Published online May 15, 2025. doi: 10.4251/wjgo.v17.i5.102459
Revised: February 17, 2025
Accepted: March 10, 2025
Published online: May 15, 2025
Processing time: 208 Days and 0.8 Hours
Stage IV pancreatic cancer (PC) has a poor prognosis and lacks individualized prognostic tools. Current survival prediction models are limited, and there is a need for more accurate, personalized methods. The Surveillance, Epidemiology, and End Results (SEER) database offers a valuable resource for studying large patient cohorts, yet machine learning-based nomograms for stage IV PC prognosis remain underexplored. This study hypothesizes that a machine learning-based nomogram can predict cancer-specific survival (CSS) and overall survival (OS) with high accuracy in stage IV PC patients.
To construct and validate a machine learning-based nomogram for predicting survival in stage IV PC patients using real-world data.
Clinical data from stage IV PC patients diagnosed via pathology from 2000 to 2019 were extracted from the SEER database. Patients were randomly divided into a training set and a validation set in a 7:3 ratio. Multivariate Cox proportional hazards, Least Absolute Shrinkage and Selection Operator regression, and Random Survival Forest models were used to identify prognostic variables. A nomogram was constructed to predict CSS and OS at 6, 12, and 18 months. The C-index, receiver operating characteristic curves, and calibration curves were used to evaluate the model’s predictive performance.
A total of 1662 patients were included (1163 in the training set, 499 in the validation set). The median follow-up times were 4 months [interquartile range (IQR): 1-10 months] for the training set and 4 months (IQR: 1-11 months) for the validation set. Key independent prognostic factors identified included age, race, marital status, tumor location, N stage, grade, surgery, chemotherapy, and liver metastasis. The nomogram accurately predicted OS and CSS at 6, 12, and 18 months, with a C-index of 0.727 (OS) and 0.727 (CSS) in the training set, and 0.719 (OS) and 0.716 (CSS) in the validation set. Calibration curves demonstrated excellent model accuracy.
The nomogram developed using age, grade, chemotherapy, surgery, and liver metastasis as predictors can reliably estimate survival outcomes for stage IV PC patients and offers a potential tool for individualized clinical decision-making.
Core Tip: This study develops and validates a machine learning-based nomogram to predict survival in patients with stage IV pancreatic ductal adenocarcinoma. Using data from the Surveillance, Epidemiology, and End Results program, the model integrates prognostic factors and demonstrates high accuracy in predicting cancer-specific survival and overall survival at various time points. This nomogram offers a personalized approach to survival prediction, potentially guiding clinical decision-making and treatment strategies for advanced pancreatic cancer patients.
- Citation: Huang K, Chen Z, Yuan XZ, He YS, Lan X, Du CY. Development and validation of machine learning nomograms for predicting survival in stage IV pancreatic cancer: A retrospective study. World J Gastrointest Oncol 2025; 17(5): 102459
- URL: https://www.wjgnet.com/1948-5204/full/v17/i5/102459.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i5.102459
Pancreatic cancer (PC) is a significant human health issue and, by 2025, is projected to surpass breast cancer as the third leading cause of cancer-related deaths[1]. In the United States, an estimated 66440 new cases and 51750 deaths due to PC were reported in 2024. PC is often asymptomatic in its early stages, with more than half of patients presenting with distant organ metastasis at the time of initial diagnosis[2]. Consequently, the prognosis is very poor, with a 5-year relative survival rate of only 12.8%[2] In clinical practice, considerable heterogeneity in survival outcomes has been observed among patients with stage IV PC, highlighting the need for an individualized survival prediction tool for this population.
Nomograms, which are visual tools incorporating multiple prognostic factors to predict patient survival, aid in personalized treatment planning and clinical decision-making and are widely used in cancer prognosis evaluation[3-6].
Machine learning, a core technique within artificial intelligence, employs algorithms to analyze data, learn from patterns, and predict real-world events with high accuracy, and is increasingly applied in health assessment, medical decision-making, prognosis, and personalized treatment[7-9].
This study leverages the large sample size and comprehensive clinical data from the United State Surveillance, Epidemiology, and End Results (SEER) database to develop a prognostic nomogram for stage IV PC patients using machine learning, with the aim of providing individualized prognostic assessments to improve clinical decision-making.
Utilizing SEER*Stat v8.3.9 software, clinical data of patients with stage IV PC diagnosed pathologically between 2000 and 2019 were extracted from the SEER database.
Patients were included in the study if they met all of the following criteria: (1) Had a diagnosis of primary PC at stage IV according to the tumor-node-metastasis (TNM) classification; (2) Were diagnosed pathologically; (3) Were diagnosed between 2000 and 2019; and (4) Had International Classification of Diseases-O-3 codes of 8576/3, 8560/3, 8500/3, 8490/3, 8480/3, 8035/3, or 8020/3. Exclusion criteria were as follows: (1) Patients with multifocal tumors; (2) Cases diagnosed posthumously via autopsy or death certificate; (3) Patients with inaccessible study metrics; and (4) Individuals with incomplete clinical or follow-up information.
Data extracted included patients' age at diagnosis, race, gender, primary tumor location, treatment information, survival time, and survival outcomes.
The primary endpoints of this study were cancer-specific survival (CSS) and overall survival (OS). CSS was defined as the time (in months) from the start of follow-up to death due to PC, with deaths from other causes, censoring at the conclusion of follow-up, or loss to follow-up considered censored events. OS was defined as the time (in months) from the start of follow-up to death from any cause, with similar criteria for censored events applied[3]. Statistical analyses were conducted using Stata/MP 16.0 and R (version 4.2.3). Normally distributed data are expressed as mean ± SD, while non-normal data are presented as median [interquartile range (IQR)]. Categorical variables are presented as rates (percentages) and compared using the chi-square test. The dataset was randomly divided into a training set (70%) for nomogram development and internal validation, and a validation set (30%) for external validation[10,11]. Model accuracy was validated by constructing calibration curves through 1000 bootstrap resamples. The model's discriminative ability was assessed by calculating the C-index, and survival rates were estimated using the Kaplan-Meier method, with survival rate comparisons made using the log-rank test. The variable selection process for model construction involved an initial univariate Cox proportional hazards analysis, followed by the integration of significant factors into a multivariate Cox proportional hazards model based on the results of the univariate analysis and their clinical significance[10,11] This process ultimately identified the independent prognostic factors. The multivariate Cox proportional hazards model was used to calculate hazard ratios (HR) and their corresponding 95%CI[11]. In this analysis, multicollinearity was evaluated using variance inflation factors (VIFs) and pairwise Pearson correlation coefficients[12,13]. A VIF value exceeding 5 to 10 indicated significant multicollinearity, necessitating exclusion of the associated variables prior to further analysis[14]. Pairwise Pearson correlation coefficients were computed to assess collinearity among variables, with a threshold of < 0.7 considered as no significant collinearity[15,16]. To refine variable selection and prevent overfitting, Least Absolute Shrinkage and Selection Operator (LASSO) regression was applied, with a 10-fold cross-validation approach used to determine the optimal λ parameter. Variables with nonzero coefficients were retained to ensure that only the most relevant prognostic factors were included in the final model.
In parallel, a random survival forest model was employed for independent prognostic analysis. The random survival forest, a machine learning algorithm known for its robustness, operates independently of the proportional hazards assumption or log-linearity and utilizes two-stage random sampling to mitigate overfitting. Variable importance scores were computed to further assess the contribution of each predictor to OS and CSS[10]. Model evaluation included receiver operating characteristic (ROC) curve analysis, where area under the curve (AUC) values at 6, 12, and 18 months were calculated to assess the model’s discriminative ability.
The optimal cutoff values for risk stratification were determined using X-tile software (version 3.6.1), and patients were categorized into low-risk and high-risk groups accordingly. Kaplan-Meier curves were generated to estimate survival probabilities, and the log-rank test was used to compare survival differences between risk groups. To evaluate the potential clinical benefit of the nomogram in guiding individualized treatment decisions, decision curve analysis (DCA) was performed, comparing its net benefit to that of the traditional TNM staging system. All statistical tests were two-tailed, with P < 0.05 considered statistically significant.
A total of 1662 patients were enrolled in this study, with 1163 in the training set and 499 in the validation set. The clinicopathological characteristics of the two groups are presented in Table 1. As shown in Table 1, baseline characteristics were well-balanced between the two groups (all P > 0.05).
| Variables | Total cohort (n = 1662) | Training cohort (n = 1163) | Validation cohort (n = 499) | P value |
| Age (yeas) | 0.281 | |||
| < 60 | 445 (26.77) | 313 (26.91) | 132 (26.45) | |
| 60-75 | 838 (50.42) | 597 (51.33) | 241 (48.30) | |
| > 75 | 379 (22.80) | 253 (21.75) | 126 (25.25) | |
| Sex | 0.062 | |||
| Female | 803 (48.32) | 544 (46.78) | 259 (51.90) | |
| Male | 859 (51.68) | 619 (53.22) | 240 (48.10) | |
| Race | 0.191 | |||
| White | 1326 (79.78) | 937 (80.57) | 389 (77.96) | |
| Black | 172 (10.35) | 110 (9.46) | 62 (12.42) | |
| Other | 164 (9.87) | 116 (9.97) | 48 (9.62) | |
| Marital status | 0.935 | |||
| Married | 895 (53.85) | 630 (54.17) | 265 (53.11) | |
| Unmarried | 273 (16.43) | 189 (16.25) | 84 (16.83) | |
| Divorced | 429 (25.81) | 297 (25.54) | 132 (26.45) | |
| Unknown | 65 (3.91) | 47 (4.04) | 18 (3.61) | |
| Tumor location | 0.806 | |||
| Head | 621 (37.36) | 426 (36.63) | 195 (39.08) | |
| Body | 258 (15.52) | 182 (15.65) | 76 (15.23) | |
| Tail | 334 (20.10) | 235 (20.21) | 99 (19.84) | |
| Other | 449 (27.02) | 320 (27.52) | 129 (25.85) | |
| T stage | 0.174 | |||
| T1/T2 | 411 (24.73) | 303 (26.05) | 108 (21.64) | |
| T3 | 575 (34.60) | 386 (33.19) | 189 (37.88) | |
| T4 | 337 (20.28) | 236 (20.29) | 101 (20.24) | |
| TX | 339 (20.40) | 238 (20.46) | 101 (20.24) | |
| N stage | 0.833 | |||
| N0 | 781 (46.99) | 547 (47.03) | 234 (46.89) | |
| N1 | 599 (36.04) | 415 (35.68) | 184 (36.87) | |
| NX | 282 (16.97) | 201 (17.28) | 81 (16.23) | |
| Grade | 0.701 | |||
| Grade I/II | 296 (17.81) | 204 (17.54) | 92 (18.44) | |
| Grade III/IV | 292 (17.57) | 200 (17.20) | 92 (18.44) | |
| Unknown | 1074 (64.62) | 759 (65.26) | 315 (63.13) | |
| Surgery | 0.672 | |||
| No | 1482 (89.17) | 1040 (89.42) | 442 (88.58) | |
| Yes | 180 (10.83) | 123 (10.58) | 57 (11.42) | |
| Chemotherapy | 0.706 | |||
| No | 686 (41.28) | 484 (41.62) | 202 (40.48) | |
| Yes | 976 (58.72) | 679 (58.38) | 297 (59.52) | |
| Radiotherapy | 0.822 | |||
| No | 1557 (93.68) | 1088 (93.55) | 469 (93.99) | |
| Yes | 105 (6.32) | 75 (6.45) | 30 (6.01) | |
| Bone metastasis | 0.172 | |||
| No | 1549 (93.20) | 1077 (92.61) | 472 (94.59) | |
| Yes | 113 (6.80) | 86 (7.39) | 27 (5.41) | |
| Liver metastasis | 0.384 | |||
| No | 615 (37.00) | 422 (36.29) | 193 (38.68) | |
| Yes | 1047 (63.00) | 741 (63.71) | 306 (61.32) | |
| Lung metastasis | 0.462 | |||
| No | 1278 (76.90) | 888 (76.35) | 390 (78.16) | |
| Yes | 384 (23.10) | 275 (23.65) | 109 (21.84) | |
Univariate Cox proportional hazards model: Among the 1163 patients in the training set, the median follow-up duration was 4 months (IQR, 1-10 months). The results of the univariate Cox proportional hazards model analysis are detailed in Table 2. The analysis indicated that age, race, marital status, tumor location, T stage, N stage, grade, surgery, che
| Variables | OS | CSS | ||
| HR (95%CI) | P value | HR (95%CI) | P value | |
| Age (years) | ||||
| < 60 | 1 (reference) | 1 (reference) | ||
| 60-75 | 1.26 (1.10, 1.45) | 0.001 | 1.25 (1.08, 1.44) | 0.002 |
| > 75 | 1.70 (1.44, 2.02) | < 0.001 | 1.71 (1.44, 2.03) | < 0.001 |
| Sex | ||||
| Female | 1 (reference) | 1 (reference) | ||
| Male | 0.97 (0.87, 1.09) | 0.645 | 0.98 (0.87, 1.10) | 0.730 |
| Race | ||||
| White | 1 (reference) | 1 (reference) | ||
| Black | 1.26 (1.03, 1.54) | 0.024 | 1.20 (0.98, 1.48) | 0.078 |
| Other | 0.88 (0.72, 1.07) | 0.195 | 0.89 (0.73, 1.08) | 0.237 |
| Marital status | ||||
| Married | 1 (reference) | 1 (reference) | ||
| Unmarried | 1.17 (0.99, 1.38) | 0.069 | 1.16 (0.98, 1.38) | 0.079 |
| Divorced | 1.54 (1.34, 1.77) | < 0.001 | 1.52 (1.32, 1.76) | < 0.001 |
| Unknown | 1.72 (1.28, 2.33) | < 0.001 | 1.71 (1.26, 2.32) | 0.001 |
| Tumor location | ||||
| Head | 1 (reference) | 1 (reference) | ||
| Body | 1.34 (1.13, 1.60) | 0.001 | 1.35 (1.13, 1.61) | 0.001 |
| Tail | 1.20 (1.02, 1.41) | 0.027 | 1.22 (1.03, 1.43) | 0.020 |
| Other | 1.42 (1.22, 1.64) | < 0.001 | 1.42 (1.22, 1.65) | < 0.001 |
| T stage | ||||
| T1/T2 | 1 (reference) | 1 (reference) | ||
| T3 | 0.76 (0.65, 0.89) | < 0.001 | 0.77 (0.66, 0.90) | 0.001 |
| T4 | 0.92 (0.77, 1.09) | 0.339 | 0.93 (0.78, 1.11) | 0.413 |
| TX | 1.13 (0.96, 1.35) | 0.148 | 1.10 (0.92, 1.31) | 0.288 |
| N stage | ||||
| N0 | 1 (reference) | 1 (reference) | ||
| N1 | 0.96 (0.84, 1.09) | 0.488 | 0.98 (0.86, 1.12) | 0.789 |
| NX | 1.44 (1.22, 1.69) | < 0.001 | 1.42 (1.20, 1.68) | < 0.001 |
| Grade | ||||
| Grade I/II | 1 (reference) | 1 (reference) | ||
| Grade III/IV | 1.68 (1.37, 2.05) | < 0.001 | 1.69 (1.38, 2.08) | < 0.001 |
| Unknown | 1.74 (1.48, 2.04) | < 0.001 | 1.72 (1.46, 2.02) | < 0.001 |
| Surgery | ||||
| No | 1 (reference) | 1 (reference) | ||
| Yes | 0.46 (0.38, 0.56) | < 0.001 | 0.47 (0.38, 0.57) | < 0.001 |
| Chemotherapy | ||||
| No | 1 (reference) | 1 (reference) | ||
| Yes | 0.33 (0.30, 0.38) | < 0.001 | 0.34 (0.30, 0.38) | < 0.001 |
| Radiotherapy | ||||
| No | 1 (reference) | 1 (reference) | ||
| Yes | 0.76 (0.60, 0.97) | 0.027 | 0.78 (0.61, 0.99) | 0.040 |
| Bone metastasis | ||||
| No | 1 (reference) | 1 (reference) | ||
| Yes | 1.10 (0.88, 1.37) | 0.397 | 1.15 (0.92, 1.44) | 0.206 |
| Liver metastasis | ||||
| No | 1 (reference) | 1 (reference) | ||
| Yes | 1.49 (1.32, 1.68) | < 0.001 | 1.50 (1.32, 1.70) | < 0.001 |
| Lung metastasis | ||||
| No | 1 (reference) | 1 (reference) | ||
| Yes | 1.02 (0.89, 1.17) | 0.819 | 1.01 (0.88, 1.16) | 0.910 |
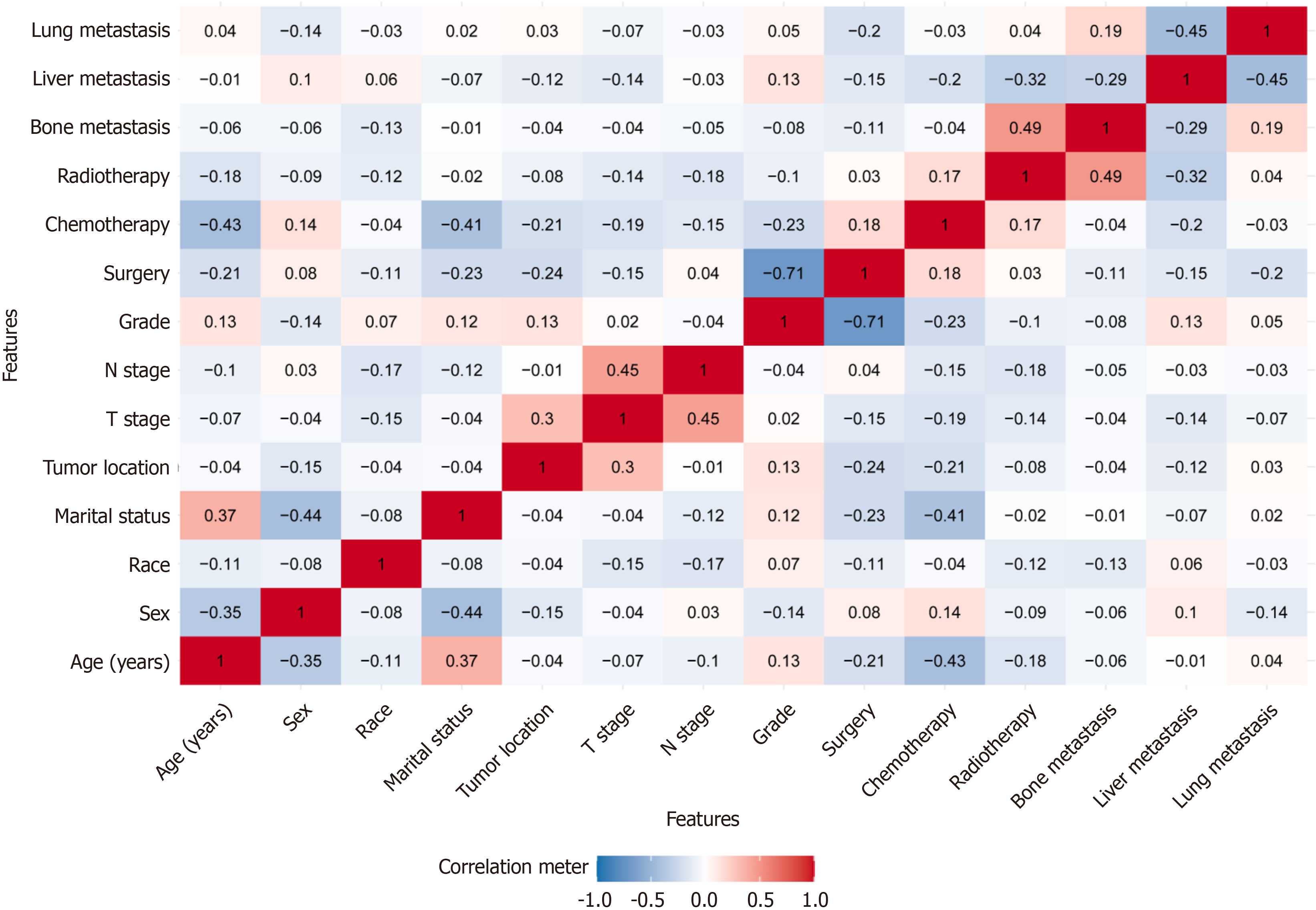
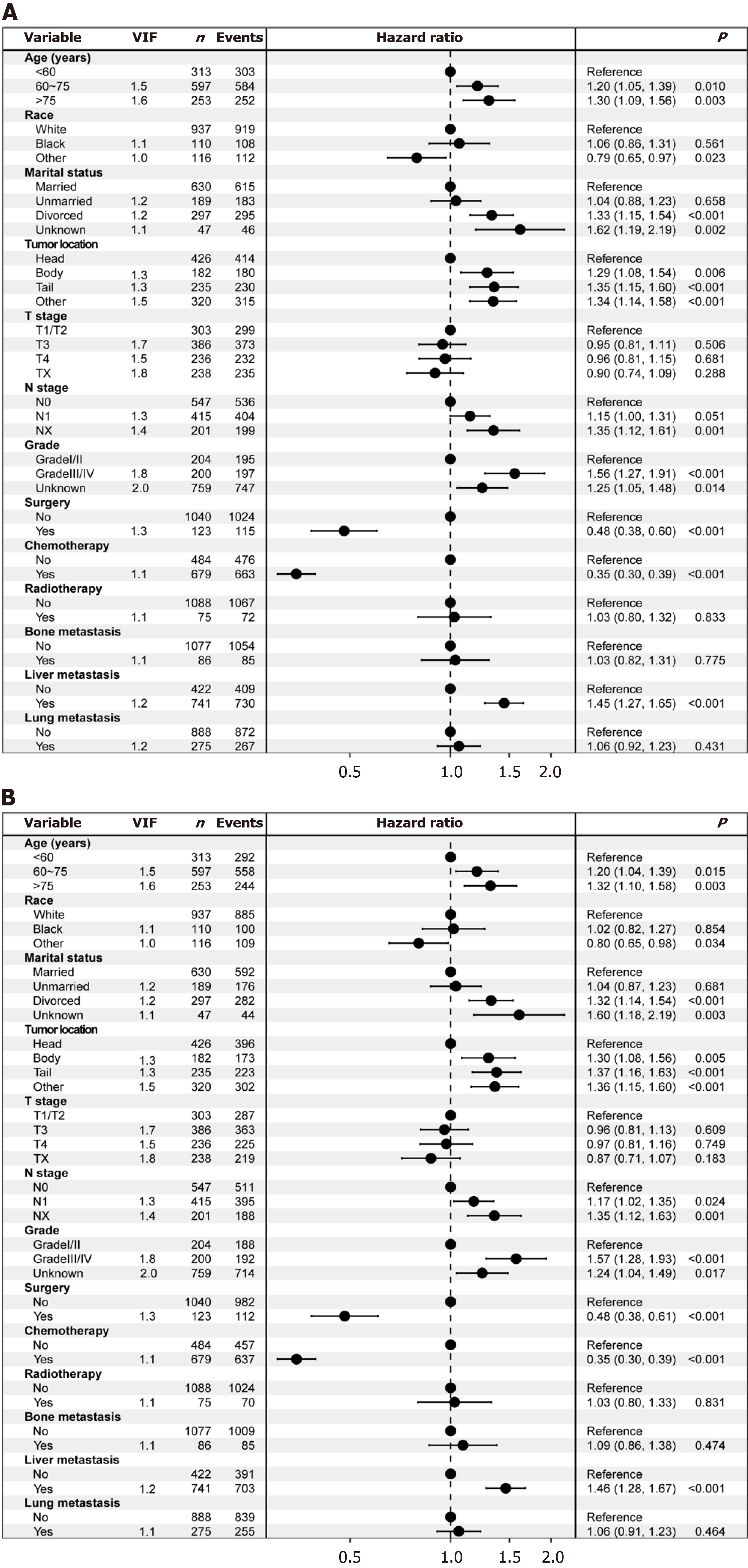
Multivariate Cox proportional hazards model: Before initiating multivariate Cox proportional hazards modeling, we assessed potential multicollinearity among the variables. The results indicated that correlation coefficients for all variable pairs (except surgery and grade) were below 0.7 (Figure 1), and VIF values were all less than 5, suggesting that multicollinearity was not a concern among the independent variables. The multivariate Cox proportional hazards model analysis identified age, race, marital status, tumor location, N stage, grade, surgery, chemotherapy, and liver metastasis as independent predictors of OS (all P < 0.05); additionally, these factors were also independently associated with CSS (Figure 2).
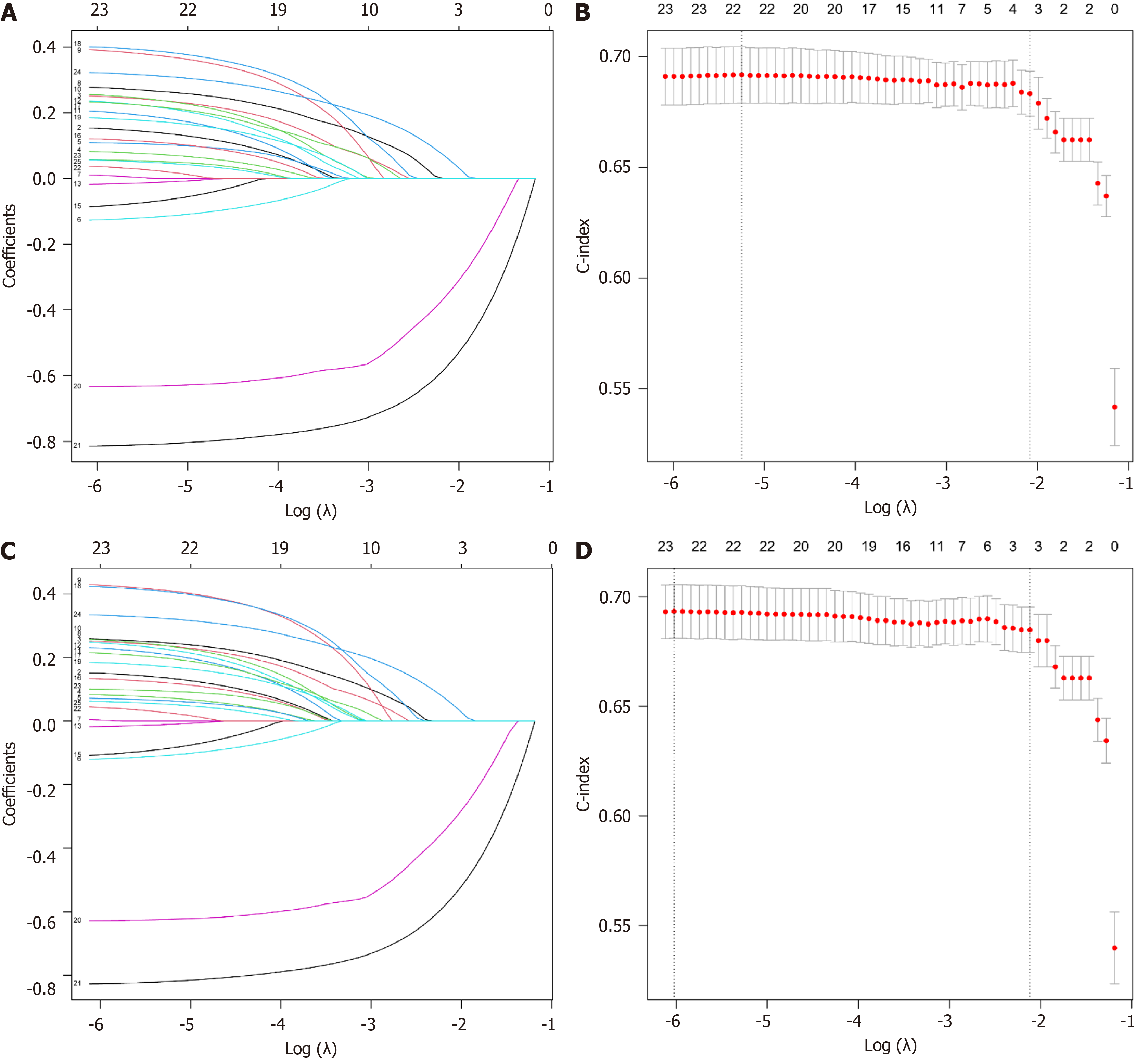
LASSO regression model: To accurately and comprehensively identify independent factors affecting the prognosis of patients with stage IV PC and to minimize the impact of variable collinearity on the results, we concurrently employed LASSO regression analysis with a 10-fold cross-validation approach to further refine the variables. The results indicated that surgery, chemotherapy, and liver metastasis were associated with both OS and CSS (Figure 3).
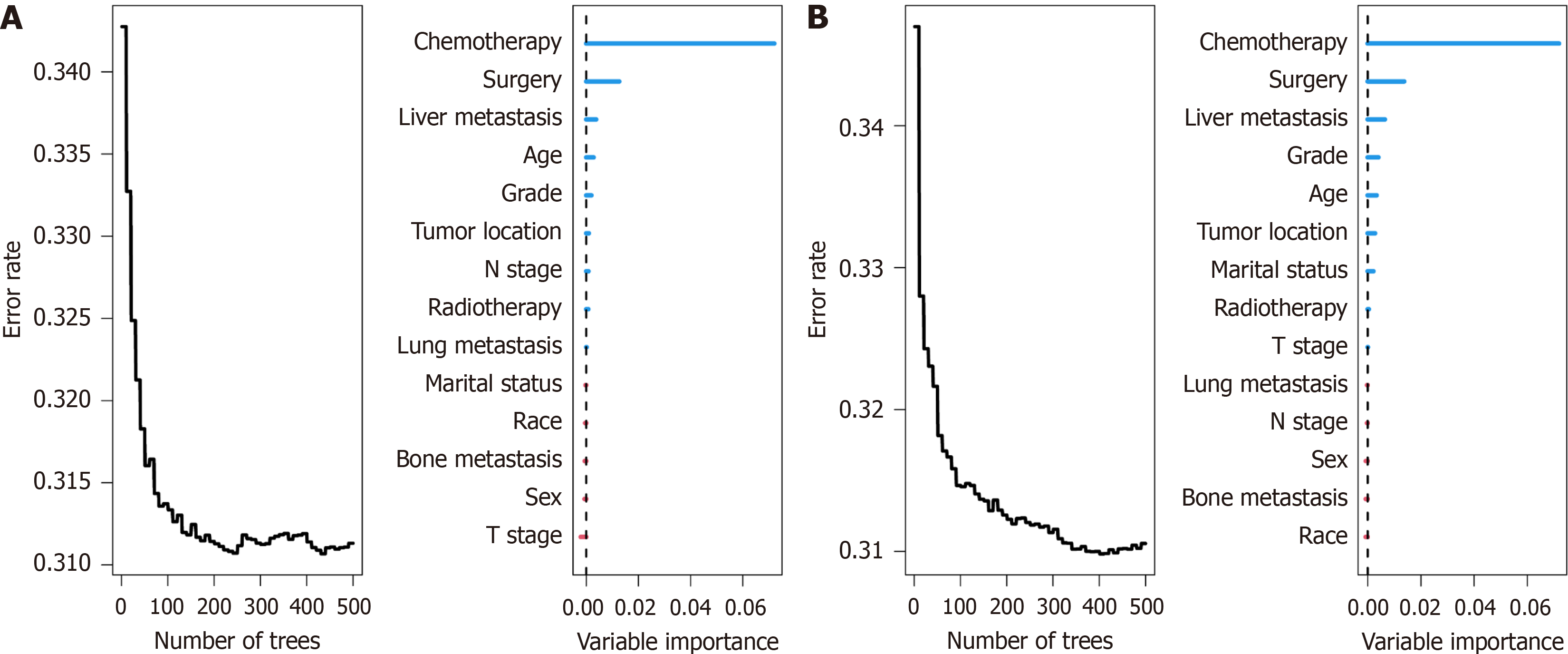
Random survival forest: The random survival forest is a machine learning algorithm characterized by high robustness, as it does not rely on assumptions such as the proportional hazards assumption or log-linearity. It employs two random sampling processes to mitigate issues of overfitting within the algorithm. In this study, we further evaluated the importance of variables for OS and CSS using this method. The results revealed that the top five variables ranked by their importance for OS were chemotherapy, surgery, liver metastasis, age, and grade (Figure 4A), while for CSS, the top five variables were chemotherapy, surgery, liver metastasis, grade, and age (Figure 4B).
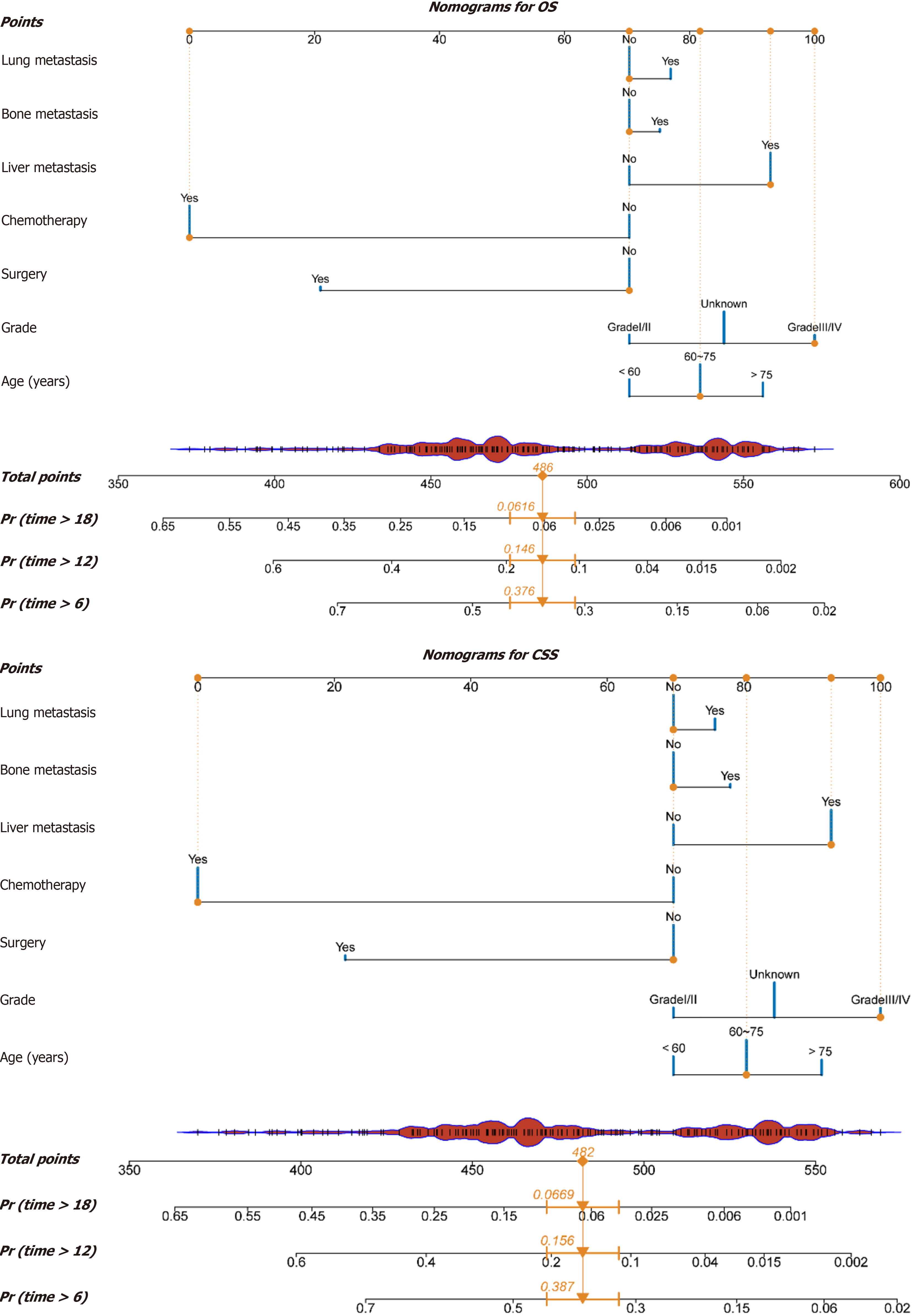
Based on the results from the multivariate Cox proportional hazards model, the LASSO regression model, and the random survival forest, combined with clinical relevance, we ultimately selected seven variables - age, tumor grade, surgical resection, chemotherapy, liver metastasis, bone metastasis, and lung metastasis - to construct a nomogram for predicting the prognosis of PC patients. This tool was used to predict 6-month, 12-month, and 18-month OS and CSS. Each variable in the figure was assigned a corresponding score, and the sum of all variable scores represented the total score (Total Points). A lower score indicated a better prognosis. The Total Points could be used to predict the OS and CSS of PC patients at different time points (Figure 5).
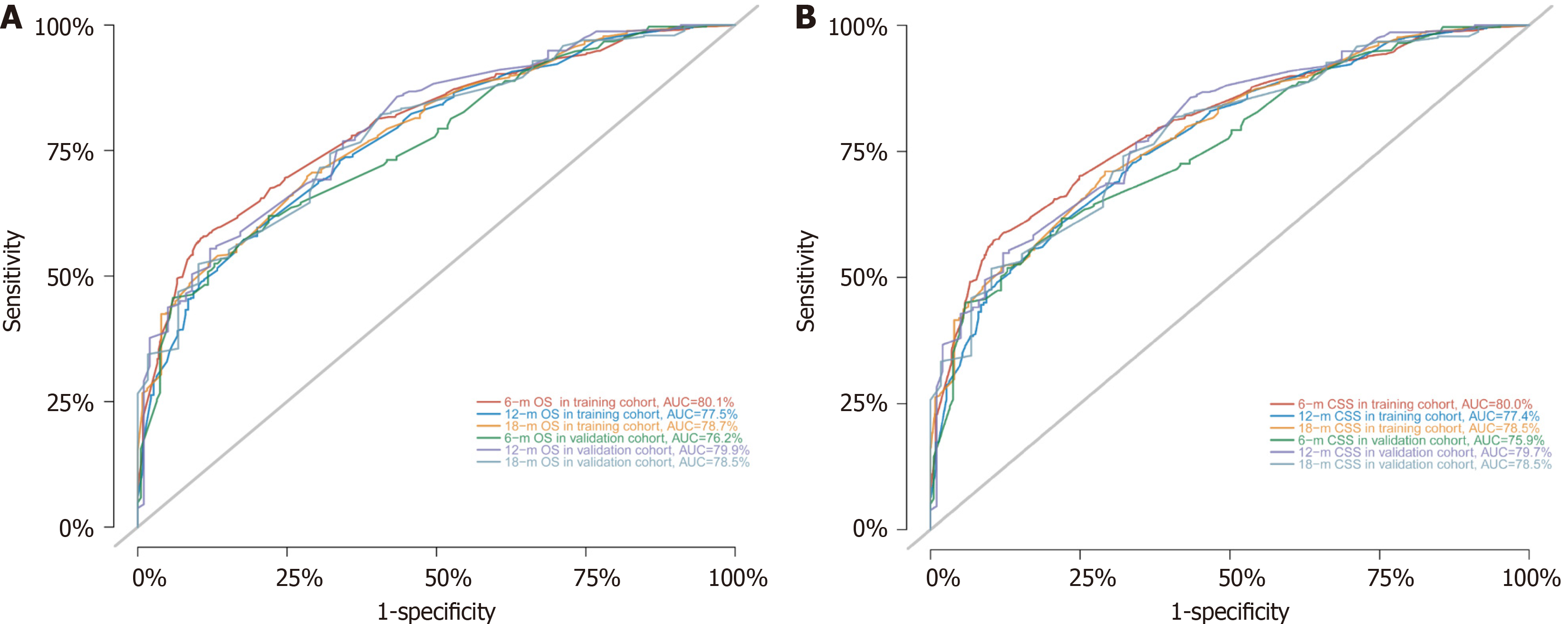
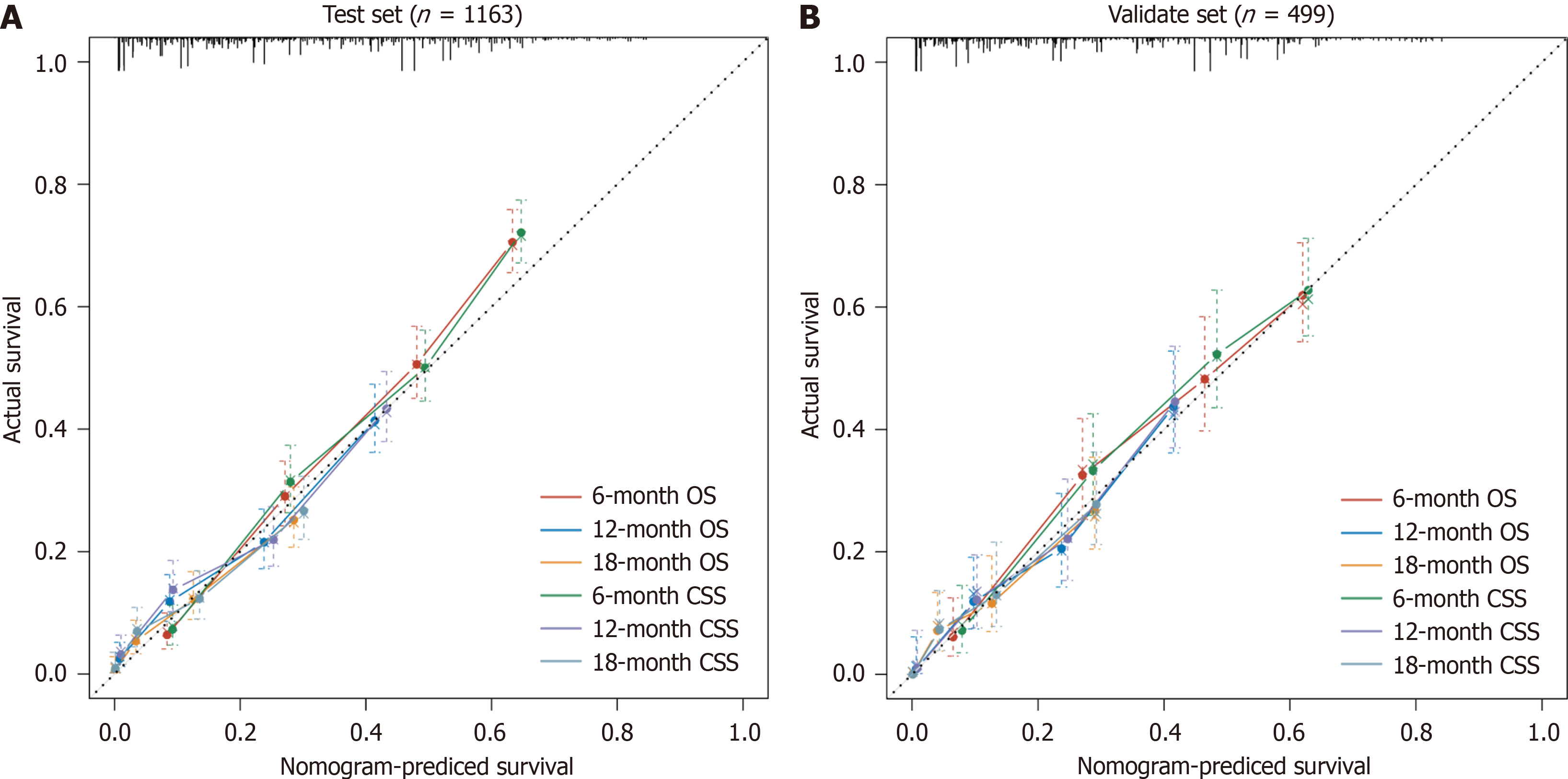
To assess the discriminatory power of the model, the C-index and AUC values were calculated for both the training and validation sets (Table 3), and ROC curves were plotted (Figure 6). The results indicated that the model exhibited strong predictive value in both the training and validation sets. To evaluate model accuracy, internal and external validations were performed using the bootstrap method with B = 1000 resamples, and calibration curves were drawn. The validation results showed that, in both the training set (internal) and validation set (external), the calibration curves for 6-, 12-, and 18-month OS and CSS closely aligned with the ideal 45-degree reference line, suggesting good consistency between the model's predicted values and the actual observed values (Figure 7).
| Data set | OS | CSS | ||||||||
| C-index (95%CI) | AUC | C-index (95%CI) | AUC | |||||||
| 6-month | 12-month | 18-month | 6-month | 12-month | 18-month | |||||
| Training set | 0.727 | 0.711-0.743 | 0.801 | 0.775 | 0.787 | 0.727 | 0.711-0.743 | 0.800 | 0.774 | 0.785 |
| Validation set | 0.719 | 0.695-0.744 | 0.762 | 0.799 | 0.785 | 0.716 | 0.691-0.741 | 0.759 | 0.797 | 0.785 |
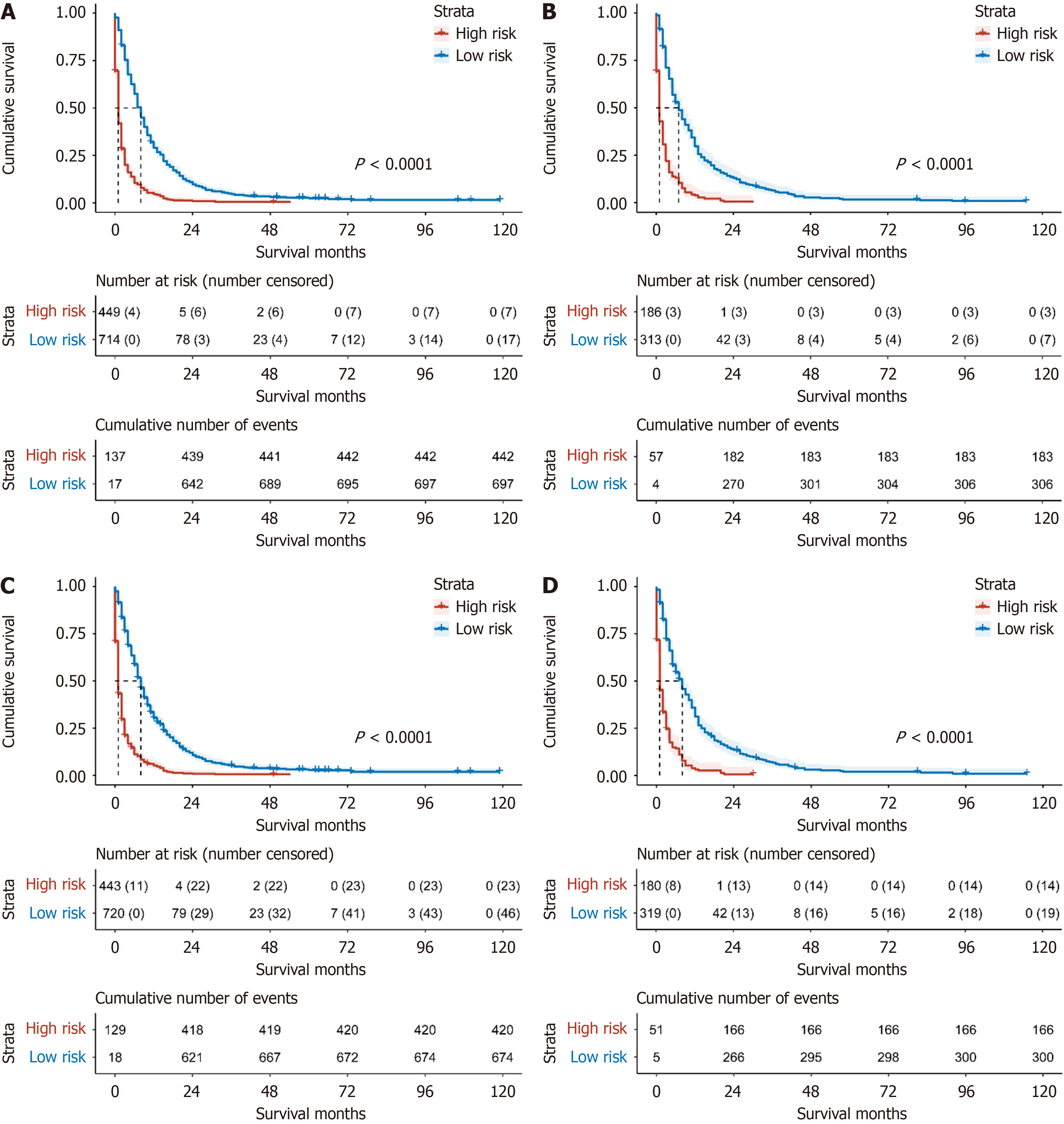
To further validate the clinical utility of the model in practice, we calculated each patient's Total Points based on the constructed nomogram in both the training and validation sets and used X-tile software to stratify risk within the training set. For both OS and CSS, the cutoff values for the low-risk and high-risk groups were determined to be 148 and 189.7 points, respectively. Specifically, for OS, patients with Total Points less than 148 were considered low risk, while those with Total Points greater than 148 were categorized as high risk. For CSS, patients with Total Points less than 189.7 were classified as low risk, whereas those with Total Points greater than 189.7 were classified as high risk. The results demonstrated that the model effectively differentiated the survival prognosis of stage IV PC patients in both the validation and training sets (both P < 0.0001; Figure 8).
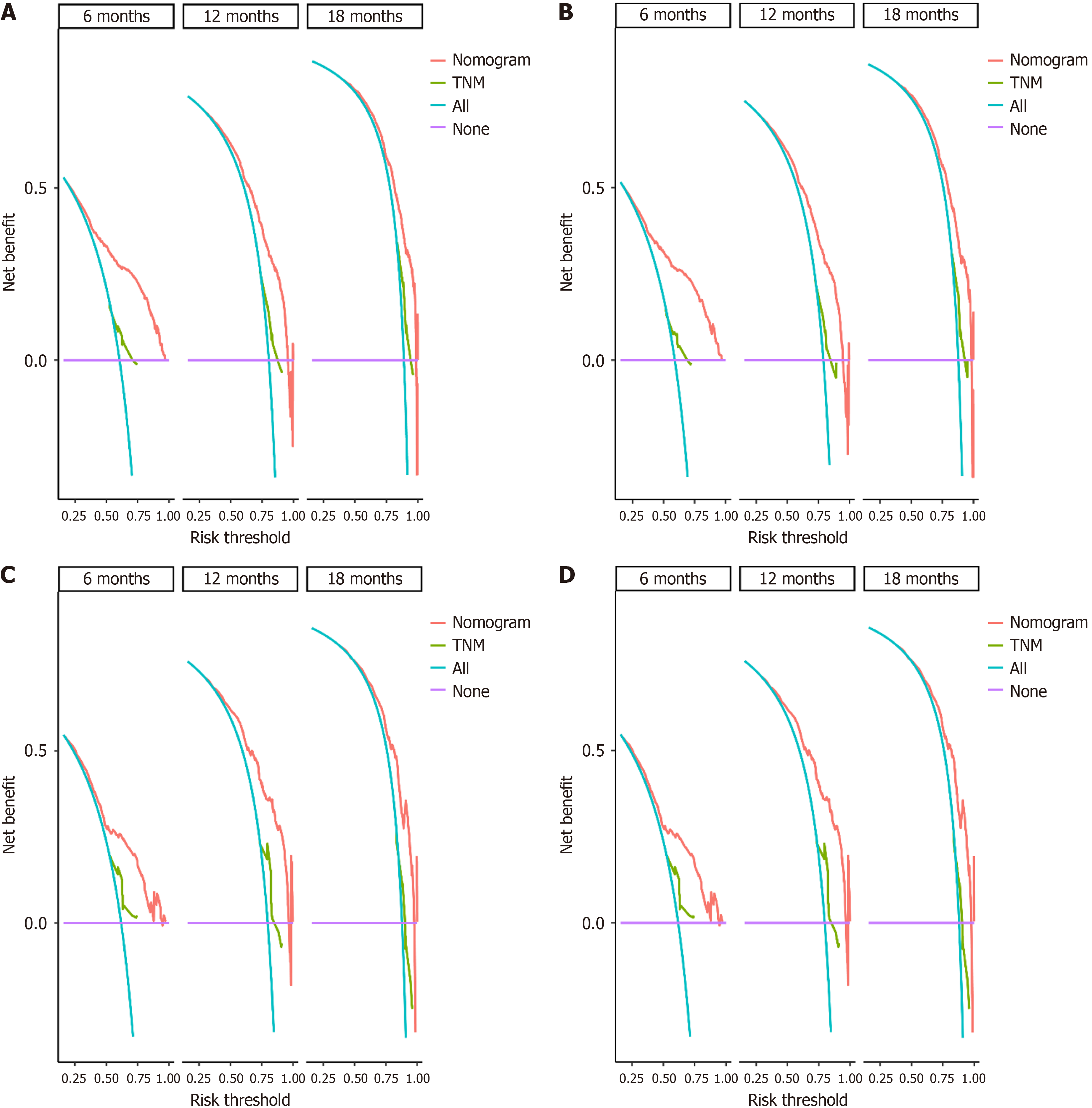
DCA, a novel method for evaluating predictive models, places greater emphasis on assessing clinical benefit. In this study, we employed DCA to determine whether the model could provide benefits in clinical practice. The results revealed that, compared with the TNM staging system, the nomogram model demonstrated a greater net benefit in predicting patients' OS and CSS (Figure 9).
PC, a highly aggressive solid tumor affecting the digestive organs, is associated with a grim prognosis. At the time of initial presentation, approximately 80% of patients exhibit locally advanced disease or distant metastases, precluding them from surgical intervention[17,18]. In clinical practice, we have observed that survival outcomes for patients with stage IV PC at initial diagnosis can vary significantly. However, there are few clinical prognostic assessment tools specifically designed for this population. This study utilized the SEER database to construct a prognostic nomogram using machine learning algorithms, aiming to provide precise and personalized prognostic assessments for stage IV PC patients and offer a reference for clinical decision-making.
In this study, we integrated results from the multivariate Cox proportional hazards model, LASSO regression model, and random survival forest, along with clinical relevance, to select seven variables: Age, tumor grade, surgical resection, chemotherapy, liver metastasis, bone metastasis, and lung metastasis. These variables were used to construct a nomogram for predicting patient prognosis. The nomogram was employed to predict 6-month, 12-month, and 18-month OS and CSS. Further validation demonstrated good discrimination and accuracy in both the training and validation sets, providing valuable guidance for clinical decision-making. A study by Shi et al[19] indicated that patients aged 65 years and older had significantly poorer OS. Consistent with Shi et al[19], our findings revealed a 20% increase in HR (OS) and HR (CSS) for patients aged 60-75 years compared to those under 60 years, with respective increases of 30% and 32% for patients over 75 years. Tumor grade was also identified as an independent risk factor for prognosis[20,21]. Shrikhande et al[22] reported that among 129 pancreatic ductal adenocarcinoma patients with liver metastases, only 11 who underwent surgery had a median OS of 11.4 months, compared to 5.9 months for those without surgery. Multiple studies have confirmed that patients with liver metastasis who undergo surgery experience better outcomes[2,23-26]. Our study reached a similar conclusion. Evidence indicates that over half of PC patients present with distant organ metastasis at diagnosis[2], and distant metastasis is an independent risk factor for prognosis[27]. Improving the survival prognosis of these patients remains a critical clinical challenge. Currently, the TNM staging system is widely used to predict prognosis in clinical practice; however, it has limitations, such as disregarding other factors affecting survival and lacking the ability to visualize individual survival outcomes[19].
Nomograms are extensively utilized in predicting tumor prognosis, assisting clinicians in devising individualized treatment plans[3,28-31]. In this study, we developed a predictive model based on independent prognostic factors for stage IV PC, which exhibited strong accuracy and consistency. This model can assist clinicians in making optimal clinical decisions. For example, a 65-year-old patient with grade III PC and liver metastases - but without bone or lung me
Moreover, this tool can aid clinicians in making medical decisions. Using the aforementioned patient as an example, we utilized the model to estimate OS and CSS with and without surgery and chemotherapy. This provides personalized, intuitive, and comprehensible objective parameters for clinical decision-making. Additionally, risk stratification based on the model's total score may guide postoperative follow-up strategies. Given the complex factors influencing OS, CSS is often considered more critical[32]. Consequently, we stratified patient risk based on CSS. With a total score of 482, exceeding the cutoff value of 189.7, this patient was identified as high-risk, necessitating more vigilant monitoring and a stringent treatment and follow-up plan. To our knowledge, there are no large-scale real-world studies on prognostic models for stage IV PC in the literature. This study has several strengths. First, we employed the random survival forest model, a robust machine learning algorithm within ensemble learning that is not constrained by assumptions such as proportional hazards or log-linearity, and mitigates overfitting through dual random sampling. Consequently, this algorithm yields more reliable and stable conclusions. Second, we independently applied LASSO and classical univariate and multivariate Cox proportional hazards models, while quantitatively assessing multicollinearity in the models using VIFs and pairwise Pearson correlation coefficients. Finally, this study included a total of 1662 patients, of whom 1628 (97.95%) experienced mortality events, and 1560 (93.86%) had cancer-specific deaths, providing ample statistical power for analysis.
However, this study has inherent limitations. As a retrospective study, despite stringent inclusion and exclusion criteria, selection bias is inevitable. Additionally, the SEER database, being a large-scale cancer registry, is subject to coding errors and missing values. Furthermore, due to data access restrictions, certain information (such as specific treatment protocols for chemotherapy, surgery, and radiation therapy, as well as data on tumor recurrence) is un
In conclusion, age, tumor grade, surgical resection, chemotherapy, and metastases to the liver, bone, and lung were identified as independent prognostic factors. The model developed from these factors offers a practical reference for clinical use.
This study developed a machine learning-based nomogram that effectively predicts the survival outcomes of stage IV PC patients, utilizing independent prognostic factors such as age, tumor grade, chemotherapy, surgery, and liver, bone, and lung metastasis. The model demonstrates strong accuracy in predicting CSS and OS at 6, 12, and 18 months.
| 1. | Dalmartello M, La Vecchia C, Bertuccio P, Boffetta P, Levi F, Negri E, Malvezzi M. European cancer mortality predictions for the year 2022 with focus on ovarian cancer. Ann Oncol. 2022;33:330-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 86] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 2. | Wang L, Yang L, Chen L, Chen Z. Do Patients Diagnosed with Metastatic Pancreatic Cancer Benefit from Primary Tumor Surgery? A Propensity-Adjusted, Population-Based Surveillance, Epidemiology and End Results (SEER) Analysis. Med Sci Monit. 2019;25:8230-8241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Chen MS, Liu PC, Yi JZ, Xu L, He T, Wu H, Yang JQ, Lv Q. Development and validation of nomograms for predicting survival in patients with de novo metastatic triple-negative breast cancer. Sci Rep. 2022;12:14659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 4. | Huang C, Yu QP, Li H, Ding Z, Zhou Z, Shi X. A novel nomogram model to predict the overall survival of patients with retroperitoneal leiomyosarcoma: a large cohort retrospective study. Sci Rep. 2022;12:11851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 5. | Zhang SL, Wang ZM, Wang WR, Wang X, Zhou YH. Novel nomograms individually predict the survival of patients with soft tissue sarcomas after surgery. Cancer Manag Res. 2019;11:3215-3225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Wang W, Hong J, Meng J, Wu H, Shi M, Yan S, Huang Y. Nomograms Predict Cancer-Specific and Overall Survival of Patients With Primary Limb Leiomyosarcoma. J Orthop Res. 2019;37:1649-1657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Tian H, Ning Z, Zong Z, Liu J, Hu C, Ying H, Li H. Application of Machine Learning Algorithms to Predict Lymph Node Metastasis in Early Gastric Cancer. Front Med (Lausanne). 2021;8:759013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 8. | Sun D, Peng H, Wu Z. Establishment and Analysis of a Combined Diagnostic Model of Alzheimer's Disease With Random Forest and Artificial Neural Network. Front Aging Neurosci. 2022;14:921906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 9. | Liu W, Zhang L, Xin Z, Zhang H, You L, Bai L, Zhou J, Ying B. A Promising Preoperative Prediction Model for Microvascular Invasion in Hepatocellular Carcinoma Based on an Extreme Gradient Boosting Algorithm. Front Oncol. 2022;12:852736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Huang K, Yuan X, Zhao P, He Y. Effect of chemotherapy on prognosis in patients with primary pancreatic signet ring cell carcinoma: A large real-world study based on machine learning. PLoS One. 2024;19:e0302685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 11. | Kim Y, Margonis GA, Prescott JD, Tran TB, Postlewait LM, Maithel SK, Wang TS, Evans DB, Hatzaras I, Shenoy R, Phay JE, Keplinger K, Fields RC, Jin LX, Weber SM, Salem AI, Sicklick JK, Gad S, Yopp AC, Mansour JC, Duh QY, Seiser N, Solorzano CC, Kiernan CM, Votanopoulos KI, Levine EA, Poultsides GA, Pawlik TM. Nomograms to Predict Recurrence-Free and Overall Survival After Curative Resection of Adrenocortical Carcinoma. JAMA Surg. 2016;151:365-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 12. | Katsuyama E, Miyawaki Y, Sada KE, Asano Y, Hayashi K, Yamamura Y, Hiramatsu-Asano S, Morishita M, Ohashi K, Watanabe H, Katsuyama T, Narazaki M, Matsumoto Y, Wada J. Association of explanatory histological findings and urinary protein and serum creatinine levels at renal biopsy in lupus nephritis: a cross-sectional study. BMC Nephrol. 2020;21:208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Kalantari S, Khalili D, Asgari S, Fahimfar N, Hadaegh F, Tohidi M, Azizi F. Predictors of early adulthood hypertension during adolescence: a population-based cohort study. BMC Public Health. 2017;17:915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Kim JH. Multicollinearity and misleading statistical results. Korean J Anesthesiol. 2019;72:558-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 890] [Article Influence: 148.3] [Reference Citation Analysis (0)] |
| 15. | Mo X, Zhou M, Yan H, Chen X, Wang Y. Competing risk analysis of cardiovascular/cerebrovascular death in T1/2 kidney cancer: a SEER database analysis. BMC Cancer. 2021;21:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 16. | Liao Y, Yin G, Fan X. The Positive Lymph Node Ratio Predicts Survival in T(1-4)N(1-3)M(0) Non-Small Cell Lung Cancer: A Nomogram Using the SEER Database. Front Oncol. 2020;10:1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 17. | Chen H, Kong Y, Yao Q, Zhang X, Fu Y, Li J, Liu C, Wang Z. Three hypomethylated genes were associated with poor overall survival in pancreatic cancer patients. Aging (Albany NY). 2019;11:885-897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 18. | Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1481] [Cited by in RCA: 1545] [Article Influence: 73.6] [Reference Citation Analysis (0)] |
| 19. | Shi H, Chen Z, Dong S, He R, Du Y, Qin Z, Zhou W. A nomogram for predicting survival in patients with advanced (stage III/IV) pancreatic body tail cancer: a SEER-based study. BMC Gastroenterol. 2022;22:279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Yao Q, Jia W, Chen S, Wang Q, Liu Z, Liu D, Ji X. Machine learning was used to predict risk factors for distant metastasis of pancreatic cancer and prognosis analysis. J Cancer Res Clin Oncol. 2023;149:10279-10291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Reference Citation Analysis (0)] |
| 21. | Zhang W, Ji L, Wang X, Zhu S, Luo J, Zhang Y, Tong Y, Feng F, Kang Y, Bi Q. Nomogram Predicts Risk and Prognostic Factors for Bone Metastasis of Pancreatic Cancer: A Population-Based Analysis. Front Endocrinol (Lausanne). 2021;12:752176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 22. | Shrikhande SV, Kleeff J, Reiser C, Weitz J, Hinz U, Esposito I, Schmidt J, Friess H, Büchler MW. Pancreatic resection for M1 pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2007;14:118-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 171] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 23. | Strobel O, Neoptolemos J, Jäger D, Büchler MW. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol. 2019;16:11-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 603] [Article Influence: 100.5] [Reference Citation Analysis (0)] |
| 24. | Timmer FEF, Geboers B, Nieuwenhuizen S, Schouten EAC, Dijkstra M, de Vries JJJ, van den Tol MP, Meijerink MR, Scheffer HJ. Locoregional Treatment of Metastatic Pancreatic Cancer Utilizing Resection, Ablation and Embolization: A Systematic Review. Cancers (Basel). 2021;13:1608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (1)] |
| 25. | Tsitskari M, Filippiadis D, Kostantos C, Palialexis K, Zavridis P, Kelekis N, Brountzos E. The role of interventional oncology in the treatment of colorectal cancer liver metastases. Ann Gastroenterol. 2019;32:147-155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 26. | Chow FC, Chok KS. Colorectal liver metastases: An update on multidisciplinary approach. World J Hepatol. 2019;11:150-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 154] [Article Influence: 25.7] [Reference Citation Analysis (3)] |
| 27. | Han R, Tian Z, Jiang Y, Guan G, Wang X, Sun X, Yu Y, Jing X. Prognostic significance of the systemic immune inflammation index in patients with metastatic and unresectable pancreatic cancer. Front Surg. 2022;9:915599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 28. | Zhang H, Dong H, Pan Z, Du X, Liu S, Xu W, Zhang Y. Risk factors and predictive nomograms for early death of patients with pancreatic cancer liver metastasis: A large cohort study based on the SEER database and Chinese population. Front Oncol. 2022;12:998445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 29. | Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, Zhao HW, Chen W, He YL, Wang HY, Xie D, Luo JH. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 452] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 30. | Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol. 2016;34:2157-2164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 896] [Cited by in RCA: 1320] [Article Influence: 146.7] [Reference Citation Analysis (0)] |
| 31. | Zhou G, Xiao K, Gong G, Wu J, Zhang Y, Liu X, Jiang Z, Ma C. A novel nomogram for predicting liver metastasis in patients with gastrointestinal stromal tumor: a SEER-based study. BMC Surg. 2020;20:298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Xu L, Wen N, Qiu J, He T, Tan Q, Yang J, Du Z, Lv Q. Predicting Survival Benefit of Sparing Sentinel Lymph Node Biopsy in Low-Risk Elderly Patients With Early Breast Cancer: A Population-Based Analysis. Front Oncol. 2020;10:1718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |