Published online May 15, 2020. doi: 10.4239/wjd.v11.i5.155
Peer-review started: December 11, 2019
First decision: January 13, 2020
Revised: February 10, 2020
Accepted: March 22, 2020
Article in press: March 22, 2020
Published online: May 15, 2020
Processing time: 135 Days and 19.9 Hours
The progressive aging of populations has resulted in an increased prevalence of chronic pathologies, especially of metabolic, neurodegenerative and movement disorders. In particular, type 2 diabetes (T2D), Alzheimer’s disease (AD) and Parkinson’s disease (PD) are among the most prevalent age-related, multifactorial pathologies that deserve particular attention, given their dramatic impact on patient quality of life, their economic and social burden as well the etiopathogenetic mechanisms, which may overlap in some cases. Indeed, the existence of common triggering factors reflects the contribution of mutual genetic, epigenetic and environmental features in the etiopathogenetic mechanisms underlying T2D and AD/PD. On this subject, this review will summarize the shared (epi)genomic features that characterize these complex pathologies. In particular, genetic variants and gene expression profiles associated with T2D and AD/PD will be discussed as possible contributors to determine the susceptibility and progression to these disorders. Moreover, potential shared epigenetic modifications and factors among T2D, AD and PD will also be illustrated. Overall, this review shows that findings from genomic studies still deserves further research to evaluate and identify genetic factors that directly contribute to the shared etiopathogenesis. Moreover, a common epigenetic background still needs to be investigated and characterized. The evidences discussed in this review underline the importance of integrating large-scale (epi)genomic data with additional molecular information and clinical and social background in order to finely dissect the complex etiopathogenic networks that build up the “disease interactome” characterizing T2D, AD and PD.
Core tip: Populations’ progressive aging raises important challenges to be faced, including the increased prevalence of metabolic, neurodegenerative and movement disorders, especially of type 2 diabetes, Alzheimer’s disease and Parkinson’s disease. These disorders are characterized by a multifactorial etiology, involving genetic and non-genetic factors, which may overlap. This review will discuss the shared (epi)genomic features, the role of mutually-associated genetic variants, common gene expression profiles and epigenetic background leading to development and progression of such disorders. Overall, this review highlights the importance of characterizing the “disease interactome” in order to establish adequate personalized and preventative healthcare approaches for the ageing populations.
- Citation: Caputo V, Termine A, Strafella C, Giardina E, Cascella R. Shared (epi)genomic background connecting neurodegenerative diseases and type 2 diabetes. World J Diabetes 2020; 11(5): 155-164
- URL: https://www.wjgnet.com/1948-9358/full/v11/i5/155.htm
- DOI: https://dx.doi.org/10.4239/wjd.v11.i5.155
The recent progress in medicine and the improvement of health conditions have contributed to the rise of life expectancy, on the one hand. On the other hand, the better healthcare conditions and the availability of several therapeutic approaches have run in parallel to the progressive aging of populations, which raised novel challenges to be faced by the healthcare systems and the scientific communities. In fact, the progressive aging population has resulted in the increased prevalence of chronic pathologies, especially of metabolic, neurodegenerative and movement disorders[1]. In particular, type 2 diabetes (T2D), Alzheimer’s disease (AD) and Parkinson’s disease (PD) are among the most prevalent chronic, age-related pathologies that deserve particular attention given their dramatic impact on patient quality of life, their economic and social burden as well the etiopathogenetic mechanisms, which may overlap in some cases.
In fact, T2D accounts for 90% of cases of diabetes mellitus, which affects 285 million people worldwide[2]. It is mainly caused by a combination of insulin resistance and relative insulin deficiency[3], which results in glucose dyshomeostasis and other concomitant conditions, including hypertension and dyslipidemia[4]. AD is characterized by progressive loss of memory and cognitive domains responsible for functional independence[5,6]. This pathology accounts for the 60%-80% of overall forms of dementia and represents the sixth cause of death in the world. It affects about 30-46 million people[5,7-9], with an increasing prevalence depending on age (ranging from 0.3%-0.5% at age 60 to 11%-15% at age 80)[10-12]. PD affects 0.3% of the worldwide population, with prevalence increasing by age; in fact, it is estimated to be 1% in people over 60 years of age and 3%-5% in individuals over 85[13,14]. The clinical features of PD include typical motor symptomatology (bradykinesia, resting tremor, postural instability, and gait difficulties) and non-motor symptoms (dysautonomia, sleep disturbances, mood, and cognitive disorders)[15,16].
T2D, AD and PD are all characterized by a multifactorial etiology, involving the interplay among genetic, epigenetic and environmental factors[17,18]. Interestingly, there are lines of evidence at the epidemiological, cognitive and neuropathological levels that seem to link T2D to AD and PD[19]. In particular, brain insulin resistance could represent the bridge linking metabolic disorders to neurodegenerative /movement pathological conditions[20]. Insulin is transported via the blood brain barrier to the central nervous system, where it regulates local blood and cerebrospinal fluid glucose levels. Nevertheless, it is thought that the principal activity in the brain may be related to the regulation of synaptic plasticity and cognitive functions[7,21]. Moreover, a little proportion of insulin may be produced in the brain, as well. Indeed, insulin levels detected in humans and rodents have been found to be lower than those in the systemic circulation. However, differences in the levels of insulin among AD brains and age-matched controls have not been established[7]. Insulin Receptors (IRs) are well distributed in the brain, especially in the cortex, hippocampus and hypothalamus, corroborating the importance of brain insulin signaling[7,21]. The “diabetic brain” may suffer of the hyperglycemia and insulin resistance arising from the decrease in insulin receptor expression or activity[7,21]. This alteration may lead to the activation of pathogenic processes, namely enhanced production of reactive oxygen species (ROS) and pro-inflammatory cytokines, that trigger inflammatory responses also in the brain, advanced glycation products and dysfunctions of autophagic functions. Moreover, insulin resistance is able to increase the production and secretion of beta amyloid (Aβ) and alter the molecular pathways involved in the phosphorylation of Tau protein: both Aβ and hyperphosphorilated-Tau are known to misfold, aggregate and accumulate leading to the loss of synapses and death of neurons, which are typical of neurodegeneration processes[7,22-28]. Indeed, the increased neuroinflammation represents a pathological feature shared by all of the three age-related pathologies[29]. Thus, the existence of common triggering factors reflects the contribution of mutual genetic and epigenetic features in the etiopathogenetic mechanisms underlying AD, PD and T2D. On this subject, this review will summarize the shared (epi)genomic features that characterize these complex pathologies.
Several studies have attempted to dissect the contributing genetic background(s) to determine the susceptibility to T2D, AD and PD. Concerning T2D, most of the identified genetic risk factors are mainly involved in the maintenance of β-cell homeostasis and in the modulation of insulin metabolism[2,30,31]. As previously mentioned, insulin resistance has been reported to likely influence brain functions and neuronal activity. Concerning the genetic susceptibility factors of AD and PD, several genome-wide association studies (commonly referred to as GWAS) have identified many genetic polymorphisms associated with the onset and progression of sporadic forms of AD and PD. Most of them have been found to be located within genes involved in dopamine metabolic process, apoptosis, autophagy-related pathways, Aβ cascade, Tau pathology, neuroinflammation, regulation of neuronal transmission, and survival[17,32-35]. The availability of GWAS and bioinformatic approaches has allowed for the identification of 927 single nucleotide polymorphisms (SNPs) associated with both T2D and AD in populations of European ancestry. Intriguingly, 395 of these SNPs have been reported to share the same risk allele between T2D and AD[36]. These SNPs are involved in immunity/inflammation-related pathways, cell-cell communication and neuronal plasticity, whose dysregulation may lead to increase in the neuroinflammation typically occurring in T2D and AD[7,37,38]. Polymorphisms within the IDE/HHEX region have also been investigated as combined susceptibility factors for T2D and AD (Table 1)[39-41]. Notably, IDE codes for the enzyme responsible for insulin clearance, although it is also able to degrade Aβ peptide in neurons and glia cells[7,39].
| Gene symbol | Gene name | Genomic location | SNP | Biological function | Potential associated diseases |
| IDE | Insulin degrading enzyme | 10q23.33 | rs6583817 | Insulin clearance | T2D/AD |
| IDE/HHEX | Insulin degrading enzyme/hematopoietically expressed homeobox | rs1544210 | Insulin clearance/transcriptional repression | ||
| TP53INP1 | Tumor protein P53 inducible nuclear protein 1 | 8q22.1 | rs896854 | Cell stress response, autophagy activation, cell cycle regulation | |
| TP53INP1/NDUFAF6 | Tumor protein P53 inducible nuclear protein 1/NADH:Ubiquinone oxidoreductase complex assembly factor 6 | rs6982393 | Cell stress response, autophagy activation, cell cycle regulation Mitochondrial function | ||
| rs4734295 | |||||
| NDUFAF6 | NADH:Ubiquinone oxidoreductase complex assembly factor 6 | rs7812465 | Mitochondrial function | ||
| TOMM40 | Translocase of outer mitochondrial membrane 40 | 19q13.32 | rs2075650 | ||
| BTBD16/PLEKHA1 | BTB domain containing 16/pleckstrin homology domain containing A1 | 10q26.13 | rs10510109 | Apoptosis regulation/plasma membrane function | |
| PLEKHA1 | Pleckstrin homology domain containing A1 | rs2421016 | Plasma membrane function | ||
| PVRL2 | Poliovirus receptor-like 2 | 19q13.32 | rs6859 | Cell junctions, inflammation | |
| APOC1 | Apolipoprotein C1 | rs111789331 | Lipid metabolism | ||
| rs12721046 | |||||
| rs12721051 | |||||
| rs4420638 | |||||
| rs56131196 | |||||
| rs66626994 | |||||
| DNM3 | Dynamin 3 | 1q24.3 | rs4504922 | Vesicle transport, phagocytosis | |
| rs7539972 | |||||
| ADCY5 | Adenylate cyclase 5 | 3q21.1 | rs2877709 | Chemokine signaling, insulin secretion | |
| CDC123 | Cell division cycle 123 | 10p14-p13 | rs11257655 | Cell cycle regulation | T2D/PD |
| CDKN2B | Cyclin dependent kinase inhibitor 2B | 9p21.3 | rs2383208 | ||
| rs10965250 | |||||
| rs10811661 | |||||
| KANSL1 | KAT8 regulatory NSL complex subunit 1 | 17q21.31 | rs17661428 | Transcriptional activation | T1D/PD |
| CXCR4 | C-X-C motif chemokine receptor 4 | 2q22.1 | rs2011946 | Inflammation, neuronal development | |
| MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 17q21.31 | rs2867316 | ||
| CRHR1 | Corticotropin releasing hormone receptor 1 | rs393152 | Hormonal signaling, stress and immune response |
A recent study performed on populations of European ancestry has described the association of 14 common SNPs with both T2D and AD; these are located in TP53INP1, NDUFAF6, TOMM40, BTBD16, PLEKHA1, PVRL2 and APOC1 genes[42,43] (Table 1). Interestingly, these genes encode proteins involved in the regulation of autophagy, apoptosis, response to oxidative stress, mitochondrial function and lipid metabolism, and their overall dysregulation can contribute to the etiopathogenetic pathways underlying T2D and AD[7,22]. Of note, Hao et al[34], 2015 and Wang et al[39], 2017 found that both disorders shared the same risk variant in SNPs (rs10510109 and rs2421016) located in BTBD16 and PLEKHA1 genes (Table 1). This is of particular interest, as different SNPs within PLEKHA1 have been associated with age-related macular degeneration (an ocular neurodegenerative complex disease)[44-46] and they map on the 10q26.13 locus, which also contains another age-related macular degeneration-associated gene (ARMS2/HTRA1)[47,48]. Given these data, the genetic architecture of the 10q26.13 region may be investigated for its potential contribution to neurodegeneration and could be addressed as a shared susceptibility locus for T2D and AD. Moreover, the presence of shared genetic polymorphisms associated with both diseases may also be exploited to predict the risk of developing AD in individuals already suffering from T2D.
Less information is available concerning the genetic overlap between T2D and PD. The possible link between T2D-associated genetic loci and AD/PD has been investigated in a study involving 500 PD and 400 AD patients of Asian ancestry. The authors reported four SNPs located in CDC123 and CDKN2B genes mutually associated with T2D and PD. However, this association was not confirmed after correction for multiple testing[49] (Table 1). CDC123 and CDKN2B exert a role in cell cycle regulation, and their dysfunction leads to alterations in cell homeostasis, suggesting that the genetic association with T2D and PD should be further investigated in larger cohorts and different populations. Furthermore, four different genes, namely KANSL1, CXCR4, MAP3K14 and CRHR1, were found to be shared between PD and type 1 diabetes in a study aiming to evaluate the common risk factors between PD and autoimmune disorders[50] (Table 1). Intriguingly, CXCR4 and MAP3K14 are involved in the regulation of neuronal inflammatory responses. In particular, CXCR4 is involved in microglia recruitment, neuronal guidance and neurodevelopmental processes[51], whereas MAP3K14 mediates NFκB signaling (involved in immunological cytotoxicity) in brain neurons[52]. Moreover, the CXCR4 protein has been found to be overexpressed in a rodent model of diabetic neuropathic pain[53]. KANSL1 has been found to be associated with AD, thus suggesting that the encoded protein may take part in neuronal development. Indeed, KANSL1, as part of the NLS1 complex which regulates histone acetylation, is mainly involved in the epigenetic regulation of chromatin[54]. Interestingly, mutations within KANSL1 are able to cause intellectual disability and developmental delay[54,55]. CRHR1 is known to be involved in the activation of hypothalamic-pituitary-adrenal axis, leading to secretion of cortisol that, in turn, causes insulin resistance. Notably, the chronic stress activated by the adrenal secretion of cortisol represents a risk factor for AD onset and progression[56]. Given these lines of evidence, the association between this set of genes and their potential involvement in etiopathogenetic pathways leading to T2D, AD and PD should be further elucidated.
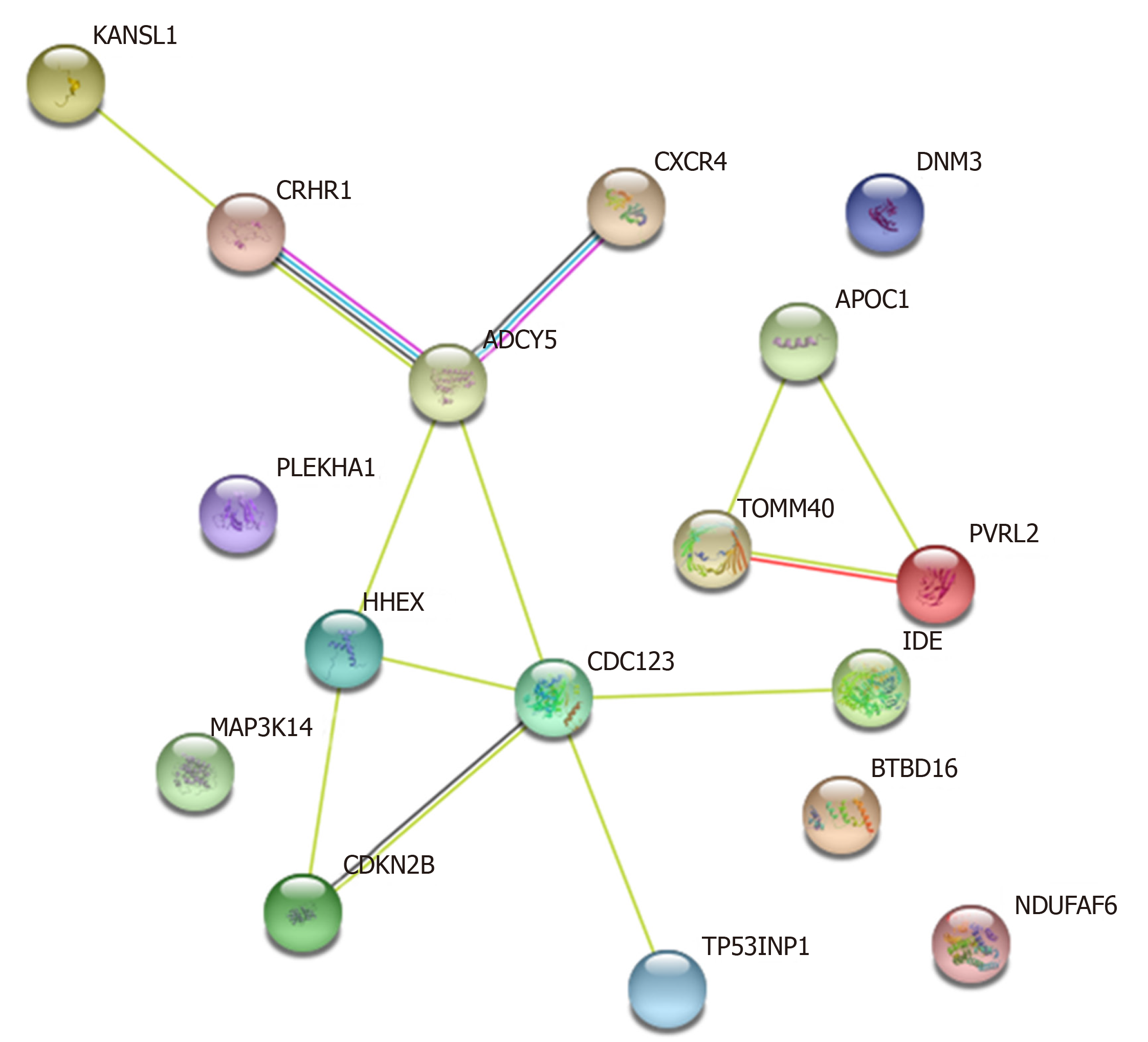
In addition to the identification of shared genetic variants, the investigation of common gene expression profiles may facilitate the discovery and exploration of molecular pathways that are deregulated in T2D, AD and PD. On this subject, Rahman et al[57], 2018 reported intriguing insights, exploiting human gene expression datasets. Among the significant Gene Ontologies (known as GOs) and Kyoto Encyclopedia Genes and Genomes (known as KEGG) pathways shared by T2D and AD, pathways involved in glycosphingolipid biosynthesis, immune/inflammatory response, regulation of neurotransmitter transports, synaptic vesicle formation, lipid metabolism and apoptosis have been identified. T2D and PD share genes involved in immune-related networks, cell adhesion, mitochondrial activity, connective tissue/extracellular matrix organization, and synaptic maturation. Indeed, neuroinflammation may represent a common hallmark among T2D, AD and PD, given that most of the shared genes are implicated in the regulation of inflammatory networks. Interestingly, Santiago et al[58], 2013 found that APP mRNA was overexpressed in the whole blood of both T2D and PD patients. Therefore, the knowledge of shared genetic factors and gene expression profiles may help to further dissect the molecular network characterizing and linking T2D, AD and PD (Figure 1).
The human genome is able to dynamically interact with the environment through epigenetic modifications, which altogether create the complex machinery designated to regulate lifetime and aging processes. In fact, epigenetics modulate gene expression without altering the DNA sequence. This is possible by means of different kinds of epigenetic modifications, including DNA methylation and histone modifications (which might affect gene transcription), and noncoding (nc)RNAs (which might change gene expression at the post-transcriptional level)[59]. Given the crucial role of epigenetics in the modulation of gene expression, its alteration can contribute to pathogenesis and progression of several age-related diseases, including metabolic, neurodegenerative and movement disorders[17,60]. The existence of a shared epigenetic background among T2D and neurodegenerative diseases deserves to be investigated. As a matter of fact, the gene expression signatures shared among T2D and AD/PD[57,58] may also be related to the presence of common epigenetic alterations[61]. On this subject, there are intriguing hypotheses that could be evaluated. For instance, the analysis of long-range chromatin contacts among regulatory regions and their target genes will provide insights into how epigenetic background(s) may modify chromatin conformation[62] and thus gene expression profiles in the context of T2D, AD and PD. Moreover, an interaction between micro (mi)RNA-661 and BACE1 mRNA was found to cause a reduced expression of the resultant protein in pancreatic islets and contribute, thereby, to the development of T2D[63]. Of note, BACE1 is involved not only in the regulation of insulin biogenesis but also in the formation of Aβ so that it could be also investigated in the etiopathogenesis of AD[64].
Furthermore, sirtuins are a family of histone deacetylases, playing critical roles in the physiology of metabolism, central nervous system, and immune system. In fact, these epigenetic modifiers are involved in a variety of molecular pathways underlying different complex diseases (cancer, diabetes, and neurodegenerative disorders)[65]. Given their role, sirtuins may be addressed as potential therapeutic targets able to counteract the progression of T2D, AD and PD through their epigenetic activity[66].
The study of DNA methylation affecting mitochondrial genes could unveil interesting insights into the pathogenesis of T2D, AD and PD. In fact, alteration of DNA methylation status has been supposed to be responsible for the reduction of complex I and IV subunits in AD and PD human brain samples[67]. Moreover, it has been demonstrated that the alteration of miR-181a/b levels impacts mitochondrial biogenesis and turnover in the brain, through the modulation of autophagy and mitophagy-related pathways[68]. These miRNAs could be, therefore, investigated for their potential role in the common pathogenetic processes leading to T2D and AD/PD. Furthermore, the study of other miRNAs and ncRNAs related to these disorders could be helpful for designing innovative class of drugs (epidrugs).
A growing body of evidence suggests the existence of multilevel networks of pathogenetic pathways which mutually contribute to the onset and progression of metabolic, neurodegenerative and movement disorders. However, few shared genetic contributors have been well characterized and a common epigenetic landscape needs to be explored. Of note, in T2D, an impairment of glucose metabolism in brain generates oxidative stress, leading to the alteration of autophagy-related pathways, mitochondrial dysfunction, increased neuronal apoptosis and, eventually, depletion of synapsis[7]. Overall, these alterations contribute to the formation of amyloid plaques and neurofibrillary tangles in AD and to the deterioration of dopaminergic neurons in different brain regions in PD[1]. As mentioned, despite a plethora of data highlighting a possible overlapping of disease mechanisms involved in T2D, AD and PD, the critical molecular and genetic features remain to be clarified. The genetic polymorphisms (Table 1) shared with T2D, AD and PD are located within genes involved not only in brain insulin signaling but also in neuroinflammation-related pathways[34-52]. This evidence is also corroborated by the expression data obtained by the investigation of human patients and animal models presenting these pathologies[57].
Understanding the contribution of genetics, epigenetics and environment in determining the susceptibility, onset and progression of T2D, AD and PD will be crucial to achieve a deeper knowledge of metabolic, neurodegenerative and movement disorders. On this subject, the enhancement of social and cognitive activities in the high-income countries seems to strengthen the resilience against neurodegeneration, leading to a stable or reduced incidence of dementia in these regions[69,70]. On the other hand, T2D prevalence is also rising in the more developed areas[71]. Given this data and considering that T2D is regarded overall as a risk factor for dementia[72], the contribution of T2D to the development of neurodegeneration needs to be monitored. Moreover, these lines of evidence encourage the further exploration of gene-environment interactions in order to understand the similarities and the differences in the etiopathogenesis underlying AD, PD and T2D. Indeed, more comprehensive and higher resolution (epi)genomic studies should be implemented in order to collect information on genome architecture, DNA methylation, histone modifications, ncRNAs and three-dimensional genome organization. These large-scale data should be exploited to integrate genomic, epigenomic, transcriptomic, metabolomic and proteomic information with the clinical phenotype and draw the network of interactions which build up a “disease-interactome”.
Indeed, the fine knowledge of the disease interactome could highlight the molecular relationships existing among T2D, AD, PD which, thereby, could be exploited to treat these conditions through a network medicine approach, able to integrate all these interactions to understand the molecular and cellular perturbations underlying diseases, providing insights and targets for the accurate diagnosis and treatment[73]. By this way, the patient could benefit from a healthcare approach based on a multilevel characterization of his condition, derived not only by clinical and molecular testing but also by his environmental and social backgrounds.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gillberg L, Qi X S-Editor: Gong ZM L-Editor: A E-Editor: Ma YJ
| 1. | Fiory F, Perruolo G, Cimmino I, Cabaro S, Pignalosa FC, Miele C, Beguinot F, Formisano P, Oriente F. The Relevance of Insulin Action in the Dopaminergic System. Front Neurosci. 2019;13:868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 2. | Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus--present and future perspectives. Nat Rev Endocrinol. 2011;8:228-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1333] [Cited by in RCA: 1445] [Article Influence: 103.2] [Reference Citation Analysis (0)] |
| 3. | Bullard KM, Cowie CC, Lessem SE, Saydah SH, Menke A, Geiss LS, Orchard TJ, Rolka DB, Imperatore G. Prevalence of Diagnosed Diabetes in Adults by Diabetes Type - United States, 2016. MMWR Morb Mortal Wkly Rep. 2018;67:359-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 314] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 4. | Jeong SU, Kang DG, Lee DH, Lee KW, Lim DM, Kim BJ, Park KY, Chin HJ, Koh G. Clinical Characteristics of Type 2 Diabetes Patients according to Family History of Diabetes. Korean Diabetes J. 2010;34:222-228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Erkkinen MG, Kim MO, Geschwind MD. Clinical Neurology and Epidemiology of the Major Neurodegenerative Diseases. Cold Spring Harb Perspect Biol. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 679] [Cited by in RCA: 687] [Article Influence: 98.1] [Reference Citation Analysis (0)] |
| 6. | Pierce AL, Bullain SS, Kawas CH. Late-Onset Alzheimer Disease. Neurol Clin. 2017;35:283-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 7. | Chatterjee S, Mudher A. Alzheimer's Disease and Type 2 Diabetes: A Critical Assessment of the Shared Pathological Traits. Front Neurosci. 2018;12:383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 177] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 8. | Lanoiselée HM, Nicolas G, Wallon D, Rovelet-Lecrux A, Lacour M, Rousseau S, Richard AC, Pasquier F, Rollin-Sillaire A, Martinaud O, Quillard-Muraine M, de la Sayette V, Boutoleau-Bretonniere C, Etcharry-Bouyx F, Chauviré V, Sarazin M, le Ber I, Epelbaum S, Jonveaux T, Rouaud O, Ceccaldi M, Félician O, Godefroy O, Formaglio M, Croisile B, Auriacombe S, Chamard L, Vincent JL, Sauvée M, Marelli-Tosi C, Gabelle A, Ozsancak C, Pariente J, Paquet C, Hannequin D, Campion D; collaborators of the CNR-MAJ project. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017;14:e1002270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 410] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 9. | Condello C, Stöehr J. Aβ propagation and strains: Implications for the phenotypic diversity in Alzheimer's disease. Neurobiol Dis. 2018;109:191-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 10. | Naj AC, Schellenberg GD; Alzheimer's Disease Genetics Consortium (ADGC). Genomic variants, genes, and pathways of Alzheimer's disease: An overview. Am J Med Genet B Neuropsychiatr Genet. 2017;174:5-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 140] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 11. | Cimler R, Maresova P, Kuhnova J, Kuca K. Predictions of Alzheimer's disease treatment and care costs in European countries. PLoS One. 2019;14:e0210958. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Bloom BS, de Pouvourville N, Straus WL. Cost of illness of Alzheimer's disease: how useful are current estimates? Gerontologist. 2003;43:158-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Tysnes OB, Storstein A. Epidemiology of Parkinson's disease. J Neural Transm (Vienna). 2017;124:901-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1008] [Cited by in RCA: 1489] [Article Influence: 186.1] [Reference Citation Analysis (0)] |
| 14. | Reeve A, Simcox E, Turnbull D. Ageing and Parkinson's disease: why is advancing age the biggest risk factor? Ageing Res Rev. 2014;14:19-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 607] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 15. | Verma M, Steer EK, Chu CT. ERKed by LRRK2: a cell biological perspective on hereditary and sporadic Parkinson's disease. Biochim Biophys Acta. 2014;1842:1273-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Shapira AL, Handzel R, Korczyn AD. The Lived Experience of Parkinson's Disease: A Content Analysis of Parkinson's Patients' Blogs. Isr Med Assoc J. 2017;19:685-690. [PubMed] |
| 17. | Strafella C, Caputo V, Galota MR, Zampatti S, Marella G, Mauriello S, Cascella R, Giardina E. Application of Precision Medicine in Neurodegenerative Diseases. Front Neurol. 2018;9:701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 18. | Kumar B, Sharma D. Recent Patent Advances for Neurodegenerative Disorders and its Treatment. Recent Pat Drug Deliv Formul. 2017;11:158-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | Bharadwaj P, Wijesekara N, Liyanapathirana M, Newsholme P, Ittner L, Fraser P, Verdile G. The Link between Type 2 Diabetes and Neurodegeneration: Roles for Amyloid-β, Amylin, and Tau Proteins. J Alzheimers Dis. 2017;59:421-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 148] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 20. | Ahmed RM, Devenney EM, Irish M, Ittner A, Naismith S, Ittner LM, Rohrer JD, Halliday GM, Eisen A, Hodges JR, Kiernan MC. Neuronal network disintegration: common pathways linking neurodegenerative diseases. J Neurol Neurosurg Psychiatry. 2016;87:1234-1241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 99] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 21. | Grillo CA, Woodruff JL, Macht VA, Reagan LP. Insulin resistance and hippocampal dysfunction: Disentangling peripheral and brain causes from consequences. Exp Neurol. 2019;318:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 22. | Pagano G, Polychronis S, Wilson H, Giordano B, Ferrara N, Niccolini F, Politis M. Diabetes mellitus and Parkinson disease. Neurology. 2018;90:e1654-e1662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 178] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 23. | Biosa A, Outeiro TF, Bubacco L, Bisaglia M. Diabetes Mellitus as a Risk Factor for Parkinson's Disease: a Molecular Point of View. Mol Neurobiol. 2018;55:8754-8763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 24. | Athauda D, Foltynie T. Insulin resistance and Parkinson's disease: A new target for disease modification? Prog Neurobiol. 2016;145-146:98-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 235] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 25. | Ashraghi MR, Pagano G, Polychronis S, Niccolini F, Politis M. Parkinson's Disease, Diabetes and Cognitive Impairment. Recent Pat Endocr Metab Immune Drug Discov. 2016;10:11-21. [PubMed] |
| 26. | Umeno A, Biju V, Yoshida Y. In vivo ROS production and use of oxidative stress-derived biomarkers to detect the onset of diseases such as Alzheimer's disease, Parkinson's disease, and diabetes. Free Radic Res. 2017;51:413-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 186] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 27. | De Pablo-Fernandez E, Sierra-Hidalgo F, Benito-León J, Bermejo-Pareja F. Association between Parkinson's disease and diabetes: Data from NEDICES study. Acta Neurol Scand. 2017;136:732-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 28. | de Matos AM, de Macedo MP, Rauter AP. Bridging Type 2 Diabetes and Alzheimer's Disease: Assembling the Puzzle Pieces in the Quest for the Molecules With Therapeutic and Preventive Potential. Med Res Rev. 2018;38:261-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 29. | McKenzie JA, Spielman LJ, Pointer CB, Lowry JR, Bajwa E, Lee CW, Klegeris A. Neuroinflammation as a Common Mechanism Associated with the Modifiable Risk Factors for Alzheimer's and Parkinson's Diseases. Curr Aging Sci. 2017;10:158-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 30. | Wang Q, Li WX, Dai SX, Guo YC, Han FF, Zheng JJ, Li GH, Huang JF. Meta-Analysis of Parkinson's Disease and Alzheimer's Disease Revealed Commonly Impaired Pathways and Dysregulation of NRF2-Dependent Genes. J Alzheimers Dis. 2017;56:1525-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 31. | Nolan CJ, Damm P, Prentki M. Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet. 2011;378:169-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 626] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 32. | Giri M, Zhang M, Lü Y. Genes associated with Alzheimer's disease: an overview and current status. Clin Interv Aging. 2016;11:665-681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 220] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 33. | Green H, Tsitsi P, Markaki I, Aarsland D, Svenningsson P. Novel Treatment Opportunities Against Cognitive Impairment in Parkinson's Disease with an Emphasis on Diabetes-Related Pathways. CNS Drugs. 2019;33:143-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | International Parkinson Disease Genomics Consortium, Nalls MA Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, Simón-Sánchez J, Schulte C, Lesage S, Sveinbjörnsdóttir S, Stefánsson K, Martinez M, Hardy J, Heutink P, Brice A, Gasser T, Singleton AB, Wood NW. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641-649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 764] [Cited by in RCA: 783] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 35. | Nicolas G, Charbonnier C, Campion D. From Common to Rare Variants: The Genetic Component of Alzheimer Disease. Hum Hered. 2016;81:129-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Hao K, Di Narzo AF, Ho L, Luo W, Li S, Chen R, Li T, Dubner L, Pasinetti GM. Shared genetic etiology underlying Alzheimer's disease and type 2 diabetes. Mol Aspects Med. 2015;43-44:66-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 37. | Bae CS, Song J. The Role of Glucagon-Like Peptide 1 (GLP1) in Type 3 Diabetes: GLP-1 Controls Insulin Resistance, Neuroinflammation and Neurogenesis in the Brain. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 38. | Pugazhenthi S, Qin L, Reddy PH. Common neurodegenerative pathways in obesity, diabetes, and Alzheimer's disease. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 481] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 39. | Xu WL, Pedersen NL, Keller L, Kalpouzos G, Wang HX, Graff C, Winblad B, Bäckman L, Fratiglioni L. HHEX_23 AA Genotype Exacerbates Effect of Diabetes on Dementia and Alzheimer Disease: A Population-Based Longitudinal Study. PLoS Med. 2015;12:e1001853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | McFall GP, Wiebe SA, Vergote D, Westaway D, Jhamandas J, Dixon RA. IDE (rs6583817) polymorphism and type 2 diabetes differentially modify executive function in older adults. Neurobiol Aging. 2013;34:2208-2216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Carrasquillo MM, Belbin O, Zou F, Allen M, Ertekin-Taner N, Ansari M, Wilcox SL, Kashino MR, Ma L, Younkin LH, Younkin SG, Younkin CS, Dincman TA, Howard ME, Howell CC, Stanton CM, Watson CM, Crump M, Vitart V, Hayward C, Hastie ND, Rudan I, Campbell H, Polasek O, Brown K, Passmore P, Craig D, McGuinness B, Todd S, Kehoe PG, Mann DM, Smith AD, Beaumont H, Warden D, Holmes C, Heun R, Kölsch H, Kalsheker N, Pankratz VS, Dickson DW, Graff-Radford NR, Petersen RC, Wright AF, Younkin SG, Morgan K. Concordant association of insulin degrading enzyme gene (IDE) variants with IDE mRNA, Abeta, and Alzheimer's disease. PLoS One. 2010;5:e8764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Gao L, Cui Z, Shen L, Ji HF. Shared Genetic Etiology between Type 2 Diabetes and Alzheimer's Disease Identified by Bioinformatics Analysis. J Alzheimers Dis. 2016;50:13-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 43. | Wang XF, Lin X, Li DY, Zhou R, Greenbaum J, Chen YC, Zeng CP, Peng LP, Wu KH, Ao ZX, Lu JM, Guo YF, Shen J, Deng HW. Linking Alzheimer's disease and type 2 diabetes: Novel shared susceptibility genes detected by cFDR approach. J Neurol Sci. 2017;380:262-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 44. | SanGiovanni JP, SanGiovanni PM, Sapieha P, De Guire V. miRNAs, single nucleotide polymorphisms (SNPs) and age-related macular degeneration (AMD). Clin Chem Lab Med. 2017;55:763-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | Wang G. Chromosome 10q26 locus and age-related macular degeneration: a progress update. Exp Eye Res. 2014;119:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Ricci F, Zampatti S, D'Abbruzzi F, Missiroli F, Martone C, Lepre T, Pietrangeli I, Sinibaldi C, Peconi C, Novelli G, Giardina E. Typing of ARMS2 and CFH in age-related macular degeneration: case-control study and assessment of frequency in the Italian population. Arch Ophthalmol. 2009;127:1368-1372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Cascella R, Strafella C, Longo G, Ragazzo M, Manzo L, De Felici C, Errichiello V, Caputo V, Viola F, Eandi CM, Staurenghi G, Cusumano A, Mauriello S, Marsella LT, Ciccacci C, Borgiani P, Sangiuolo F, Novelli G, Ricci F, Giardina E. Uncovering genetic and non-genetic biomarkers specific for exudative age-related macular degeneration: significant association of twelve variants. Oncotarget. 2018;9:7812-7821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 48. | Cascella R, Strafella C, Caputo V, Errichiello V, Zampatti S, Milano F, Potenza S, Mauriello S, Novelli G, Ricci F, Cusumano A, Giardina E. Towards the application of precision medicine in Age-Related Macular Degeneration. Prog Retin Eye Res. 2018;63:132-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 49. | Chung SJ, Kim MJ, Kim J, Ryu HS, Kim YJ, Kim SY, Lee JH. Association of type 2 diabetes GWAS loci and the risk of Parkinson's and Alzheimer's diseases. Parkinsonism Relat Disord. 2015;21:1435-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 50. | Witoelar A, Jansen IE, Wang Y, Desikan RS, Gibbs JR, Blauwendraat C, Thompson WK, Hernandez DG, Djurovic S, Schork AJ, Bettella F, Ellinghaus D, Franke A, Lie BA, McEvoy LK, Karlsen TH, Lesage S, Morris HR, Brice A, Wood NW, Heutink P, Hardy J, Singleton AB, Dale AM, Gasser T, Andreassen OA, Sharma M; International Parkinson’s Disease Genomics Consortium (IPDGC), North American Brain Expression Consortium (NABEC), and United Kingdom Brain Expression Consortium (UKBEC) Investigators. Genome-wide Pleiotropy Between Parkinson Disease and Autoimmune Diseases. JAMA Neurol. 2017;74:780-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 257] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 51. | Bonham LW, Karch CM, Fan CC, Tan C, Geier EG, Wang Y, Wen N, Broce IJ, Li Y, Barkovich MJ, Ferrari R, Hardy J, Momeni P, Höglinger G, Müller U, Hess CP, Sugrue LP, Dillon WP, Schellenberg GD, Miller BL, Andreassen OA, Dale AM, Barkovich AJ, Yokoyama JS, Desikan RS; International FTD-Genomics Consortium (IFGC); International Parkinson’s Disease Genetics Consortium (IPDGC); International Genomics of Alzheimer’s Project (IGAP). CXCR4 involvement in neurodegenerative diseases. Transl Psychiatry. 2018;8:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 52. | Mao X, Phanavanh B, Hamdan H, Moerman-Herzog AM, Barger SW. NFκB-inducing kinase inhibits NFκB activity specifically in neurons of the CNS. J Neurochem. 2016;137:154-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 53. | Zhu D, Fan T, Huo X, Cui J, Cheung CW, Xia Z. Progressive Increase of Inflammatory CXCR4 and TNF-Alpha in the Dorsal Root Ganglia and Spinal Cord Maintains Peripheral and Central Sensitization to Diabetic Neuropathic Pain in Rats. Mediators Inflamm. 2019;2019:4856156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 54. | Koolen DA, Kramer JM, Neveling K, Nillesen WM, Moore-Barton HL, Elmslie FV, Toutain A, Amiel J, Malan V, Tsai AC, Cheung SW, Gilissen C, Verwiel ET, Martens S, Feuth T, Bongers EM, de Vries P, Scheffer H, Vissers LE, de Brouwer AP, Brunner HG, Veltman JA, Schenck A, Yntema HG, de Vries BB. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat Genet. 2012;44:639-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 173] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 55. | Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, Kunkle BW, Wang LS, Bis JC, Bellenguez C, Harold D, Lunetta KL, Destefano AL, Grenier-Boley B, Sims R, Beecham GW, Smith AV, Chouraki V, Hamilton-Nelson KL, Ikram MA, Fievet N, Denning N, Martin ER, Schmidt H, Kamatani Y, Dunstan ML, Valladares O, Laza AR, Zelenika D, Ramirez A, Foroud TM, Choi SH, Boland A, Becker T, Kukull WA, van der Lee SJ, Pasquier F, Cruchaga C, Beekly D, Fitzpatrick AL, Hanon O, Gill M, Barber R, Gudnason V, Campion D, Love S, Bennett DA, Amin N, Berr C, Tsolaki M, Buxbaum JD, Lopez OL. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108-117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 221] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 56. | Gragnoli C. Hypothesis of the neuroendocrine cortisol pathway gene role in the comorbidity of depression, type 2 diabetes, and metabolic syndrome. Appl Clin Genet. 2014;7:43-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 57. | Rahman MH, Peng S, Chen C, Lio’ P, Moni MA, Deramecourt V, Fox NC, Cantwell LB, Tárraga L, Dufouil C, Hardy J, Crane PK, Eiriksdottir G, Hannequin D, Clarke R, Evans D, Mosley TH, Letenneur L, Brayne C, Maier W, De Jager P, Emilsson V, Dartigues JF, Hampel H, Kamboh MI, de Bruijn RF, Tzourio C, Pastor P, Larson EB, Rotter JI, O'Donovan MC, Montine TJ, Nalls MA, Mead S, Reiman EM, Jonsson PV, Holmes C, St George-Hyslop PH, Boada M, Passmore P, Wendland JR, Schmidt R, Morgan K, Winslow AR, Powell JF, Carasquillo M, Younkin SG, Jakobsdóttir J, Kauwe JS, Wilhelmsen KC, Rujescu D, Nöthen MM, Hofman A, Jones L; IGAP Consortium, Haines JL, Psaty BM, Van Broeckhoven C, Holmans P, Launer LJ, Mayeux R, Lathrop M, Goate AM, Escott-Price V, Seshadri S, Pericak-Vance MA, Amouyel P, Williams J, van Duijn CM, Schellenberg GD, Farrer LA. Genetic effect of type 2 Diabetes to the progression of Neurological Diseases. bioRxiv. 2018;480400. [DOI] [Full Text] |
| 58. | Santiago JA, Potashkin JA. Integrative network analysis unveils convergent molecular pathways in Parkinson's disease and diabetes. PLoS One. 2013;8:e83940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 59. | Silberman DM. Metabolism, neurodegeneration and epigenetics: Emerging role of Sirtuins. Neural Regen Res. 2018;13:417-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 60. | Cheng Z, Zheng L, Almeida FA. Epigenetic reprogramming in metabolic disorders: nutritional factors and beyond. J Nutr Biochem. 2018;54:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 61. | Tzika E, Dreker T, Imhof A. Epigenetics and Metabolism in Health and Disease. Front Genet. 2018;9:361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 62. | Umlauf D, Mourad R. The 3D genome: From fundamental principles to disease and cancer. Semin Cell Dev Biol. 2019;90:128-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 63. | Hoffmeister A, Tuennemann J, Sommerer I, Mössner J, Rittger A, Schleinitz D, Kratzsch J, Rosendahl J, Klöting N, Stahl T, Rossner S, Paroni F, Maedler K, Kovacs P, Blüher M. Genetic and biochemical evidence for a functional role of BACE1 in the regulation of insulin mRNA expression. Obesity (Silver Spring). 2013;21:E626-E633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 64. | Koelsch G. BACE1 Function and Inhibition: Implications of Intervention in the Amyloid Pathway of Alzheimer's Disease Pathology. Molecules. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 65. | Poulose N, Raju R. Sirtuin regulation in aging and injury. Biochim Biophys Acta. 2015;1852:2442-2455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 189] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 66. | Foolad F, Khodagholi F, Javan M. Sirtuins in Multiple Sclerosis: The crossroad of neurodegeneration, autoimmunity and metabolism. Mult Scler Relat Disord. 2019;34:47-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 67. | Blanch M, Mosquera JL, Ansoleaga B, Ferrer I, Barrachina M. Altered Mitochondrial DNA Methylation Pattern in Alzheimer Disease-Related Pathology and in Parkinson Disease. Am J Pathol. 2016;186:385-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 145] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 68. | Indrieri A, Carrella S, Romano A, Spaziano A, Marrocco E, Fernandez-Vizarra E, Barbato S, Pizzo M, Ezhova Y, Golia FM, Ciampi L, Tammaro R, Henao-Mejia J, Williams A, Flavell RA, De Leonibus E, Zeviani M, Surace EM, Banfi S, Franco B. miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol Med. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 69. | Perneczky R. Dementia prevention and reserve against neurodegenerative disease. Dialogues Clin Neurosci. 2019;21:53-60. [PubMed] |
| 70. | Roehr S, Pabst A, Luck T, Riedel-Heller SG. Secular trends in the incidence of dementia in high-income countries: a protocol of a systematic review and a planned meta-analysis. BMJ Open. 2017;7:e013630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 71. | Sattar N. Advances in the clinical management of type 2 diabetes: a brief history of the past 15 years and challenges for the future. BMC Med. 2019;17:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 72. | Moran C, Beare R, Wang W, Callisaya M, Srikanth V; Alzheimer's Disease Neuroimaging Initiative (ADNI). Type 2 diabetes mellitus, brain atrophy, and cognitive decline. Neurology. 2019;92:e823-e830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 73. | Barabási AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3846] [Cited by in RCA: 3035] [Article Influence: 216.8] [Reference Citation Analysis (0)] |