Published online Jun 15, 2025. doi: 10.4251/wjgo.v17.i6.105140
Revised: April 1, 2025
Accepted: April 22, 2025
Published online: June 15, 2025
Processing time: 152 Days and 3.5 Hours
Hepatobiliary carcinoma is a frequently occurring and highly invasive cancer within the digestive tract, known for its rapid progression. Due to its difficult diagnosis and treatment in clinical practice, hepatobiliary carcinoma is a serious threat to human life and health. In recent years, the incidence of hepatobiliary carcinoma has gradually increased. N6-methyladenosine (m6A) modification, as a reversible post-transcriptional modification of the adenosine N6 site, is one of the most important RNA modifications in eukaryotes. Emerging research indicates that m6A affects the biological process of cells through the regulation of gene expression. m6A modification also plays a key role in the occurrence and deve
Core Tip: N6-methyladenine (m6A) modifications is one of the most common RNA modifications in eukaryotes, and it has been reported to affect biological process of cells via regulation of gene expression, and play a crucial role in occurrence and development in various cancers by regulating RNA stability, decay, spicing and transport. This article summarized the role and mechanism of m6A modification in hepatobiliary carcinoma, and discussed its potential clinical application in an attempt to provide theoretical evidences for the individualized treatment of hepatobiliary carcinoma.
- Citation: Jia C, Lang QF, Yin ZJ, Sun J, Meng QH, Pei TM. Role, mechanism, and application of N6-methyladenosine in hepatobiliary carcinoma. World J Gastrointest Oncol 2025; 17(6): 105140
- URL: https://www.wjgnet.com/1948-5204/full/v17/i6/105140.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i6.105140
Hepatobiliary carcinoma is a common malignant disease of the digestive system, and mainly includes cholangiocarcinoma (CCA) and gallbladder cancer. Its onset is hidden and progress is rapid, which is a serious threat to human life and health[1]. Since early hepatobiliary carcinoma has no significant clinical symptoms, most patients are diagnosed at an advanced stage and have a high recurrence rate after radical surgery. The 5-year overall survival rate is < 10%[1-5].
RNA modification is important in epigenetics and is closely related to the occurrence and development of cancer. As a reversible post-transcriptional modification, N6-methyladenosine (m6A) is the most common and important modification of eukaryotic cells[6,7]. m6A modification plays a significant role in many biological processes such as cell proliferation, differentiation and apoptosis[8,9]. Emerging research indicates that m6A modification plays a crucial role in the initiation and development of tumors by regulating RNA stability, decay, splicing and transport[7-12].
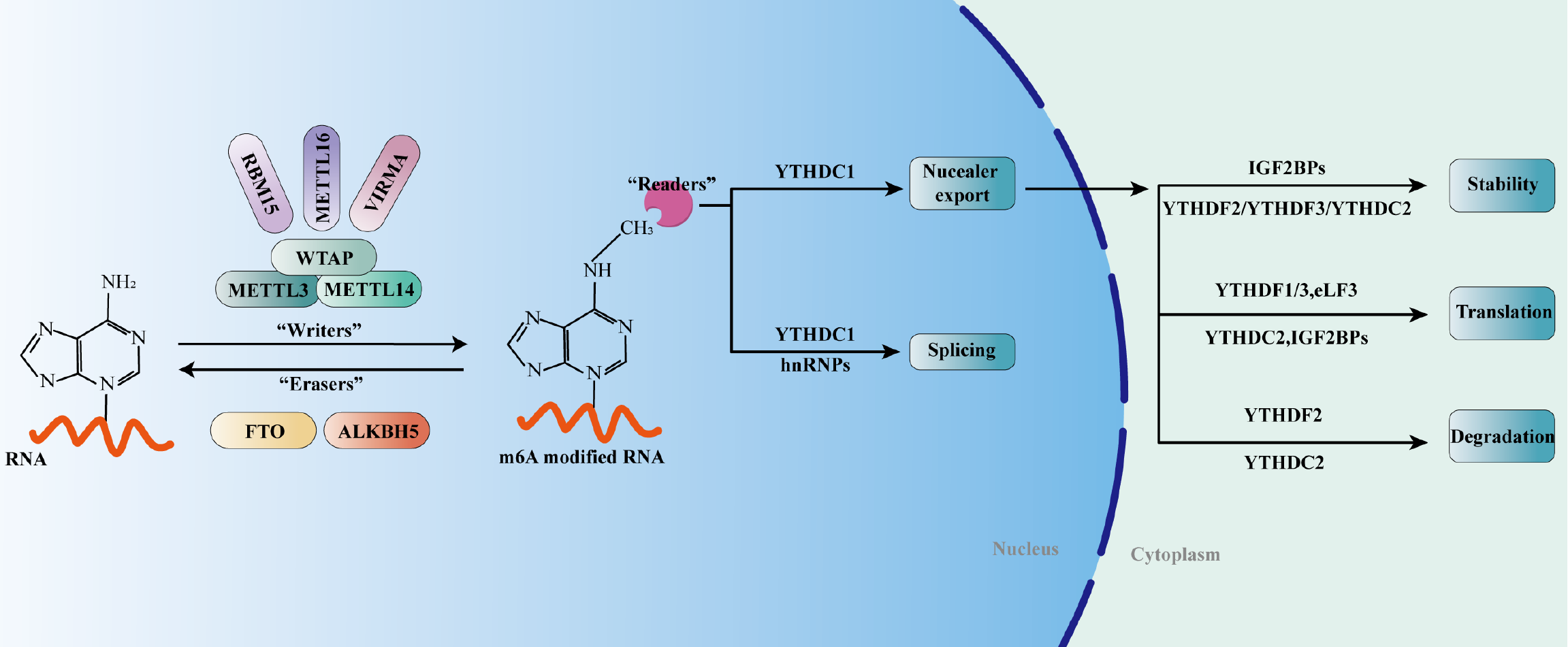
As a reversible post-transcriptional modification, the level of m6A modification depends on the activity of m6A methyltransferase ("writer") and demethylase ("eraser")[13-15]. m6A writers include but are not limited to methyltransferase-like (METTL)3, METTL14, WT1-associated protein (WTAP) and RNA-binding motif protein (RBM)15, and erasers include fat mass and obesity-associated protein (FTO) and AlkB homolog (ALKBH)5[6]. RNA-binding proteins ("readers") speci
Emerging research indicates that the ability of writers to regulate m6A modification is achieved through a multicomponent methyltransferase complex[6,18]. As the first identified m6A methyltransferase, METTL3 was able to catalyze the m6A modification process by forming a stable methyltransferase complex with METTL14[19-22]. WTAP, as an adaptor protein, can interact with the above dimer complex to determine its location in the nuclear spots and play a catalytic role[20,23,24]. Subsequently, members of m6A methyltransferase such as RBM15 and METTL16 were successively found, which played an irreplaceable role in the process of m6A modification[25-27].
Erasers, namely m6A demethylase, can reverse m6A methylation modification, and co-regulate m6A RNA modification with writers[28]. These demethylases promote the conversion of m6A to N6-hydroxymethyl adenosine, thereby hydro
Although the level of m6A modification depends on the activities of writers and erasers, its biological function also requires different readers to induce a variety of biological phenotypes by recognizing and combining m6A methylation targets[34]. The readers mainly include the YTH domain family (including YTHDC1-2 and YTHDF1-3)[35-37], insulin-like growth factor 2 mRNA binding protein (IGF2BP)1-3[38,39], heterogeneous nuclear RNA protein family (HNRNP, including HNRNPC and HNRNPG)[40-42], and eukaryotic initiation factor (eIF)3[43]. YTHDF1-3 and YTHDC1-2 can directly recognize the conserved m6A binding domain, thereby directly reading and binding to m6A modified RNA[6]. IGF2BPs possess the ability to recognize m6A modifications and thus modulate the abundance and function of m6A-modified RNA[38]. HNRNP, also known as the m6A switch, can directly bind to mRNA possessing m6A recognition sites, indirectly regulating its abundance, translation, and stability by affecting the secondary structure of RNA[44,45].
Since the biological function of m6A modification depends on readers, different reader groups can regulate mRNA transcription by influencing the biological function of m6A modification, thus affecting cell function and regulating physiological status. Once there is an error in regulation, it may lead to the initiation and development of various diseases, including carcinomas. Therefore, blocking the binding sites of certain specific readers may become a new method for cancer treatment.
As a reversible post-transcriptional modification, m6A methylation is modulated by writers, erasers and readers, which is a dynamic balancing process. Through m6A methylation modification, various RNA splicing, translation, transport and stability can be regulated in an orderly manner[46-48].
Splicing mRNA precursors into mature mRNA can affect the biological functions of eukaryotes, which is a complex and dynamically balanced process. The m6A site overlaps spatially with the splicing enhancer binding region of the serine/arginine-rich (SR) protein exon, and FTO can preferentially bind the alternative splicing exons and polyA sites, thereby modulating the RNA binding ability of SR splicing factor (SRSF)2 and further controlling mRNA splicing[7,29,49]. ALKBH5 colocalizes with ASF/SF2; it is able to alter the phosphorylation status of ASF/SF2, and the hyperphosphorylated ASF/SF2 is involved in mRNA splicing[50]. Previous studies have confirmed that loss of the m6A reader protein HNRNPG regulates alternative splicing in an m6A-dependent way[7,40]. Also, downregulation of m6A writers has been found to interfere with the processes of splicing and gene expression[51,52].
It has been reported that ALKBH5 can regulate the nuclear export process, thereby influencing the subcellular distribution of mRNAs[7]. Mechanistically, ALKBH5 colocalizes with ASF/SF2; it is able to alter the phosphorylation status of ASF/SF2, thereby facilitating TAP/p15-complex-mediated mRNA export[50]. YTHDC1 is able to promote RNA binding to both nuclear RNA export factor 1 and export adaptor protein SRSF3, thus facilitating nuclear export[53]. Previous studies have confirmed that fragile X mental retardation protein, as an m6A reader, is able to facilitate nuclear export mediated by nuclear export protein chromosome region maintenance 1[54].
The function of METTL3 in regulating translation is enabled by the specific binding of different readers to the m6A sites. METTL3 can interact with eIF3h to modulate and facilitate translation, which is independent of methyltransferase activity[55,56]. Previous studies have confirmed that METTL16 and YTHDF proteins play pivotal roles in regulating translation[57-60]. YTHDF1, which is spatially adjacent to the translation start site bridged by eIF4G, plays a pivotal role in constructing loop structures with eIF4G and eIF3, as well as in the recruitment of ribosomes, thereby facilitating cap-dependent translation initiation[60]. YTHDC2 can interact with the 5'→3' exoribonuclease XRN1 and enhance helicase activity to promote translation, which is independent of m6A modification[61]. YTHDF3, in cooperation with YTHDF1, can interact with the 40S and 60S ribosome subunits to promote translation[59]. Besides, YTHDF3 facilitate the malignant development of bladder carcinoma through improving the translation of ITGA6[62].
As a reversible post-transcriptional modification, m6A modification bidirectionally regulates mRNA stability. For instance, YTHDF1 destabilizes MAT2A mRNA through binding to the m6A site 3’-untranslated region[63]. YTHDF2 is able to directly recruit the CCR4-NOT deadenylase complex to accelerate the process of deadenylation, thus promoting RNA degradation[64]. In synergism with YTHDF2, YTHDF3 promotes decay of m6A-modified RNA[65]. YTHDF2 can directly identify the methylation of Arrestin domain containing 4 and suppressor of cytokine signaling (SOCS)2 and destabilize their mRNAs, thereby facilitating the malignant progression of tumors[66,67]. It has been reported that the IGF2BPs family of proteins can significantly bolster the stability of mRNAs through the binding of their KH domains to specific m6A sites[38].
Modification of m6A plays a significant role in various biological processes, such as cell proliferation, differentiation and apoptosis, and it is crucial in the initiation and progression of tumors. The function of m6A modification depends on its readers, which means that the same m6A modification can have completely opposite effects when interacting with different readers. In this context, we have comprehensively summarized the research progress on the role of m6A in hepatobiliary carcinoma (Table 1).
| Tumor typed role | Regulators | m6A-relate | Functions | Mechanisms | Ref. |
| Hepatocellular carcinoma | METTL3 | m6A writers | Promotes HCC cell proliferation, migration, and tumorigenicity | Accelerates the degradation of SOCS2 | [66] |
| Promotes HCC progression | Modulates MAPK cascade | [78] | |||
| Promotes the formation of vasculogenic mimicry | Enhances the translation efficiency of YAP1 mRNA | [133] | |||
| METTL14 | m6A writers | Inhibits tumor invasion and metastasis | Regulates the pri-microRNA 126 processing | [69] | |
| Inhibits HCC progression | Inhibits the EGFR/PI3K/AKT signaling pathway | [74] | |||
| Inhibits HCC tumor growth | Regulates degradation of SLC7A11 mRNA | [75] | |||
| Boosts the differentiation of liver CSCs and repress HCC growth | Maintains the stability of HNF3γ mRNA | [76] | |||
| Impairs the metabolic reprogramming of HCC cells | Stabilizes USP48 mRNA | [77] | |||
| Pancreatic cancer | METTL3 | m6A writers | Promotes tumorigenesis and chemoradiation tolerance | Modulates MAPK cascade | [52] |
| METTL14 | m6A writers | Promotes PC proliferation and migration | Induces PERP mRNA degradation | [81,82] | |
| Inhibits PC cells apoptosis and autophagy, and enhances chemotherapy tolerance | Facilitates AMPKα, ERK1/2, and mTOR pathways | [83] | |||
| Enhances PC cells chemotherapy tolerance | Maintains CDA mRNA stability | [84] | |||
| Inhibits PC progression | Induces PIK3CB mRNA degradation | [85] | |||
| ALKBH5 | m6A erasers | Inhibits the initiation and progression of PC | Reduce WIF-1 mRNA methylation and down-regulate the Wnt pathway | [86] | |
| Prevents PC tumorigenesis | Activates PER1 via YTHDF2-dependent manner | [87] | |||
| FTO | m6A erasers | Promotes PC progression | Stabilizes proto-oncogene MYC mRNA | [88] | |
| YTHDF2 | m6A readers | Inhibits PC progression | Curtails cell invasion and migration via the YAP pathway | [89] | |
| Accelerating the growth of PC cells | Activates Akt/GSK3b/CyclinD1 pathway | [89] | |||
| Gallbladder carcinoma | IGF2BP3 | m6A readers | Promotes the aggressive progression of GBC | Stabilizes CLDN4 mRNA | [93] |
| Promotes GBC progression | Stabilizes KLK5 mRNA | [94] | |||
| YTHDF2 | m6A readers | Promotes GBC progression and chemoresistance | Decreases DAPK3 mRNA stability | [95] | |
| METTL3 | m6A writers | Promotes GBC cell proliferation, migration, and tumorigenicity | Destabilizes DUSP5 mRNA | [96] | |
| Promotes GBC progression | Regulates miR-92b-3p expression | [97] | |||
| Cholangiocarcinoma | METTL3 | m6A writers | Facilitates glycolysis and CCA progression | Increase AKR1B10 expression via m6A-dependent manner | [101] |
| Promotes CCA progression | Mediates IFIT2 mRNA degradation | [102] | |||
| Facilitates iCCA proliferation and metastasis | Stabilizes NFAT5 mRNA | [103] | |||
| METTL5 | m6A writers | Promotes CCA progression | Mediating 18S rRNA m6A methylation | [104] | |
| METTL14 | m6A writers | Promotes CCA progression | Disrupts Siah2 mRNA stability | [105] | |
| METTL16 | m6A writers | Promotes ICC proliferation and metastasis | Up-regulates PRDM15-mediated FGFR4 expression | [106] | |
| VIRMA | m6A writers | Facilitates the malignant development of iCCA | Enhances the abundance of TMED2 | [107] | |
| Facilitates the malignant development of iCCA | Enhances the abundance of PARD3B | [107] | |||
| Promotes CCA progression | Activates the downstream target SIRT1 | [108] | |||
| IGF2BP1 | m6A readers | Promotes iCCA progression | Activates the c-Myc/p16 and ZIC2/PAK4/AKT/MMP2 pathways | [109] | |
| YTHDF1 | m6A readers | Induces CCA growth and metastasis | Regulates EGFR mRNA translation | [110] | |
| Induces CCA growth and metastasis | Mediating the m6A methylation of lncRNA CTBP1-AS2 | [111] | |||
| YTHDF2 | m6A readers | Facilitates iCCA progression and chemoresistance | Destabilizes CDKN1B mRNA | [112] | |
| ALKBH5 | m6A erasers | Promotes the malignant development of CCA | Increases the expression of PD-L1 to facilitate immune evasion | [113] |
Hepatocellular carcinoma (HCC), as the main type of primary liver cancer, is the sixth most common malignancy worldwide and the third most common cause of cancer-related deaths[68]. Previous studies have confirmed that abnor
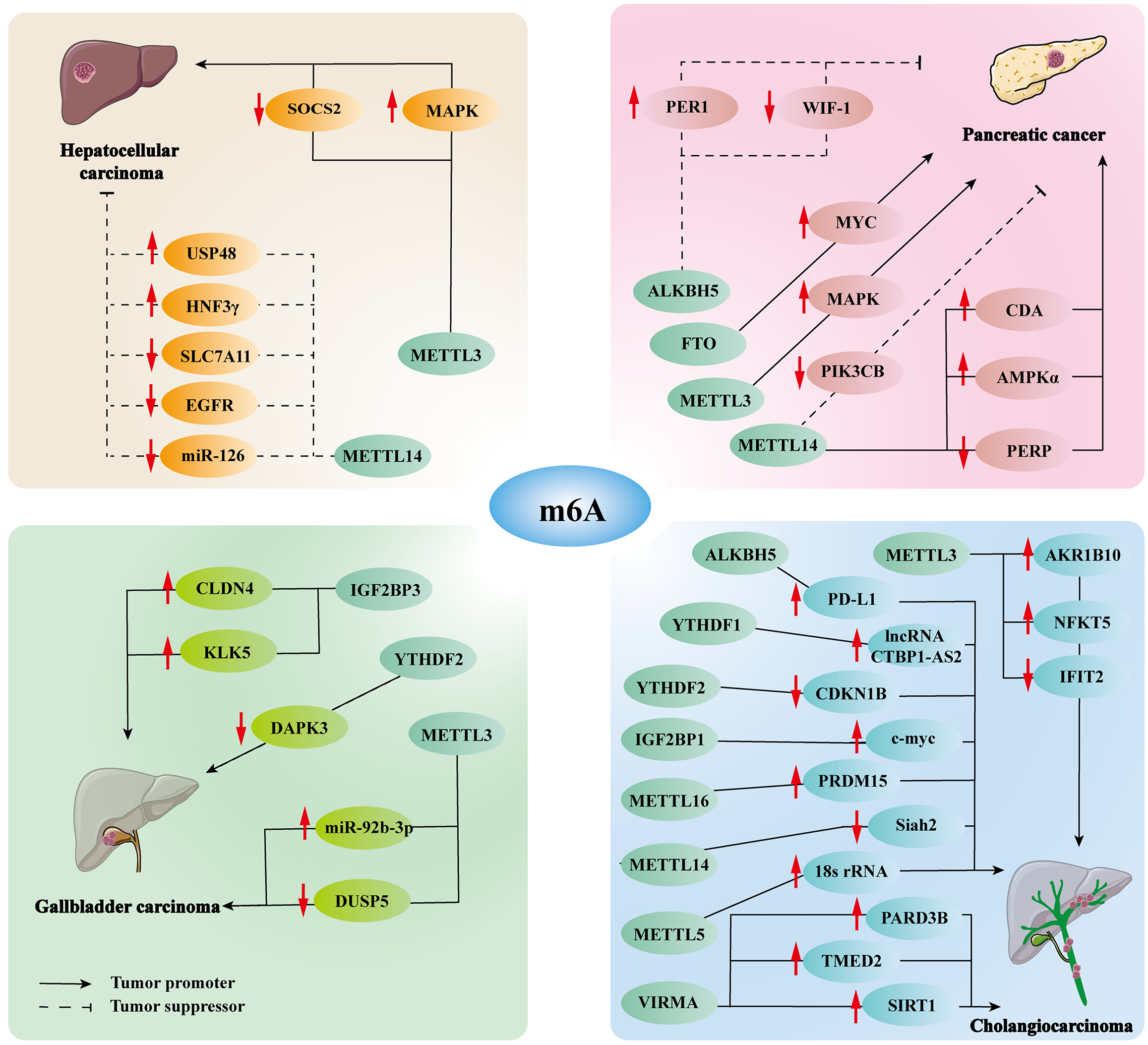
Likewise, up-regulated the expression of m6A is also closely related to HCC. For instance, the expression and prognosis of METTL3 are absolutely inverse to METTL14[70]. As a writer of m6A, excessive METTL3 accelerates the degradation of tumor suppressor SOCS2 through a YTHDF2-dependent mechanism, and ultimately promotes tumorigenesis[66]. METTL3 affects extracellular signaling pathways and the cell cycle through regulating the mitogen-activated protein kinase (MAPK) cascade, thus facilitating HCC malignant progression[78]. Recent studies have demonstrated that METTL3 enhanced beaded filament structural protein 1 stability by upregulating m6A modification, ultimately promoting tumorigenesis[79]. Therefore, upregulated m6A modification also plays a vital role in the initiation and malignant progression of HCC.
Pancreatic cancer (PC) could become the world’s third-most lethal cancer, according to projections, and the 5-year survival rate remains < 10% due to its late diagnosis, chemotherapy resistance and metastasis[80]. Recent studies have confirmed that abnormal m6A modification plays a pivotal role in the progression of PC (Figure 2). The MAPK cascade, as a ubiquitin-dependent modification in PC, can be modulated by m6A writer METTL3, thereby facilitating tumorigenesis and chemoradiation tolerance[52]. In addition, METTL14 is highly expressed in PC tissues, which can induce p53 effector related to PMP-22 mRNA degradation in an m6A-dependent manner, significantly promoting PC proliferation and migration[81,82]. Another study showed that downregulation of METTL14 facilitates apoptosis and autophagy in PC cells, while also enhancing sensitivity to chemotherapy. This effect is achieved through the inhibition of key signaling pathways, including AMP-activated protein kinase α, extracellular signal-regulated kinase (ERK)1/2 and mammalian target of rapamycin (mTOR)[83]. Similarly, downregulated METTL14 reduces cytidine deaminase mRNA stability through m6A-dependent manner, thereby enhancing the sensitivity of PC cells to chemotherapy[84]. Emerging research indicates that the m6A modification, catalyzed by METTL14 and recognized by YTHDF2, can enhance the rs142933486[G] allele variation of oncogene PIK3CB, thereby promoting the degradation of mRNA and decreasing PIK3CB expression, which is significant in PC, especially pancreatic ductal adenocarcinoma[85]. The eraser ALKBH5 can reduce WIF-1 mRNA methylation and downregulate the Wnt pathway, further inhibiting the initiation and progression of PC[86]. ALKBH5 also activates period circadian regulator 1 via YTHDF2-dependent manner, which activates the ATM/CHK2/P53/CDC25C pathway and ultimately prevents PC tumorigenesis[87]. Another type of eraser, FTO, is highly expressed in PC. Previous studies have confirmed that FTO reduces the methylation level of the proto-oncogene MYC mRNA and improves its stability, which ultimately promotes PC progression[88]. Moreover, YTHDF2 functions as an inhibitor, curtailing cell invasion and migration via the YAP signaling pathway. Conversely, it also acts as a promoter, accelerating the growth of PC cells through the Akt/GSK3b/CyclinD1 pathway[89]. Recent studies have confirmed that Proteasome 26S subunit non-ATPase 14 enhanced spondin 2 mRNA stability through RBM15B-mediated m6A modification, thereby facilitating PC proliferation, migration, and invasion[90]. Overall, all aspects of m6A modification, including writers, readers and erasers, can affect the initiation and development of PC, and it may be of importance to target one of these aspects as a therapeutic target for PC.
Gallbladder carcinoma (GBC) is the most common and deadly type of hepatobiliary carcinoma, with 5-year survival < 5% in the advanced stages, and most patients already present with metastases at initial diagnosis, frequently to the liver[91,92]. At present, research on the role of m6A in GBC is still in the initial stage, and the m6A aspects that are involved in the malignant progression of GBC include IGF2BP3, YTHDF2 and METTL3 (Figure 2). IGF2BP3 is expressed at elevated levels in GBC, which implies poor prognosis. IGF2BP3 stabilizes claudin 4 mRNA via an m6A-dependent manner, thereby activating the nuclear factor-kB pathway and ultimately promoting the aggressive progression of GBC[93]. Another study showed that IGF2BP3 stabilizes KLK5 mRNA via an m6A-dependent manner to activate the PAR2/AKT pathway, ultimately promoting the progression of GBC. It also confirmed that let-7 g-5p negatively regulates IGF2BP3 and the let-7 g-5p/IGF2BP3/KLK5/PAR2/AKT pathway is a potential treatment strategy for GBC[94]. Another type of reader, YTHDF2, decreases tumor suppressor death-associated protein kinase 3 mRNA stability and abundance via an m6A-dependent manner, ultimately promoting GBC malignant progression and chemoresistance[95]. METTL3 des
CCA is a primary liver cancer with an incidence second only to that of HCC, and is anatomically divided into intrahepatic CCA (iCCA) and extrahepatic CCA (eCCA). The early symptoms are not obvious; therefore, most patients are diagnosed at an advanced stage, and the 5-year survival rate is only 5%-15%[98-100]. Many studies have indicated that m6A methylation modification is critical in CCA (Figure 2). As a writer, METTL3 expression levels are elevated in CCA cells, which means poor prognosis. Highly expressed METTL3 bind to aldo-keto reductase family 1 (AKR1)B10 at m6A modification sites, and increase the expression level of AKR1B10 in an m6A-dependent manner, thereby facilitating glycolysis and CCA progression[101]. METTL3 also mediates interferon-induced protein with tetratricopeptide repeats (IFIT)2 mRNA degradation via a YTHDF2-dependent manner, thereby downregulating IFIT2 expression levels and ultimately promoting malignant prognosis of iCCA[102]. Importantly, METTL3-mediated nuclear factor of activated T-cells (NFAT)5 m6A modification stabilizes NFAT5 mRNA in the m6A-IGF2BP1-dependent pathway, thereby increasing the abundance of GLUT1 and PGK1, resulting in iCCA proliferation and metastasis[103]. The above views suggest that down-regulation of METTL3 may be a potential therapeutic target for CCA. Similarly, as methyltransferases, METTL5, METTL14, and METTL16 also play significant roles in CCA. Mechanistically, METTL5 facilitates ribosome synthesis and oncogenic mRNA translation by mediating 18S rRNA m6A methylation, and ultimately promoting the malignant prognosis of CCA[104]. METTL14 inhibits Siah2-mediated antitumor T-cell activity by disrupting Siah2 mRNA stability in an m6A-YTHDF2-dependent way, ultimately leading to CCA progression[105]. METTL16 upregulates members of the PRDI-BF1 and RIZ homology domain 15-mediated fibroblast growth factor receptor 4 expression in a YTHDF1-dependent manner to promote ICC proliferation and metastasis[106]. As another type of writer, vir-Like m6A methyltransferase associated (VIRMA) enhances the abundance of transmembrane effector 2 and par-3 homolog B through an m6A-HuR-mediated manner, activates the Akt/GSK/β-catenin and MEK/ERK/Slug pathways, thus facilitating malignant development of iCCA[107]. The pro-oncogenic effect of VIRMA-mediated m6A modification is also achieved by activating the downstream target SIRT1[108]. Readers and erasers also participate in the progression of CCA. For instance, IGF2BP1-mediated m6A modification activates the c-Myc/p16 and ZIC2/PAK4/AKT/MMP2 pathways, promoting iCCA progression[109]. YTHDF1 induces CCA growth and metastasis through several pathways, such as mediating the m6A methylation of lncRNA CTBP1-AS2 and regulation of EGFR mRNA translation[110,111]. YTHDF2 facilitates iCCA progression and chemoresistance by destabilizing cyclin-dependent kinase inhibitor 1B mRNA in an m6A-dependent manner[112]. As an important m6A demethylase, ALKBH5 increases the expression of programmed death protein ligand (PD-L)1 through Ya THDF2-dependent manner to facilitate immune evasion and ultimately promote malignant development of CCA[113]. To summarize, the role of m6A methylation modifications is pivotal and irrepla
Tumor immune microenvironment (TIME) is crucial in the progression of hepatobiliary carcinoma. In recent years, the important role of epigenetic regulation in TIME has gradually emerged, among which m6A has received special attention as the most abundant post-transcriptional modification[114]. As an important writer protein, METTL3-mediated m6A modification increases SHP-2 expression to activate interleukin (IL)-15-dependent pathways AKT/mTOR and MAPK/ERK, ultimately promoting the antitumor immunity of natural killer (NK) cells[115]. METTL3 also facilitates immunosuppression of tumor-infiltrating myeloid cells by elevating Janus kinase 1 expression in a YTHDF1-dependent manner[116]. Emerging research indicates that METTL3 increases the expression of protumor chemokines such as CXCL1, CXCL5 and 20, and promotes the degradation of PD-L1 mRNA through m6A methylation, resulting in the formation of non-inflamed tumor microenvironment[117]. Targeting METTL3 is a potential method to improve immunotherapy in hepatobiliary carcinoma patients. m6A modification mediated by YTHDF1 plays a pivotal role in facilitating hypoxia-induced auto
Presently, an accumulating body of evidence suggests that abnormal m6A modification is critically involved in the progression of various carcinomas. Consequently, targeting m6A regulators with precision might offer a promising therapeutic approach for treating hepatobiliary carcinoma.
As FTO inhibitors, natural product rhein, meclofenamic acid and fluorescein derivatives were among the first small-molecule identified to target m6A regulators, but their activity and specificity are relatively limited[122-124]. In recent years, a new generation of more potent and selective FTO inhibitors, including FTO inhibitor (FB)23, FB23-2, compounds from NCI DTP library NSC337766 (CS1) and NSC368390 (CS2), has been discovered[125-127]. FB23 and FB23-2, in particular, are known to positively regulate m6A modification, playing a pivotal role in leukemia treatment[125]. CS1 and CS2 impede tumor progression via the FTO/m6A/LILRB4 pathway[126]. Emerging research highlights the significant impact of FTO-04 in glioblastoma treatment through its inhibition of FTO[127]. At present, there are few studies on small-molecule inhibitors of demethylase ALKBH5, and two compounds have been discovered including 2-[(1-hydroxy-2-oxo-2-phenylethyl)sulfanyl]acetic acid(3) and 4-{[(furan-2-yl)methyl]amino}-1,2-diazinane-3,6-dione(6), which can inhibit the proliferation of cancer cells and play a considerable role in the progression of leukemia[128]. Although these small-molecule inhibitors have been reported to have significant effects in hematological tumors, these inhibitors may also become potential therapeutic modalities in hepatobiliary carcinoma overexpressing FTO or ALKBH5. The current small-molecule modulators of m6A writers are poorly studied, and STM2457, as the earliest bioavailable METTL3 small-molecule inhibitor, which has significant antitumor effects[102,129,130]. UZH1a, an emerging small-molecule METTL3 inhibitor, also plays a significant role in anticancer treatment[131,132].
Overall, RNA epigenetics-based tumor-targeted treatment is still in its infancy, and research on small-molecule drugs targeting m6A regulators is insufficient. It is necessary to further explore the mechanism of m6A modification, which is the basis for the development of new antitumor drugs.
Hepatobiliary carcinoma is a prevalent malignancy in the digestive system, and presents with a stealthy onset and swift progression, posing a significant threat to human life and health. m6A methylation, the most copious post-transcriptional modification, is currently the focus of extensive research. It is a dynamic and balanced process, intricately regulated by a trio of elements known as writers, erasers and readers. Accumulating evidence indicates that aberrations in m6A modification play a pivotal role in the pathophysiological development of the hepatobiliary system, substantially influencing the advancement of hepatobiliary carcinoma. This review systematically elucidates the dual roles of m6A modification in hepatobiliary carcinoma, revealing its molecular mechanisms in either promoting or suppressing tumor progression through dynamic regulation of oncogene expression, cancer stem cell properties, and tumor microenvironment interactions. Notably, the spatiotemporal specificity of m6A regulation leads to opposing effects in similar tumors, a phenomenon that may stem from: (1) Site-specific methylation differences (e.g., distinct functions of m6A in coding sequences vs 3'UTRs); (2) Variable expression profiles of reader proteins; and (3) Context-dependent integration of downstream signaling pathways. Although targeting m6A regulators (such as METTL3 inhibitors or FTO activators) has shown promise in preclinical models, current research faces three major challenges: First, at the technical level, existing m6A detection methods are challenging to resolve methylation dynamics at single-cell resolution, limiting our under
| 1. | Pant K, Gradilone SA. Hepatobiliary Cancers: Progress in Diagnosis, Pathogenesis, and Treatment. Technol Cancer Res Treat. 2022;21:15330338221097203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 2. | Moris D, Palta M, Kim C, Allen PJ, Morse MA, Lidsky ME. Advances in the treatment of intrahepatic cholangiocarcinoma: An overview of the current and future therapeutic landscape for clinicians. CA Cancer J Clin. 2023;73:198-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 254] [Article Influence: 127.0] [Reference Citation Analysis (0)] |
| 3. | Ruff SM, Shannon AH, Pawlik TM. Advances in Targeted Immunotherapy for Hepatobiliary Cancers. Int J Mol Sci. 2022;23:13961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, Calvisi DF, Perugorria MJ, Fabris L, Boulter L, Macias RIR, Gaudio E, Alvaro D, Gradilone SA, Strazzabosco M, Marzioni M, Coulouarn C, Fouassier L, Raggi C, Invernizzi P, Mertens JC, Moncsek A, Ilyas SI, Heimbach J, Koerkamp BG, Bruix J, Forner A, Bridgewater J, Valle JW, Gores GJ. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1555] [Cited by in RCA: 1524] [Article Influence: 304.8] [Reference Citation Analysis (0)] |
| 5. | Moffat GT, Hu ZI, Meric-Bernstam F, Kong EK, Pavlick D, Ross JS, Murugesan K, Kwong L, De Armas AD, Korkut A, Javle M, Knox JJ. KRAS Allelic Variants in Biliary Tract Cancers. JAMA Netw Open. 2024;7:e249840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 6. | Gao R, Ye M, Liu B, Wei M, Ma D, Dong K. m6A Modification: A Double-Edged Sword in Tumor Development. Front Oncol. 2021;11:679367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 7. | Shi B, Liu WW, Yang K, Jiang GM, Wang H. The role, mechanism, and application of RNA methyltransferase METTL14 in gastrointestinal cancer. Mol Cancer. 2022;21:163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 8. | Liu J, Harada BT, He C. Regulation of Gene Expression by N(6)-methyladenosine in Cancer. Trends Cell Biol. 2019;29:487-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 9. | Pan Y, Ma P, Liu Y, Li W, Shu Y. Multiple functions of m(6)A RNA methylation in cancer. J Hematol Oncol. 2018;11:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 242] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 10. | Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell. 2019;74:640-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1299] [Article Influence: 216.5] [Reference Citation Analysis (0)] |
| 11. | Pinello N, Sun S, Wong JJ. Aberrant expression of enzymes regulating m(6)A mRNA methylation: implication in cancer. Cancer Biol Med. 2018;15:323-334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 12. | Ye H, Chen T, Zeng Z, He B, Yang Q, Pan Q, Chen Y, Wang W. The m6A writers regulated by the IL-6/STAT3 inflammatory pathway facilitate cancer cell stemness in cholangiocarcinoma. Cancer Biol Med. 2021;19:343-357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Roignant JY, Soller M. m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017;33:380-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 327] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 14. | Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. Int J Biol Sci. 2020;16:1929-1940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 15. | Lou X, Wang JJ, Wei YQ, Sun JJ. Emerging role of RNA modification N6-methyladenosine in immune evasion. Cell Death Dis. 2021;12:300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 16. | Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1100] [Cited by in RCA: 1609] [Article Influence: 268.2] [Reference Citation Analysis (0)] |
| 17. | Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 1275] [Article Influence: 318.8] [Reference Citation Analysis (0)] |
| 18. | Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697-17704. [PubMed] |
| 19. | Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1590] [Cited by in RCA: 2513] [Article Influence: 209.4] [Reference Citation Analysis (0)] |
| 20. | Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63:306-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 931] [Article Influence: 103.4] [Reference Citation Analysis (0)] |
| 21. | Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 392] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 22. | Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 862] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 23. | Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, Huang Y, Zheng R, Yu H, Wang J, Hu M, Miao J, Li J. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6)A-dependent manner. Cell Death Dis. 2020;11:659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 24. | Chen Y, Peng C, Chen J, Chen D, Yang B, He B, Hu W, Zhang Y, Liu H, Dai L, Xie H, Zhou L, Wu J, Zheng S. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 447] [Cited by in RCA: 453] [Article Influence: 75.5] [Reference Citation Analysis (0)] |
| 25. | Visvanathan A, Somasundaram K. mRNA Traffic Control Reviewed: N6-Methyladenosine (m(6) A) Takes the Driver's Seat. Bioessays. 2018;40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 26. | Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, Lan F, Shi YG, He C, Shi Y, Diao J. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018;69:1028-1038.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 679] [Article Influence: 113.2] [Reference Citation Analysis (0)] |
| 27. | Ruszkowska A, Ruszkowski M, Dauter Z, Brown JA. Structural insights into the RNA methyltransferase domain of METTL16. Sci Rep. 2018;8:5311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 28. | Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 714] [Article Influence: 142.8] [Reference Citation Analysis (0)] |
| 29. | Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, Jin KX, Wang X, Huang CM, Fu Y, Ge XM, Song SH, Jeong HS, Yanagisawa H, Niu Y, Jia GF, Wu W, Tong WM, Okamoto A, He C, Rendtlew Danielsen JM, Wang XJ, Yang YG. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403-1419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 656] [Cited by in RCA: 915] [Article Influence: 83.2] [Reference Citation Analysis (0)] |
| 30. | Zuidhof HR, Calkhoven CF. Oncogenic and Tumor-Suppressive Functions of the RNA Demethylase FTO. Cancer Res. 2022;82:2201-2212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 31. | Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K, Tsujikawa K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7:42271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 237] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 32. | Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, Lu Z, Zheng Z, Dai Q, Wang H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533-2545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 260] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 33. | Liu H, Yang M, Zhang C, Zhang Y, Wang Y, Chen Y. m(6)A transferase KIAA1429 mediates the upregulation of LncRNA LINC00968 promoting the progression of gastric cancer cells. Hereditas. 2025;162:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 34. | Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1401] [Cited by in RCA: 1777] [Article Influence: 197.4] [Reference Citation Analysis (0)] |
| 35. | Zaccara S, Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell. 2020;181:1582-1595.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 518] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 36. | Liu J, Gao M, He J, Wu K, Lin S, Jin L, Chen Y, Liu H, Shi J, Wang X, Chang L, Lin Y, Zhao YL, Zhang X, Zhang M, Luo GZ, Wu G, Pei D, Wang J, Bao X, Chen J. The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. 2021;591:322-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 223] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 37. | Fu Y, Zhuang X. m(6)A-binding YTHDF proteins promote stress granule formation. Nat Chem Biol. 2020;16:955-963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 222] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 38. | Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Hüttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 928] [Cited by in RCA: 2016] [Article Influence: 288.0] [Reference Citation Analysis (0)] |
| 39. | Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res. 2020;39:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 190] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 40. | Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN, He C, Parisien M, Pan T. Regulation of Co-transcriptional Pre-mRNA Splicing by m(6)A through the Low-Complexity Protein hnRNPG. Mol Cell. 2019;76:70-81.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 277] [Article Influence: 46.2] [Reference Citation Analysis (1)] |
| 41. | Wang LC, Chen SH, Shen XL, Li DC, Liu HY, Ji YL, Li M, Yu K, Yang H, Chen JJ, Qin CZ, Luo MM, Lin QX, Lv QL. M6A RNA Methylation Regulator HNRNPC Contributes to Tumorigenesis and Predicts Prognosis in Glioblastoma Multiforme. Front Oncol. 2020;10:536875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 42. | Huang XT, Li JH, Zhu XX, Huang CS, Gao ZX, Xu QC, Zhao W, Yin XY. HNRNPC impedes m(6)A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. 2021;518:196-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 43. | Lin Y, Li F, Huang L, Polte C, Duan H, Fang J, Sun L, Xing X, Tian G, Cheng Y, Ignatova Z, Yang X, Wolf DA. eIF3 Associates with 80S Ribosomes to Promote Translation Elongation, Mitochondrial Homeostasis, and Muscle Health. Mol Cell. 2020;79:575-587.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 44. | Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1074] [Cited by in RCA: 1535] [Article Influence: 153.5] [Reference Citation Analysis (0)] |
| 45. | Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299-1308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 715] [Cited by in RCA: 1169] [Article Influence: 116.9] [Reference Citation Analysis (0)] |
| 46. | Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 346] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 47. | Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 872] [Article Influence: 174.4] [Reference Citation Analysis (0)] |
| 48. | Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 499] [Cited by in RCA: 598] [Article Influence: 85.4] [Reference Citation Analysis (0)] |
| 49. | Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017;45:11356-11370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 237] [Cited by in RCA: 350] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 50. | Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1763] [Cited by in RCA: 2673] [Article Influence: 205.6] [Reference Citation Analysis (0)] |
| 51. | Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2600] [Cited by in RCA: 3628] [Article Influence: 279.1] [Reference Citation Analysis (0)] |
| 52. | Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, Doki Y, Mori M, Ishii H, Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 189] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 53. | Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, He E, Shen B, He C. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 501] [Cited by in RCA: 919] [Article Influence: 114.9] [Reference Citation Analysis (0)] |
| 54. | Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, Miller N, Rojas Ringeling F, Ming GL, He C, Song H, Ma YC. FMRP Modulates Neural Differentiation through m(6)A-Dependent mRNA Nuclear Export. Cell Rep. 2019;28:845-854.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 212] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 55. | Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, Santisteban P, George RE, Richards WG, Wong KK, Locker N, Slack FJ, Gregory RI. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556-560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 311] [Cited by in RCA: 555] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 56. | Ianniello Z, Sorci M, Ceci Ginistrelli L, Iaiza A, Marchioni M, Tito C, Capuano E, Masciarelli S, Ottone T, Attrotto C, Rizzo M, Franceschini L, de Pretis S, Voso MT, Pelizzola M, Fazi F, Fatica A. New insight into the catalytic -dependent and -independent roles of METTL3 in sustaining aberrant translation in chronic myeloid leukemia. Cell Death Dis. 2021;12:870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 57. | Su R, Dong L, Li Y, Gao M, He PC, Liu W, Wei J, Zhao Z, Gao L, Han L, Deng X, Li C, Prince E, Tan B, Qing Y, Qin X, Shen C, Xue M, Zhou K, Chen Z, Xue J, Li W, Qin H, Wu X, Sun M, Nam Y, Chen CW, Huang W, Horne D, Rosen ST, He C, Chen J. METTL16 exerts an m(6)A-independent function to facilitate translation and tumorigenesis. Nat Cell Biol. 2022;24:205-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 213] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 58. | Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, Xiao H, Li L, Rao S, Wang F, Yu J, Yu J, Zou D, Yi P. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816-3831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 502] [Article Influence: 100.4] [Reference Citation Analysis (0)] |
| 59. | Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, Bhattarai DP, Zhao YL, Sun BF, Yang YG. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 630] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 60. | Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1663] [Cited by in RCA: 2590] [Article Influence: 259.0] [Reference Citation Analysis (0)] |
| 61. | Li L, Krasnykov K, Homolka D, Gos P, Mendel M, Fish RJ, Pandey RR, Pillai RS. The XRN1-regulated RNA helicase activity of YTHDC2 ensures mouse fertility independently of m(6)A recognition. Mol Cell. 2022;82:1678-1690.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 53] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 62. | Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, Yuan Z, Qi D, Lin S, Min W, Yang M, Ji W. N(6)-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195-207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 159] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 63. | Shima H, Matsumoto M, Ishigami Y, Ebina M, Muto A, Sato Y, Kumagai S, Ochiai K, Suzuki T, Igarashi K. S-Adenosylmethionine Synthesis Is Regulated by Selective N(6)-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 2017;21:3354-3363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 261] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 64. | Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 626] [Cited by in RCA: 1085] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 65. | Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1252] [Cited by in RCA: 1347] [Article Influence: 168.4] [Reference Citation Analysis (0)] |
| 66. | Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254-2270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 672] [Cited by in RCA: 972] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 67. | Wang H, Wei W, Zhang ZY, Liu Y, Shi B, Zhong W, Zhang HS, Fang X, Sun CL, Wang JB, Liu LX. TCF4 and HuR mediated-METTL14 suppresses dissemination of colorectal cancer via N6-methyladenosine-dependent silencing of ARRDC4. Cell Death Dis. 2021;13:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 68. | Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi L, Cai X. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol. 2021;14:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 69. | Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 676] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 70. | Liu X, Qin J, Gao T, Li C, Chen X, Zeng K, Xu M, He B, Pan B, Xu X, Pan Y, Sun H, Xu T, Wang S. Analysis of METTL3 and METTL14 in hepatocellular carcinoma. Aging (Albany NY). 2020;12:21638-21659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 71. | Li Z, Li F, Peng Y, Fang J, Zhou J. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med. 2020;9:1877-1889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 72. | Wu X, Zhang X, Tao L, Dai X, Chen P. Prognostic Value of an m6A RNA Methylation Regulator-Based Signature in Patients with Hepatocellular Carcinoma. Biomed Res Int. 2020;2020:2053902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 73. | Gu Z, Du Y, Zhao X, Wang C. Diagnostic, Therapeutic, and Prognostic Value of the m(6)A Writer Complex in Hepatocellular Carcinoma. Front Cell Dev Biol. 2022;10:822011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 74. | Shi Y, Zhuang Y, Zhang J, Chen M, Wu S. METTL14 Inhibits Hepatocellular Carcinoma Metastasis Through Regulating EGFR/PI3K/AKT Signaling Pathway in an m6A-Dependent Manner. Cancer Manag Res. 2020;12:13173-13184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 75. | Fan Z, Yang G, Zhang W, Liu Q, Liu G, Liu P, Xu L, Wang J, Yan Z, Han H, Liu R, Shu M. Hypoxia blocks ferroptosis of hepatocellular carcinoma via suppression of METTL14 triggered YTHDF2-dependent silencing of SLC7A11. J Cell Mol Med. 2021;25:10197-10212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 117] [Article Influence: 29.3] [Reference Citation Analysis (1)] |
| 76. | Zhou T, Li S, Xiang D, Liu J, Sun W, Cui X, Ning B, Li X, Cheng Z, Jiang W, Zhang C, Liang X, Li L, Cheng X, Hui L, Wang H, Ding J. m6A RNA methylation-mediated HNF3γ reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct Target Ther. 2020;5:296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 77. | Du L, Li Y, Kang M, Feng M, Ren Y, Dai H, Wang Y, Wang Y, Tang B. USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 2021;81:3822-3834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 92] [Reference Citation Analysis (0)] |
| 78. | Liu Q, Qi J, Li W, Tian X, Zhang J, Liu F, Lu X, Zang H, Liu C, Ma C, Yu Y, Jiang S. Therapeutic effect and transcriptome-methylome characteristics of METTL3 inhibition in liver hepatocellular carcinoma. Cancer Cell Int. 2023;23:298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 79. | Li R, Li S, Shen L, Li J, Zhang D, Yu J, Huang L, Liu N, Lu H, Xu M. M6A-modified BFSP1 induces aerobic glycolysis to promote liver cancer growth and metastasis through upregulating tropomodulin 4. Mol Biomed. 2025;6:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 80. | Tonini V, Zanni M. Pancreatic cancer in 2021: What you need to know to win. World J Gastroenterol. 2021;27:5851-5889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 90] [Cited by in RCA: 80] [Article Influence: 20.0] [Reference Citation Analysis (7)] |
| 81. | Khan IA, Yoo BH, Masson O, Baron S, Corkery D, Dellaire G, Attardi LD, Rosen KV. ErbB2-dependent downregulation of a pro-apoptotic protein Perp is required for oncogenic transformation of breast epithelial cells. Oncogene. 2016;35:5759-5769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 82. | Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, Li X, Xu S, Miao J, Guo J, Zhang H, Gong J, Zhu F, Tian R, Shi C, Peng F, Feng Y, Yu S, Xie Y, Jiang J, Li M, Wei W, He C, Qin R. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 178] [Article Influence: 35.6] [Reference Citation Analysis (1)] |
| 83. | Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, Miao X, Yang L. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol. 2020;122:105731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 84. | Zhang C, Ou S, Zhou Y, Liu P, Zhang P, Li Z, Xu R, Li Y. m(6)A Methyltransferase METTL14-Mediated Upregulation of Cytidine Deaminase Promoting Gemcitabine Resistance in Pancreatic Cancer. Front Oncol. 2021;11:696371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 85. | Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D, Peng X, Ying P, Zhang M, Niu S, Li Y, Zhong R, Chang J, Miao X. N(6)-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Gut. 2020;69:2180-2192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 86. | Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, He S, Shimamoto F. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 233] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 87. | Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, Feng Y, Pan Q, Wan R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020;19:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 260] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 88. | Tang X, Liu S, Chen D, Zhao Z, Zhou J. The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol Lett. 2019;17:2473-2478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 89. | Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, Lu Y, Zeng J, Du F, Gong A, Xu M. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16:2259-2271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 170] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 90. | Hao YH, Yang CR, Shi WJ, Zhong XY. PSMD14 Transcriptionally Activated by MEF2A Promotes Pancreatic Cancer Development by Upregulating SPON2 Expression. Kaohsiung J Med Sci. 2025;e70007. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 91. | Rodas F, Vidal-Vidal JA, Herrera D, Brown-Brown DA, Vera D, Veliz J, Püschel P, Erices JI, Sánchez Hinojosa V, Tapia JC, Silva-Pavez E, Quezada-Monrás C, Mendoza-Soto P, Salazar-Onfray F, Carrasco C, Niechi I. Targeting the Endothelin-1 pathway to reduce invasion and chemoresistance in gallbladder cancer cells. Cancer Cell Int. 2023;23:318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 92. | Roa JC, García P, Kapoor VK, Maithel SK, Javle M, Koshiol J. Gallbladder cancer. Nat Rev Dis Primers. 2022;8:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 175] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 93. | Qin J, Cui Z, Zhou J, Zhang B, Lu R, Ding Y, Hu H, Cai J. IGF2BP3 drives gallbladder cancer progression by m6A-modified CLDN4 and inducing macrophage immunosuppressive polarization. Transl Oncol. 2023;37:101764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 94. | Zhang J, Yang K, Bu J, Yan J, Hu X, Liu K, Gao S, Tang S, Gao L, Chen W. IGF2BP3 promotes progression of gallbladder carcinoma by stabilizing KLK5 mRNA in N(6)-methyladenosine-dependent binding. Front Oncol. 2022;12:1035871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 95. | Bai X, Chen J, Zhang W, Zhou S, Dong L, Huang J, He X. YTHDF2 promotes gallbladder cancer progression and gemcitabine resistance via m6A-dependent DAPK3 degradation. Cancer Sci. 2023;114:4299-4313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 96. | Chen HD, Li F, Chen S, Zhong ZH, Gao PF, Gao WZ. METTL3-mediated N6-methyladenosine modification of DUSP5 mRNA promotes gallbladder-cancer progression. Cancer Gene Ther. 2022;29:1012-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 97. | Lin R, Zhan M, Yang L, Wang H, Shen H, Huang S, Huang X, Xu S, Zhang Z, Li W, Liu Q, Shi Y, Chen W, Yu J, Wang J. Deoxycholic acid modulates the progression of gallbladder cancer through N(6)-methyladenosine-dependent microRNA maturation. Oncogene. 2020;39:4983-5000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 98. | Deng M, Ran P, Chen L, Wang Y, Yu Z, Cai K, Feng J, Qin Z, Yin Y, Tan S, Liu Y, Xu C, Shi G, Ji Y, Zhao JY, Zhou J, Fan J, Hou Y, Ding C. Proteogenomic characterization of cholangiocarcinoma. Hepatology. 2023;77:411-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 99. | Vita F, Olaizola I, Amato F, Rae C, Marco S, Banales JM, Braconi C. Heterogeneity of Cholangiocarcinoma Immune Biology. Cells. 2023;12:846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 100. | Testa U, Pelosi E, Castelli G. Cholangiocarcinoma: Molecular Abnormalities and Cells of Origin. Technol Cancer Res Treat. 2023;22:15330338221128689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 101. | Cai J, Cui Z, Zhou J, Zhang B, Lu R, Ding Y, Hu H. METTL3 promotes glycolysis and cholangiocarcinoma progression by mediating the m6A modification of AKR1B10. Cancer Cell Int. 2022;22:385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 102. | Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ, Huang XT, Huang CS, Zhao W, Yin XY. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m(6)A-YTHDF2-dependent manner. Oncogene. 2022;41:1622-1633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 83] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 103. | Gao J, Fang Y, Chen J, Tang Z, Tian M, Jiang X, Tao C, Huang R, Zhu G, Qu W, Wu X, Zhou J, Fan J, Liu W, Shi Y. Methyltransferase like 3 inhibition limits intrahepatic cholangiocarcinoma metabolic reprogramming and potentiates the efficacy of chemotherapy. Oncogene. 2023;42:2507-2520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 104. | Dai Z, Zhu W, Hou Y, Zhang X, Ren X, Lei K, Liao J, Liu H, Chen Z, Peng S, Li S, Lin S, Kuang M. METTL5-mediated 18S rRNA m(6)A modification promotes oncogenic mRNA translation and intrahepatic cholangiocarcinoma progression. Mol Ther. 2023;31:3225-3242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 105. | Zheng H, Zheng WJ, Wang ZG, Tao YP, Huang ZP, Yang L, Ouyang L, Duan ZQ, Zhang YN, Chen BN, Xiang DM, Jin G, Fang L, Zhou F, Liang B. Decreased Expression of Programmed Death Ligand-L1 by Seven in Absentia Homolog 2 in Cholangiocarcinoma Enhances T-Cell-Mediated Antitumor Activity. Front Immunol. 2022;13:845193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 106. | Liu N, Zhang J, Chen W, Ma W, Wu T. The RNA methyltransferase METTL16 enhances cholangiocarcinoma growth through PRDM15-mediated FGFR4 expression. J Exp Clin Cancer Res. 2023;42:263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 107. | Xu H, Lin X, Li Z, He X, Li Y, Qiu L, Lu L, Liu B, Zhan M, He K. VIRMA facilitates intrahepatic cholangiocarcinoma progression through epigenetic augmentation of TMED2 and PARD3B mRNA stabilization. J Gastroenterol. 2023;58:925-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 108. | Zhou S, Yang K, Chen S, Lian G, Huang Y, Yao H, Zhao Y, Huang K, Yin D, Lin H, Li Y. CCL3 secreted by hepatocytes promotes the metastasis of intrahepatic cholangiocarcinoma by VIRMA-mediated N6-methyladenosine (m(6)A) modification. J Transl Med. 2023;21:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 109. | Xiao P, Meng Q, Liu Q, Lang Q, Yin Z, Li G, Li Z, Xu Y, Yu Z, Geng Q, Zhang Y, Liu L, Xie Y, Li L, Chen H, Pei T, Sun B. IGF2BP1-mediated N6-methyladenosine modification promotes intrahepatic cholangiocarcinoma progression. Cancer Lett. 2023;557:216075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 110. | Huang X, Zhu L, Wang L, Huang W, Tan L, Liu H, Huo J, Su T, Zhang M, Kuang M, Li X, Dai Z, Xu L. YTHDF1 promotes intrahepatic cholangiocarcinoma progression via regulating EGFR mRNA translation. J Gastroenterol Hepatol. 2022;37:1156-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 111. | Jin Z, Liu Y. The m6A reader YTHDC1-mediated lncRNA CTBP1-AS2 m6A modification accelerates cholangiocarcinoma progression. Heliyon. 2023;9:e19816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 112. | Huang CS, Zhu YQ, Xu QC, Chen S, Huang Y, Zhao G, Ni X, Liu B, Zhao W, Yin XY. YTHDF2 promotes intrahepatic cholangiocarcinoma progression and desensitises cisplatin treatment by increasing CDKN1B mRNA degradation. Clin Transl Med. 2022;12:e848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 113. | Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, Shen S, Jeong S, Li Z, Zhu Y, Wu T, Wu X, Wu R, Liu W, Wang HY, Chen L. M(6)A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res. 2021;81:4778-4793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 114. | Liu Y, Guo Q, Yang H, Zhang XW, Feng N, Wang JK, Liu TT, Zeng KW, Tu PF. Allosteric Regulation of IGF2BP1 as a Novel Strategy for the Activation of Tumor Immune Microenvironment. ACS Cent Sci. 2022;8:1102-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 89] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 115. | Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, Flavell RA, Zhu S, Li HB, Ding C, Wei H, Sun R, Peng H, Tian Z. METTL3-mediated m(6)A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun. 2021;12:5522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 137] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 116. | Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, Wang H, Song Y, Du Y, Cui B, Xue M, Zheng W, Kong X, Jiang K, Ding K, Lai L, Wang Q. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82:1660-1677.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 389] [Article Influence: 129.7] [Reference Citation Analysis (0)] |
| 117. | Yu H, Liu J, Bu X, Ma Z, Yao Y, Li J, Zhang T, Song W, Xiao X, Sun Y, Xiong W, Shi J, Dai P, Xiang B, Duan H, Yan X, Wu F, Zhang WC, Lin D, Hu H, Zhang H, Slack FJ, He HH, Freeman GJ, Wei W, Zhang J. Targeting METTL3 reprograms the tumor microenvironment to improve cancer immunotherapy. Cell Chem Biol. 2024;31:776-791.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 118. | Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, Hu Y, Qiu J, Pu L, Tang J, Wang X. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021;6:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 266] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 119. | Ma S, Yan J, Barr T, Zhang J, Chen Z, Wang LS, Sun JC, Chen J, Caligiuri MA, Yu J. The RNA m6A reader YTHDF2 controls NK cell antitumor and antiviral immunity. J Exp Med. 2021;218:e20210279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 129] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 120. | You Y, Wen D, Zeng L, Lu J, Xiao X, Chen Y, Song H, Liu Z. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int J Biol Sci. 2022;18:5001-5018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 121. | Ma E, Li J, Shen C, Gu Y, Zhang X, Li L, Zhao J, Wang Z. The m(6)A-related gene signature stratifies poor prognosis patients and characterizes immunosuppressive microenvironment in hepatocellular carcinoma. Front Immunol. 2023;14:1227593. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 122. | Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, Peng S, Chen K, Wang M, Gong S, Zhang R, Yin J, Li H, Yang Y, Liu H, Zhang J, Zhang H, Zhang A, Jiang H, Luo C, Yang CG. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134:17963-17971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 352] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 123. | Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, Gan J, Jiang H, Jia GF, Luo C, Yang CG. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373-384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 424] [Cited by in RCA: 520] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 124. | Wang T, Hong T, Huang Y, Su H, Wu F, Chen Y, Wei L, Huang W, Hua X, Xia Y, Xu J, Gan J, Yuan B, Feng Y, Zhang X, Yang CG, Zhou X. Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein. J Am Chem Soc. 2015;137:13736-13739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 125. | Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, Han L, Zhu Z, Lian F, Wei J, Deng Q, Wang Y, Wunderlich M, Gao Z, Pan G, Zhong D, Zhou H, Zhang N, Gan J, Jiang H, Mulloy JC, Qian Z, Chen J, Yang CG. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677-691.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 611] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 126. | Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, Deng X, Li H, Huang Y, Gao L, Li C, Zhao Z, Robinson S, Tan B, Qing Y, Qin X, Prince E, Xie J, Qin H, Li W, Shen C, Sun J, Kulkarni P, Weng H, Huang H, Chen Z, Zhang B, Wu X, Olsen MJ, Müschen M, Marcucci G, Salgia R, Li L, Fathi AT, Li Z, Mulloy JC, Wei M, Horne D, Chen J. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38:79-96.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 527] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 127. | Huff S, Tiwari SK, Gonzalez GM, Wang Y, Rana TM. m(6)A-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem Cells. ACS Chem Biol. 2021;16:324-333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 128. | Selberg S, Seli N, Kankuri E, Karelson M. Rational Design of Novel Anticancer Small-Molecule RNA m6A Demethylase ALKBH5 Inhibitors. ACS Omega. 2021;6:13310-13320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 129. | Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D, Hendrick AG, Webster NA, Andrews B, Fosbeary R, Guest P, Irigoyen N, Eleftheriou M, Gozdecka M, Dias JML, Bannister AJ, Vick B, Jeremias I, Vassiliou GS, Rausch O, Tzelepis K, Kouzarides T. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 796] [Article Influence: 199.0] [Reference Citation Analysis (0)] |
| 130. | Xiao H, Zhao R, Meng W, Liao Y. Effects and translatomics characteristics of a small-molecule inhibitor of METTL3 against non-small cell lung cancer. J Pharm Anal. 2023;13:625-639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 131. | Ma W, Wu T. RNA m6A modification in liver biology and its implication in hepatic diseases and carcinogenesis. Am J Physiol Cell Physiol. 2022;323:C1190-C1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 132. | Moroz-Omori EV, Huang D, Kumar Bedi R, Cheriyamkunnel SJ, Bochenkova E, Dolbois A, Rzeczkowski MD, Li Y, Wiedmer L, Caflisch A. METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes. ChemMedChem. 2021;16:3035-3043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 113] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 133. | Qiao K, Liu Y, Xu Z, Zhang H, Zhang H, Zhang C, Chang Z, Lu X, Li Z, Luo C, Liu Y, Yang C, Sun T. RNA m6A methylation promotes the formation of vasculogenic mimicry in hepatocellular carcinoma via Hippo pathway. Angiogenesis. 2021;24:83-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |