Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723-3730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 489] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 75. | Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1037] [Cited by in RCA: 1138] [Article Influence: 94.8] [Reference Citation Analysis (0)] |
| 76. | Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2963] [Cited by in RCA: 3508] [Article Influence: 250.6] [Reference Citation Analysis (0)] |
| 77. | Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4004] [Cited by in RCA: 3940] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 78. | Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 805] [Cited by in RCA: 775] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 79. | Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46:2169-2184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 219] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 80. | Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY). 2014;6:992-1009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 162] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 81. | Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH, Martindale JL, De S, Wood WH 3rd, Becker KG, Gorospe M. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. 2013;12:890-900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 82. | Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629-634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 155] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 83. | Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925-933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 755] [Cited by in RCA: 980] [Article Influence: 81.7] [Reference Citation Analysis (0)] |
| 84. | Carter G, Miladinovic B, Patel AA, Deland L, Mastorides S, Patel NA. Circulating long noncoding RNA GAS5 Levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 2015;4:102-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 85. | Jin F, Wang N, Zhu Y, You L, Wang L, De W, Tang W. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic β Cells. Cell Physiol Biochem. 2017;43:2062-2073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 86. | Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty-Colace C, Benazra M, Nakic N, Yang J, Wang H, Pasquali L, Moran I, Garcia-Hurtado J, Castro N, Gonzalez-Franco R, Stewart AF, Bonner C, Piemonti L, Berney T, Groop L, Kerr-Conte J, Pattou F, Argmann C, Schadt E, Ravassard P, Ferrer J. Human Pancreatic β Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab. 2017;25:400-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 87. | Yin DD, Zhang EB, You LH, Wang N, Wang LT, Jin FY, Zhu YN, Cao LH, Yuan QX, De W, Tang W. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic β cells. Cell Physiol Biochem. 2015;35:1892-1904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 113] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 88. | You L, Wang N, Yin D, Wang L, Jin F, Zhu Y, Yuan Q, De W. Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells. J Cell Physiol. 2016;231:852-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 123] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 89. | Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes Dev. 2016;30:502-507. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 90. | Motterle A, Gattesco S, Peyot ML, Esguerra JLS, Gomez-Ruiz A, Laybutt DR, Gilon P, Burdet F, Ibberson M, Eliasson L, Prentki M, Regazzi R. Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Mol Metab. 2017;6:1407-1418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 91. | Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1589] [Cited by in RCA: 1961] [Article Influence: 150.8] [Reference Citation Analysis (0)] |
| 92. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6052] [Article Influence: 504.3] [Reference Citation Analysis (0)] |
| 93. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6033] [Article Influence: 502.8] [Reference Citation Analysis (0)] |
| 94. | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3481] [Cited by in RCA: 3426] [Article Influence: 285.5] [Reference Citation Analysis (0)] |
| 95. | Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 96. | Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1045] [Cited by in RCA: 1511] [Article Influence: 167.9] [Reference Citation Analysis (0)] |
| 97. | Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 464] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 98. | Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 1296] [Article Influence: 144.0] [Reference Citation Analysis (0)] |
| 99. | Haque S, Harries LW. Circular RNAs (circRNAs) in Health and Disease. Genes (Basel). 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 212] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 100. | Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Venø MT, Kjems J, Laybutt DR, Regazzi R. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab. 2018;9:69-83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 160] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 101. | Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 405] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 102. | Min KH, Yang WM, Lee W. Saturated fatty acids-induced miR-424-5p aggravates insulin resistance via targeting insulin receptor in hepatocytes. Biochem Biophys Res Commun. 2018;503:1587-1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 103. | Yang WM, Jeong HJ, Park SW, Lee W. Obesity-induced miR-15b is linked causally to the development of insulin resistance through the repression of the insulin receptor in hepatocytes. Mol Nutr Food Res. 2015;59:2303-2314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 104. | Yang WM, Jeong HJ, Park SY, Lee W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 2014;588:3939-3946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 105. | Yang WM, Min KH, Lee W. Induction of miR-96 by Dietary Saturated Fatty Acids Exacerbates Hepatic Insulin Resistance through the Suppression of INSR and IRS-1. PLoS One. 2016;11:e0169039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 106. | Delibegovic M, Zimmer D, Kauffman C, Rak K, Hong EG, Cho YR, Kim JK, Kahn BB, Neel BG, Bence KK. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes. 2009;58:590-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 229] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 107. | Yang YM, Seo SY, Kim TH, Kim SG. Decrease of microRNA-122 causes hepatic insulin resistance by inducing protein tyrosine phosphatase 1B, which is reversed by licorice flavonoid. Hepatology. 2012;56:2209-2220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 115] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 108. | Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 707] [Cited by in RCA: 796] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 109. | Ryu HS, Park SY, Ma D, Zhang J, Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS One. 2011;6:e17343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 110. | Jeong HJ, Park SY, Yang WM, Lee W. The induction of miR-96 by mitochondrial dysfunction causes impaired glycogen synthesis through translational repression of IRS-1 in SK-Hep1 cells. Biochem Biophys Res Commun. 2013;434:503-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 111. | Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z, Wu J, Zhou Y. MicroRNA-145 suppresses hepatocellular carcinoma by targeting IRS1 and its downstream Akt signaling. Biochem Biophys Res Commun. 2014;446:1255-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 112. | Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suárez Y, Lai EC, Fernández-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232-9237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 536] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 113. | Tang CY, Man XF, Guo Y, Tang HN, Tang J, Zhou CL, Tan SW, Wang M, Zhou HD. IRS-2 Partially Compensates for the Insulin Signal Defects in IRS-1(-/-) Mice Mediated by miR-33. Mol Cells. 2017;40:123-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 114. | Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci. 2013;126:2678-2691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 115. | Massart J, Sjögren RJO, Lundell LS, Mudry JM, Franck N, O'Gorman DJ, Egan B, Zierath JR, Krook A. Altered miR-29 Expression in Type 2 Diabetes Influences Glucose and Lipid Metabolism in Skeletal Muscle. Diabetes. 2017;66:1807-1818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 116. | Agarwal P, Srivastava R, Srivastava AK, Ali S, Datta M. miR-135a targets IRS2 and regulates insulin signaling and glucose uptake in the diabetic gastrocnemius skeletal muscle. Biochim Biophys Acta. 2013;1832:1294-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 117. | Kurtz CL, Peck BC, Fannin EE, Beysen C, Miao J, Landstreet SR, Ding S, Turaga V, Lund PK, Turner S, Biddinger SB, Vickers KC, Sethupathy P. MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes. 2014;63:3141-3148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 118. | Fu X, Dong B, Tian Y, Lefebvre P, Meng Z, Wang X, Pattou F, Han W, Lou F, Jove R, Staels B, Moore DD, Huang W. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125:2497-2509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 203] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 119. | Ling HY, Ou HS, Feng SD, Zhang XY, Tuo QH, Chen LX, Zhu BY, Gao ZP, Tang CK, Yin WD, Zhang L, Liao DF. CHANGES IN microRNA (miR) profile and effects of miR-320 in insulin-resistant 3T3-L1 adipocytes. Clin Exp Pharmacol Physiol. 2009;36:e32-e39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 163] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 120. | Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, Cimino V, De Marinis L, Frustaci A, Catalucci D, Condorelli G. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377-2385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 313] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 121. | Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI; DIAGRAM Consortium; MAGIC Investigators, Altshuler D, Daley GQ. The Lin28/Let-7 axis regulates glucose metabolism. Cell. 2011;147:81-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 661] [Cited by in RCA: 737] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 122. | Xihua L, Shengjie T, Weiwei G, Matro E, Tingting T, Lin L, Fang W, Jiaqiang Z, Fenping Z, Hong L. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl Res. 2019;205:33-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 123. | Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019;30:656-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 532] [Cited by in RCA: 645] [Article Influence: 107.5] [Reference Citation Analysis (0)] |
| 124. | Kunej T, Jevsinek Skok D, Zorc M, Ogrinc A, Michal JJ, Kovac M, Jiang Z. Obesity gene atlas in mammals. J Genomics. 2013;1:45-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 125. | Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10:51-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 452] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 126. | Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 997] [Cited by in RCA: 1078] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 127. | Herrera BM, Lockstone HE, Taylor JM, Ria M, Barrett A, Collins S, Kaisaki P, Argoud K, Fernandez C, Travers ME, Grew JP, Randall JC, Gloyn AL, Gauguier D, McCarthy MI, Lindgren CM. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53:1099-1109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 224] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 128. | Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 490] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 129. | Jopling C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012;9:137-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 322] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 130. | Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304-2315.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 131. | Horak M, Novak J, Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev Biol. 2016;410:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 358] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 132. | Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol. 2011;11:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 142] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 133. | Ge Y, Chen J. MicroRNAs in skeletal myogenesis. Cell Cycle. 2011;10:441-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 134. | Maurizi G, Babini L, Della Guardia L. Potential role of microRNAs in the regulation of adipocytes liposecretion and adipose tissue physiology. J Cell Physiol. 2018;233:9077-9086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 135. | Vohl MC, Sladek R, Robitaille J, Gurd S, Marceau P, Richard D, Hudson TJ, Tchernof A. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res. 2004;12:1217-1222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 251] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 136. | Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, Ferrer A, Rodriguez-Hermosa JI, Ruiz B, Ricart W, Peral B, Fernández-Real JM. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One. 2010;5:e9022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 296] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 137. | Klöting N, Berthold S, Kovacs P, Schön MR, Fasshauer M, Ruschke K, Stumvoll M, Blüher M. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One. 2009;4:e4699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 267] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 138. | Keller P, Gburcik V, Petrovic N, Gallagher IJ, Nedergaard J, Cannon B, Timmons JA. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord. 2011;11:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 139. | Seeger T, Fischer A, Muhly-Reinholz M, Zeiher AM, Dimmeler S. Long-term inhibition of miR-21 Leads to reduction of obesity in db/db mice. Obesity (Silver Spring). 2014;22:2352-2360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 140. | Panic A, Stanimirovic J, Obradovic M, Sudar-Milovanovic E, Perovic M, Lackovic M, Petrovic N, Isenovic ER. Estradiol-mediated regulation of hepatic iNOS in obese rats: Impact of Src, ERK1/2, AMPKα, and miR-221. Biotechnol Appl Biochem. 2018;65:797-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 141. | Flowers E, Won GY, Fukuoka Y. MicroRNAs associated with exercise and diet: a systematic review. Physiol Genomics. 2015;47:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 142. | Rome S. Use of miRNAs in biofluids as biomarkers in dietary and lifestyle intervention studies. Genes Nutr. 2015;10:483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 143. | Safdar A, Tarnopolsky MA. Exosomes as Mediators of the Systemic Adaptations to Endurance Exercise. Cold Spring Harb Perspect Med. 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 144. | Whitham M, Parker BL, Friedrichsen M, Hingst JR, Hjorth M, Hughes WE, Egan CL, Cron L, Watt KI, Kuchel RP, Jayasooriah N, Estevez E, Petzold T, Suter CM, Gregorevic P, Kiens B, Richter EA, James DE, Wojtaszewski JFP, Febbraio MA. Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab. 2018;27:237-251.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 459] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 145. | Jiménez-Lucena R, Rangel-Zúñiga OA, Alcalá-Díaz JF, López-Moreno J, Roncero-Ramos I, Molina-Abril H, Yubero-Serrano EM, Caballero-Villarraso J, Delgado-Lista J, Castaño JP, Ordovás JM, Pérez-Martinez P, Camargo A, López-Miranda J. Circulating miRNAs as Predictive Biomarkers of Type 2 Diabetes Mellitus Development in Coronary Heart Disease Patients from the CORDIOPREV Study. Mol Ther Nucleic Acids. 2018;12:146-157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 146. | Beatty M, Guduric-Fuchs J, Brown E, Bridgett S, Chakravarthy U, Hogg RE, Simpson DA. Small RNAs from plants, bacteria and fungi within the order Hypocreales are ubiquitous in human plasma. BMC Genomics. 2014;15:933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 147. | Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, Li P, Olefsky JM. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell. 2017;171:372-384.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 877] [Article Influence: 109.6] [Reference Citation Analysis (0)] |
| 148. | Tryggestad JB, Teague AM, Sparling DP, Jiang S, Chernausek SD. Macrophage-Derived microRNA-155 Increases in Obesity and Influences Adipocyte Metabolism by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Obesity (Silver Spring). 2019;27:1856-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 149. | Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, Gorden P, Kahn CR. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1136] [Article Influence: 142.0] [Reference Citation Analysis (0)] |
| 150. | Manning P, Munasinghe PE, Bellae Papannarao J, Gray AR, Sutherland W, Katare R. Acute Weight Loss Restores Dysregulated Circulating MicroRNAs in Individuals Who Are Obese. J Clin Endocrinol Metab. 2019;104:1239-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 151. | Atkin SL, Ramachandran V, Yousri NA, Benurwar M, Simper SC, McKinlay R, Adams TD, Najafi-Shoushtari SH, Hunt SC. Changes in Blood microRNA Expression and Early Metabolic Responsiveness 21 Days Following Bariatric Surgery. Front Endocrinol (Lausanne). 2018;9:773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 152. | Raghav A, Tripathi P, Mishra BK, Jeong GB, Banday S, Gautam KA, Mateen QN, Singh P, Singh M, Singla A, Ahmad J. Mesenchymal Stromal Cell-Derived Tailored Exosomes Treat Bacteria-Associated Diabetes Foot Ulcers: A Customized Approach From Bench to Bed. Front Microbiol. 2021;12:712588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 153. | Bongiovanni L, Andriessen A, Wauben MHM, Nolte-'t Hoen ENM, de Bruin A. Extracellular Vesicles: Novel Opportunities to Understand and Detect Neoplastic Diseases. Vet Pathol. 2021;58:453-471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 154. | Raghav A, Khan ZA, Upadhayay VK, Tripathi P, Gautam KA, Mishra BK, Ahmad J, Jeong GB. Mesenchymal Stem Cell-Derived Exosomes Exhibit Promising Potential for Treating SARS-CoV-2-Infected Patients. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 155. | Pang H, Luo S, Xiao Y, Xia Y, Li X, Huang G, Xie Z, Zhou Z. Emerging Roles of Exosomes in T1DM. Front Immunol. 2020;11:593348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 156. | Chen H, Wang L, Zeng X, Schwarz H, Nanda HS, Peng X, Zhou Y. Exosomes, a New Star for Targeted Delivery. Front Cell Dev Biol. 2021;9:751079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 171] [Article Influence: 42.8] [Reference Citation Analysis (1)] |
| 157. | Raghav A, Ashraf H, Jeong GB. Engineered Extracellular Vesicles in Treatment of Type 1 Diabetes Mellitus: A Prospective Review. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Reference Citation Analysis (0)] |
| 158. | Guay C, Kruit JK, Rome S, Menoud V, Mulder NL, Jurdzinski A, Mancarella F, Sebastiani G, Donda A, Gonzalez BJ, Jandus C, Bouzakri K, Pinget M, Boitard C, Romero P, Dotta F, Regazzi R. Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic β Cell Death and May Contribute to Type 1 Diabetes Development. Cell Metab. 2019;29:348-361.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 207] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 159. | Katayama M, Wiklander OPB, Fritz T, Caidahl K, El-Andaloussi S, Zierath JR, Krook A. Circulating Exosomal miR-20b-5p Is Elevated in Type 2 Diabetes and Could Impair Insulin Action in Human Skeletal Muscle. Diabetes. 2019;68:515-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 160. | Sufianov A, Kostin A, Begliarzade S, Kudriashov V, Ilyasova T, Liang Y, Mukhamedzyanov A, Beylerli O. Exosomal non coding RNAs as a novel target for diabetes mellitus and its complications. Noncoding RNA Res. 2023;8:192-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 161. | Chang W, Wang J. Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 162. | Castaño C, Kalko S, Novials A, Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc Natl Acad Sci U S A. 2018;115:12158-12163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 293] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 163. | Improta-Caria AC, De Sousa RAL, Roever L, Fernandes T, Oliveira EM, Aras Júnior R, Souza BSF. MicroRNAs in type 2 diabetes mellitus: potential role of physical exercise. Rev Cardiovasc Med. 2022;23:29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 164. | Yamashita T, Takahashi Y, Nishikawa M, Takakura Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur J Pharm Biopharm. 2016;98:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 153] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 165. | Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7:789-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 870] [Cited by in RCA: 1389] [Article Influence: 173.6] [Reference Citation Analysis (1)] |
| 166. | Tseng YH, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov. 2010;9:465-482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 486] [Cited by in RCA: 459] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 167. | Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11:276-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 357] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 168. | Ge Q, Brichard S, Yi X, Li Q. microRNAs as a new mechanism regulating adipose tissue inflammation in obesity and as a novel therapeutic strategy in the metabolic syndrome. J Immunol Res. 2014;2014:987285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 169. | Hulsmans M, De Keyzer D, Holvoet P. MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. FASEB J. 2011;25:2515-2527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 178] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 170. | Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23:1876-1884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 185] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 171. | Arner E, Mejhert N, Kulyté A, Balwierz PJ, Pachkov M, Cormont M, Lorente-Cebrián S, Ehrlund A, Laurencikiene J, Hedén P, Dahlman-Wright K, Tanti JF, Hayashizaki Y, Rydén M, Dahlman I, van Nimwegen E, Daub CO, Arner P. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61:1986-1993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 249] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 172. | Chou WW, Wang YT, Liao YC, Chuang SC, Wang SN, Juo SH. Decreased microRNA-221 is associated with high levels of TNF-α in human adipose tissue-derived mesenchymal stem cells from obese woman. Cell Physiol Biochem. 2013;32:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 173. | Lorente-Cebrián S, Mejhert N, Kulyté A, Laurencikiene J, Åström G, Hedén P, Rydén M, Arner P. MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNF-α. PLoS One. 2014;9:e86800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 174. | Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050-1057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 455] [Cited by in RCA: 468] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 175. | Kim C, Lee H, Cho YM, Kwon OJ, Kim W, Lee EK. TNFα-induced miR-130 resulted in adipocyte dysfunction during obesity-related inflammation. FEBS Lett. 2013;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 176. | Shi C, Zhu L, Chen X, Gu N, Chen L, Yang L, Pang L, Guo X, Ji C, Zhang C. IL-6 and TNF-α induced obesity-related inflammatory response through transcriptional regulation of miR-146b. J Interferon Cytokine Res. 2014;34:342-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 177. | Karkeni E, Bonnet L, Marcotorchino J, Tourniaire F, Astier J, Ye J, Landrier JF. Vitamin D limits inflammation-linked microRNA expression in adipocytes in vitro and in vivo: A new mechanism for the regulation of inflammation by vitamin D. Epigenetics. 2018;13:156-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 178. | Zhu L, Chen L, Shi CM, Xu GF, Xu LL, Zhu LL, Guo XR, Ni Y, Cui Y, Ji C. MiR-335, an adipogenesis-related microRNA, is involved in adipose tissue inflammation. Cell Biochem Biophys. 2014;68:283-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 179. | Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 219] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 180. | Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 712] [Cited by in RCA: 863] [Article Influence: 66.4] [Reference Citation Analysis (2)] |
| 181. | Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003-5008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2643] [Article Influence: 188.8] [Reference Citation Analysis (0)] |
| 182. | Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223-7233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1356] [Cited by in RCA: 1525] [Article Influence: 108.9] [Reference Citation Analysis (0)] |
| 183. | Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2292] [Cited by in RCA: 2188] [Article Influence: 156.3] [Reference Citation Analysis (0)] |
| 184. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3554] [Article Influence: 209.1] [Reference Citation Analysis (0)] |
| 185. | Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 729] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 186. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 2087] [Article Influence: 139.1] [Reference Citation Analysis (0)] |
| 187. | Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA. 2013;110:4255-4260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 284] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 188. | Assmann TS, Recamonde-Mendoza M, De Souza BM, Crispim D. MicroRNA expression profiles and type 1 diabetes mellitus: systematic review and bioinformatic analysis. Endocr Connect. 2017;6:773-790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 189. | Nielsen LB, Wang C, Sørensen K, Bang-Berthelsen CH, Hansen L, Andersen ML, Hougaard P, Juul A, Zhang CY, Pociot F, Mortensen HB. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res. 2012;2012:896362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 190. | Erener S, Marwaha A, Tan R, Panagiotopoulos C, Kieffer TJ. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight. 2017;2:e89656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 191. | Seyhan AA, Nunez Lopez YO, Xie H, Yi F, Mathews C, Pasarica M, Pratley RE. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Sci Rep. 2016;6:31479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 131] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 192. | Osipova J, Fischer DC, Dangwal S, Volkmann I, Widera C, Schwarz K, Lorenzen JM, Schreiver C, Jacoby U, Heimhalt M, Thum T, Haffner D. Diabetes-associated microRNAs in pediatric patients with type 1 diabetes mellitus: a cross-sectional cohort study. J Clin Endocrinol Metab. 2014;99:E1661-E1665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 193. | Nabih ES, Andrawes NG. The Association Between Circulating Levels of miRNA-181a and Pancreatic Beta Cells Dysfunction via SMAD7 in Type 1 Diabetic Children and Adolescents. J Clin Lab Anal. 2016;30:727-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 194. | Zhu H, Leung SW. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia. 2015;58:900-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 195. | Zhang T, Li L, Shang Q, Lv C, Wang C, Su B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem Biophys Res Commun. 2015;463:60-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 196. | Rezk NA, Sabbah NA, Saad MS. Role of MicroRNA 126 in screening, diagnosis, and prognosis of diabetic patients in Egypt. IUBMB Life. 2016;68:452-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 197. | Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1196] [Cited by in RCA: 1136] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 198. | Olivieri F, Bonafè M, Spazzafumo L, Gobbi M, Prattichizzo F, Recchioni R, Marcheselli F, La Sala L, Galeazzi R, Rippo MR, Fulgenzi G, Angelini S, Lazzarini R, Bonfigli AR, Brugè F, Tiano L, Genovese S, Ceriello A, Boemi M, Franceschi C, Procopio AD, Testa R. Age- and glycemia-related miR-126-3p levels in plasma and endothelial cells. Aging (Albany NY). 2014;6:771-787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 199. | Zhang T, Lv C, Li L, Chen S, Liu S, Wang C, Su B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. Biomed Res Int. 2013;2013:761617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 200. | Olivieri F, Spazzafumo L, Bonafè M, Recchioni R, Prattichizzo F, Marcheselli F, Micolucci L, Mensà E, Giuliani A, Santini G, Gobbi M, Lazzarini R, Boemi M, Testa R, Antonicelli R, Procopio AD, Bonfigli AR. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: relationship with type 2 diabetes complications. Oncotarget. 2015;6:35372-35382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 201. | Dangwal S, Stratmann B, Bang C, Lorenzen JM, Kumarswamy R, Fiedler J, Falk CS, Scholz CJ, Thum T, Tschoepe D. Impairment of Wound Healing in Patients With Type 2 Diabetes Mellitus Influences Circulating MicroRNA Patterns via Inflammatory Cytokines. Arterioscler Thromb Vasc Biol. 2015;35:1480-1488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 202. | Yang Z, Chen H, Si H, Li X, Ding X, Sheng Q, Chen P, Zhang H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014;51:823-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 203. | Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, Dong Q, Pang Z, Guan Q, Gao L, Zhao J, Zhao L. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 411] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 204. | Cui X, You L, Zhu L, Wang X, Zhou Y, Li Y, Wen J, Xia Y, Ji C, Guo X. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism. 2018;78:95-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 205. | Wang C, Wan S, Yang T, Niu D, Zhang A, Yang C, Cai J, Wu J, Song J, Zhang CY, Zhang C, Wang J. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci Rep. 2016;6:20032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 206. | Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SL, Wong MT, Lim SC, Sum CF, Jeyaseelan K. Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab. 2012;97:E2271-E2276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 338] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 207. | Al-Kafaji G, Al-Mahroos G, Alsayed NA, Hasan ZA, Nawaz S, Bakhiet M. Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol Med Rep. 2015;12:7485-7490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 208. | Luo M, Li R, Deng X, Ren M, Chen N, Zeng M, Yan K, Xia J, Liu F, Ma W, Yang Y, Wan Q, Wu J. Platelet-derived miR-103b as a novel biomarker for the early diagnosis of type 2 diabetes. Acta Diabetol. 2015;52:943-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 209. | Ramzan F, D'Souza RF, Durainayagam BR, Milan AM, Markworth JF, Miranda-Soberanis V, Sequeira IR, Roy NC, Poppitt SD, Mitchell CJ, Cameron-Smith D. Circulatory miRNA biomarkers of metabolic syndrome. Acta Diabetol. 2020;57:203-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 210. | Rawal S, Munasinghe PE, Nagesh PT, Lew JKS, Jones GT, Williams MJA, Davis P, Bunton D, Galvin IF, Manning P, Lamberts RR, Katare R. Down-regulation of miR-15a/b accelerates fibrotic remodelling in the Type 2 diabetic human and mouse heart. Clin Sci (Lond). 2017;131:847-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 211. | Chen Y, Tian L, Wan S, Xie Y, Chen X, Ji X, Zhao Q, Wang C, Zhang K, Hock JM, Tian H, Yu X. MicroRNA-17-92 cluster regulates pancreatic beta-cell proliferation and adaptation. Mol Cell Endocrinol. 2016;437:213-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 212. | Jaeger A, Zollinger L, Saely CH, Muendlein A, Evangelakos I, Nasias D, Charizopoulou N, Schofield JD, Othman A, Soran H, Kardassis D, Drexel H, Eckardstein AV. Circulating microRNAs -192 and -194 are associated with the presence and incidence of diabetes mellitus. Sci Rep. 2018;8:14274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 213. | Neef T, Miller SD. Tolerogenic Nanoparticles to Treat Islet Autoimmunity. Curr Diab Rep. 2017;17:84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 214. | Gutiérrez-Vázquez C, Villarroya-Beltri C, Mittelbrunn M, Sánchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev. 2013;251:125-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 254] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 215. | Petzold C, Riewaldt J, Watts D, Sparwasser T, Schallenberg S, Kretschmer K. Foxp3(+) regulatory T cells in mouse models of type 1 diabetes. J Diabetes Res. 2013;2013:940710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 216. | Milanesi A, Lee JW, Li Z, Da Sacco S, Villani V, Cervantes V, Perin L, Yu JS. β-Cell regeneration mediated by human bone marrow mesenchymal stem cells. PLoS One. 2012;7:e42177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 217. | Yu B, Zhang X, Li X. Exosomes derived from mesenchymal stem cells. Int J Mol Sci. 2014;15:4142-4157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 624] [Cited by in RCA: 589] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 218. | Gomzikova MO, James V, Rizvanov AA. Therapeutic Application of Mesenchymal Stem Cells Derived Extracellular Vesicles for Immunomodulation. Front Immunol. 2019;10:2663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 219. | Pudlarz A, Szemraj J. Nanoparticles as Carriers of Proteins, Peptides and Other Therapeutic Molecules. Open Life Sci. 2018;13:285-298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 220. | Wang P, Liu Q, Zhao H, Bishop JO, Zhou G, Olson LK, Moore A. miR-216a-targeting theranostic nanoparticles promote proliferation of insulin-secreting cells in type 1 diabetes animal model. Sci Rep. 2020;10:5302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 221. | Ling H. Non-coding RNAs: Therapeutic Strategies and Delivery Systems. Adv Exp Med Biol. 2016;937:229-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 222. | Huang CK, Kafert-Kasting S, Thum T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ Res. 2020;126:663-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 156] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 223. | Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104-1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 325] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 224. | Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 221] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 225. | Macvanin M, Obradovic M, Zafirovic S, Stanimirovic J, Isenovic ER. The Role of miRNAs in Metabolic Diseases. Curr Med Chem. 2023;30:1922-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 226. | van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851-864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 426] [Cited by in RCA: 478] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 227. | Liew CG. Generation of insulin-producing cells from pluripotent stem cells: from the selection of cell sources to the optimization of protocols. Rev Diabet Stud. 2010;7:82-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 228. | Liu J, Ashton MP, Sumer H, O'Bryan MK, Brodnicki TC, Verma PJ. Generation of stable pluripotent stem cells from NOD mouse tail-tip fibroblasts. Diabetes. 2011;60:1393-1398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 229. | Yang CS, Li Z, Rana TM. microRNAs modulate iPS cell generation. RNA. 2011;17:1451-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 230. | Lorenzo IM, Fleischer A, Bachiller D. Generation of mouse and human induced pluripotent stem cells (iPSC) from primary somatic cells. Stem Cell Rev Rep. 2013;9:435-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 231. | Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 179] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 232. | Pfaff N, Fiedler J, Holzmann A, Schambach A, Moritz T, Cantz T, Thum T. miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep. 2011;12:1153-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 233. | Kolfschoten IG, Roggli E, Nesca V, Regazzi R. Role and therapeutic potential of microRNAs in diabetes. Diabetes Obes Metab. 2009;11 Suppl 4:118-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 234. | Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233-13241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 547] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 235. | Ørom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 278] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 236. | Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3005] [Cited by in RCA: 3070] [Article Influence: 153.5] [Reference Citation Analysis (0)] |
| 237. | Regazzi R. MicroRNAs as therapeutic targets for the treatment of diabetes mellitus and its complications. Expert Opin Ther Targets. 2018;22:153-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 238. | Mirra P, Raciti GA, Nigro C, Fiory F, D'Esposito V, Formisano P, Beguinot F, Miele C. Circulating miRNAs as intercellular messengers, potential biomarkers and therapeutic targets for Type 2 diabetes. Epigenomics. 2015;7:653-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 239. | Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1612] [Cited by in RCA: 1638] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 240. | Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1268] [Cited by in RCA: 1282] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 241. | Lima JF, Cerqueira L, Figueiredo C, Oliveira C, Azevedo NF. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018;15:338-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 242. | Distel E, Barrett TJ, Chung K, Girgis NM, Parathath S, Essau CC, Murphy AJ, Moore KJ, Fisher EA. miR33 inhibition overcomes deleterious effects of diabetes mellitus on atherosclerosis plaque regression in mice. Circ Res. 2014;115:759-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 243. | Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404-407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 550] [Cited by in RCA: 588] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 244. | Chakraborty C, Sharma AR, Sharma G, Lee SS. Therapeutic advances of miRNAs: A preclinical and clinical update. J Adv Res. 2021;28:127-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 281] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 245. | Zhou B, Li C, Qi W, Zhang Y, Zhang F, Wu JX, Hu YN, Wu DM, Liu Y, Yan TT, Jing Q, Liu MF, Zhai QW. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia. 2012;55:2032-2043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 175] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 246. | Parra P, Serra F, Palou A. Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS One. 2010;5:e13005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 247. | Joven J, Espinel E, Rull A, Aragonès G, Rodríguez-Gallego E, Camps J, Micol V, Herranz-López M, Menéndez JA, Borrás I, Segura-Carretero A, Alonso-Villaverde C, Beltrán-Debón R. Plant-derived polyphenols regulate expression of miRNA paralogs miR-103/107 and miR-122 and prevent diet-induced fatty liver disease in hyperlipidemic mice. Biochim Biophys Acta. 2012;1820:894-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 248. | Corrêa TA, Rogero MM. Polyphenols regulating microRNAs and inflammation biomarkers in obesity. Nutrition. 2019;59:150-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 249. | Ortega FJ, Cardona-Alvarado MI, Mercader JM, Moreno-Navarrete JM, Moreno M, Sabater M, Fuentes-Batllevell N, Ramírez-Chávez E, Ricart W, Molina-Torres J, Pérez-Luque EL, Fernández-Real JM. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J Nutr Biochem. 2015;26:1095-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 250. | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11979] [Cited by in RCA: 11020] [Article Influence: 847.7] [Reference Citation Analysis (0)] |
| 251. | Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3671] [Cited by in RCA: 3904] [Article Influence: 354.9] [Reference Citation Analysis (0)] |
| 252. | Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 623] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 253. | Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep. 2016;6:22312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 254. | Yoshino H, Yonemori M, Miyamoto K, Tatarano S, Kofuji S, Nohata N, Nakagawa M, Enokida H. microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget. 2017;8:20881-20894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 255. | Zhao Y, Teng H, Yao F, Yap S, Sun Y, Ma L. Challenges and Strategies in Ascribing Functions to Long Noncoding RNAs. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |