Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2091
Peer-review started: December 10, 2023
First decision: January 5, 2024
Revised: January 17, 2024
Accepted: February 22, 2024
Article in press: February 22, 2024
Published online: May 15, 2024
Processing time: 151 Days and 3.8 Hours
For the first time, we investigated the oncological role of plexin domain-contain
To investigate the oncological profile of PLXDC1 in HCC.
Based on The Cancer Genome Atlas database, we analyzed the expression of PLXDC1 in HCC. Using immunohistochemistry, quantitative real-time poly
Based on immunohistochemistry, qRT-PCR, and Western blot assays, overexpression of PLXDC1 in HCC was associated with poor prognosis. Univariate and multivariate Cox analyses indicated that PLXDC1 might be an independent prognostic factor. In HCC patients with high methylation levels, the prognosis was worse than in patients with low methylation levels. Pathway enrichment analysis of HCC tissues indicated that genes upregulated in the high-PLXDC1 subgroup were enriched in mesenchymal and immune activation signaling, and TIDE assessment showed that the risk of immune evasion was significantly higher in the high-PLXDC1 subgroup compared to the low-PLXDC1 subgroup. The high-risk group had a significantly lower immune evasion rate as well as a poor prognosis, and PLXDC1-related risk scores were also associated with a poor prognosis.
As a result of this study analyzing PLXDC1 from multiple biological perspectives, it was revealed that it is a bio
Core Tip: Based on immunohistochemistry, quantitative real-time polymerase chain reaction, and Western blot assays, overexpression of plexin domain-containing 1 (PLXDC1) in hepatocellular carcinoma (HCC) was associated with poor prognosis. Univariate and multivariate Cox analyses indicated that PLXDC1 might be an independent prognostic factor. In HCC patients with high methylation levels, the prognosis was worse than in patients with low methylation levels. Pathway enrichment analysis of HCC tissues indicated that genes upregulated in the high-PLXDC1 subgroup were enriched in mesenchymal and immune activation signaling, and tumor immune dysfunction and exclusion assessment showed that the risk of immune evasion was significantly higher in the high-PLXDC1 subgroup compared to the low-PLXDC1 subgroup. The high-risk group had a significantly lower immune evasion rate as well as a poor prognosis, and PLXDC1-related risk scores were also associated with a poor prognosis.
- Citation: Tang MY, Shen X, Yuan RS, Li HY, Li XW, Jing YM, Zhang Y, Shen HH, Wang ZS, Zhou L, Yang YC, Wen HX, Su F. Plexin domain-containing 1 may be a biomarker of poor prognosis in hepatocellular carcinoma patients, may mediate immune evasion. World J Gastrointest Oncol 2024; 16(5): 2091-2112
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2091.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2091
It is the fifth most common cancer worldwide and the second leading cause of cancer-related death[1]. Hepatocellular carcinoma (HCC) accounts for 75% to 85% of primary liver cancers[2]. The high incidence and mortality rates of HCC in China remain serious threats to human health. As HCC has an insidious onset, most patients are at an advanced stage as soon as a diagnosis is made, and treatment with a surgery including resection, ablation, and chemoembolization of hepatic arteries may be used has a constrainted effect, with a 5-year survival rate of only 18% and a recurrence rate of 70%. It is the leading cause of death for patients with advanced HCC. Despite the Improved testing techniques and therapeutic strategy for HCC, relapse with recurrent intra-hepatic dissemination, and extra-liver transplantation remain causes of death from HCC[3-5]. As the development of HCC is a multistage, multistep, multifactorial, and complex process, identifying new markers of HCC and studying their mechanisms of action are essential for improving the overall treatment outcome, reducing mortality, and improving the prognosis of HCC in the clinic.
Plexin domain-containing 1 (PLXDC1), alternative name tumor endothelial cell marker 7 (TEM7), makes up of 500 amino acids and forms a cytoplasmic shorter tail, a hydrophobic transmembrane region, and a big extracellular structural realm[6,7]. PLXDC1 was originally identified as a transmembrane protein in the human tumor vascular endothelium; there are several variants of PLXDC1, including intracellular and secreted forms, in addition to transmembrane PLXDC1[8,9]. PLXDC1 is reportedly involved in the development of cancer. In 2004, TEM7 was found to bind to cortactin, promoting cancer progression[9-11]. For example, PLXDC1 promotes the migration and invasion of gastric cancer cells[7]. PLXDC1 induces the infiltrative growth of endothelial cells to facilitate glioblastoma formation[12]. High PLXDC1 expression is associated with metastasis and poor survival in patients with OS[13]. In addition, abnormal blood vessels are a sign of the majority of real tumors, unusual vasculogenesis in tumors facilitates immune evasion[14-17]. We speculate that the PLXDC1 gene may contribute to tumor progression, but there are no studies on the role of PLXDC1 in the development of HCC or its immunological aspects. The present study will focus on these aspects.
In the past, targeting cancer characteristics has been effective in treating various types of cancer. These traditional anticancer drugs have shown remarkable therapeutic effects but still have adverse side effects for patients. Cisplatin (CIS) is a broad-spectrum anticancer drug that has cytotoxic effects on both normal and cancer cells. Hibiscus extract reduced cis-induced hepatotoxicity in mice, and its use in combination therapy could improve the efficacy of CIS for cancer treatment. Several studies have reported that steroidal saponin is also effective at inhibiting tumor markers, but its poor bioavailability and insufficient preclinical studies limit its application[18,19]. With the continuous development of proteomics and bioinformatics fields, screening new anticancer peptides may become a promising strategy for the future treatment and/or prevention of different types of cancer. The approximate of DyCluster in probing protein complexes is an effective contend in support of this way. DyCluster can also probe biologically meaningful protein team[20,21].
In recent years, immunotherapy has been a landmark discovery in cancer treatment and is one of the most promising therapeutic strategies in the field of oncology; immunotherapy aims to kill tumor cells by regulation of the immunity and modification of the tumor immune microenvironment (TIME)[22]. Drugs developed to target programmed death ligand 1 (PD-L1) on the surface of tumors kill tumor cells by disrupting the tumor immune evasion mechanism, which in turn delays tumor progression and increases the survival rate of tumor patients. A growing body of research suggests that immune cell infrastructure holds a vital role in tumor prognosis[23,24]. The tumor microenvironment (TME) is the interior and exterior environment of tumorigenesis and has an important role in tumor progression, immune escape, and therapeutic tolerance[25]. Systemic therapy, including targeted therapy, systemic chemotherapy, and immunotherapy, is the mainstay treatment for advanced HCC. Systemic therapy for advanced HCC includes targeted therapy, systemic chemotherapy, immunotherapy, etc. Combination therapy with different anticancer drugs remains a core practice for overcoming the shortcomings of conventional cancer therapy. Combination therapy allows the use of multiple drugs at dose reduction, thereby increasing efficacy and reducing the likelihood of serious adverse events. Recently, a new cancer treatment option has emerged that combines conventional chemotherapy with a naturally derived chemical that is cytotoxic to cancer cells and causes limited damage to normal cells. The current findings suggest that crocin and sorafenib treatment regimens reduce hepatotoxicity, hinder the development of HCC, and improve liver function. Here, the overexpression of PLXDC1 was shown to be associated with cirrhosis, and some studies have shown that dandelion may prevent liver fibrosis, the inflammatory response and oxidative stress in rats[26-29]. With increasing research on tumor molecular signaling pathways and TMEs, targeted therapy has become a hot topic in clinical research on advanced HCC. Therefore, how to effectively improve the efficacy of immune checkpoint inhibitors (ICIs) is an important issue to be addressed in future HCC immunotherapy studies. At present, immunotherapy still lacks closely related biomarkers, except for PD-L1 and the TMB; therefore, identifying suitable targets for HCC immunotherapy is important.
We assessed PLXDC1 expression in various cancers making use of the Oncomine database (https://www.oncomine.org/resource/Login.html) and choosing the “Gene Variable Expression” modules. Later, we chose HCC as our research subject and analyzed further the differentially expressed PLXDC1 between HCC and its subtypes and normal tissues with the dataset from the Oncomine database. Then, we downloaded HCC data (normal samples = 50; cancer samples = 374) from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/) and analyzed the pair-wise and unpaired differences in PLXDC1 expression in HCC tissues and normal tissues using the “limma” R software package.
The samples were obtained from patients who received surgery for HCC between January 2022 and December 2022 at the Department of Hepatobiliary Surgery, the First Affiliated Hospital of Bengbu Medical College. A series of 8 HCC tissue samples and their collateral tumor tissue samples were gathered for Western blot experiments and quantitative real-time polymerase chain reaction (qRT-PCR) research. Patients who did not get chemotherapy, radiotherapy, or biotherapy prior to surgery, and patients with confirmed HCC before and after surgery. Completed tissue samples were preserved in a refrigerator at -80 °C immediately after surgery until protein and RNA were removed. In addition, 123 HCC tissue samples and 20 normal paraneoplastic tissue samples were collected from January 2017 to December 2018 for immunohistochemical (IHC) staining. The research was authorized by the Ethics Committee of Bengbu Medical College.
Rabbit anti-human antibody PLXDC1 (50 μg) was provided by Abcam. Primary antibodies for GAPDH and CD73+/CD134+/CD3+/CD4+/CD8+ T cell alpha rabbit monoclonal antibodies from Cell Signaling Technology (Danvers, MA, United States). Horseradish peroxidase (HRP)-conjugated anti-rabbit antibody was obtained from Jackson ImmunoResearch, Inc. (West Grove, PA, United States). Skim milk and Tween 20 were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). PrimeScriptTM First Strand cDNA Synhesis Kit was purchased from TaKaRa (Tokyo, Japan), and SYBR Green Real-Time PCR Master Mix was purchased from TOYOBO (Osaka, Japan).
A total of 4% paraformaldehyde was applied to all samples, which were embedded in petrolatum, sliced to 4 μm, and mounted on slides. Following dewaxing of the paraffin under different intensity gradients of xylene, the slides were rehydrated and soaked in citric acid buffer (pH 7.8, 0.1 M) for 24 min at about 82 °C to extract antigens. Sections were covered evenly with endogenous catalase barrier solution for 15 min at room temperature to block endogenous catalase activities. Following incubation with either primary antibody or overnight at 4 °C, sections were gently rinsed with phosphate-buffered saline, incubated with biotin-conjugated secondary antibody for 10 min at room temperature, and then with streptavidin peroxidase for 5 min. All slices were colored with hematoxylin and then cleaned. Following drying and cleaning of the slices, they were analyzed by immunohistochemistry.
Total cellular protein was withdrawn making do with RIPA buffer and protease inhibitors. Protein was electrophoresed employing a PowerPac HV high voltage power supply (Bio-Rad Laboratories, United States). Total proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then electrophoretically shifted to a PVDF membrane. Following the block with fresh 5% skimmed milk, the blot was incubated overnight at 4 °C with a primary antibody specifically in 5% BSA. Following washing with TBST, the blots were incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG for 2 h at room temperature. Finally, bands of a selected protein were examined and visualized and amplified using an ultrasensitive enhanced chemiluminescence (ECL) kit using a Bio-Rad ChemiDoc XRS imager system (Bio-Rad Laboratories, CA, United States); and the bands were visualized and clarified using the ImageJ computer software (Software Inquiry; Quebec, Canada) to optimize and analyze the bands.
PLXDC1 expression-related methylation sites had analyzed chosing MEXPRESS (https://mexpress.be) in the “HCC” part. Survival-related methylation status (http://bio-bigdata.hrbmu.edu.cn/survivalmeth/) was subsequently chosed analyze differences in survival between patients stratified according to the location of the PLXDC1 expression-related methylation site and HCC prognosis.
Total RNA was separated through the use of TRIzol reagent, separated RNA was retrotranscribed through the usage of RevertAid First Strand cDNA Synthesis Kit, and synthesized cDNA was withdrawn through the usage of SYBR Green Real-time PCR Master Mix. Below are citations for qRT-PCR: human PLXDC1 forward, 5’-ACACGCTGCCAGATAAC
We utilized “GSVA” R package (GSEA) version 4.1.0, a tool that allows analyzing genome-wide expression profile microarray data and comparison of genes to a pre-defined genome, to determine the PLXDC1-related pathways in HCC. Gene expression matrix of HCC tissues was manipulated by Perl software to get input files associated with target genes. The collection of up-regulated genes was characterized as phenotype h (h = 28/52) and the collection of low-regulated genes was defined as phenotype l (l = 4/12), and “h-versus-l” was selected to enrich for PLXDC1-related pathways. A standardized enrichment score (NES)| > 1, nominal (NOM) P < 0.05, and false detection rate (FDR) q-value < 0.25 were typically taken to demonstrate that the gene set was enriched significantly in the pathway.
The higher-expression group (n = 90) and lower-expression group (n = 284) were divided on the basis of the average expression of PLXDC1 in HCC samples acquired through the TCGA database (2.378) and from the Molecular Signatures Database (MSigDB) online website (http://www.gsea-msigdb.org/gsea/Login.jsp) Marker pathway gene collections were segmented. Genomic variant profiling with GSEA of the hallmark pathway genes was performed with the PLXDC1 high- and low-expression groups, and single-sample gene set enrichment analysis (ssGSEA) For analyzing the spread of immune cells and the functional genome in the higher- and lower-expression sets. In order to estimate the immune cell fraction score, stromal fraction score and tumor purity score of PLXDC1 high- and low-expression sets in HCC TME, stromal and immune cells in malignant tumor tissues were also examined by using the Expression Data (ESTIMATE) algorithm. Lastly, the risk of immune evasion was evaluated by the tumor immune dysfunction and exclusion (TIDE) algorithm in the PLXDC1 high-expression and low-expression sets. We then utilized the TISIDB online website (http://cis.hku.hk/TISIDB/index.php) to analysis the correlation between PLXDC1 expression and immune cell infiltration in the immune microenvironment of HCC[29]. According to the subtype panel, the immune cells present in HCC tissues consist of six subtypes [C1 (wound healing; n = 129), C2 (IFN-gamma dominant; n = 210), C3 (inflammatory; n = 36), C4 (lymphocyte depleted; n = 9), C5 (immunologically quiet), and C6 (TGF-b dominant; n = 7)] to discovering correlation between PLXDC1 expression and the C1/2/3/4/6 subtypes.
Marker genes correlated with PLXDC1 in the high- and low-PLXDC1-expressing sets were characterized by the “limma” R package. Sixty-six HCC immune cell skin marker genes were characterized by the association of HCC with PLXDC1 expression by the R package “reshape2”. The prognostic properties of PLXDC1-associated marker genes in HCC were further characterized by stepwise multivariate Cox proportional hazards regression using prognostically related genes to determine the optimal candidate genes and to build an immune-related risk model. The risk score was calculated as follows: where “config” and “Xi” stand for the factor and expression level of each PLXDC1 prognosis-related marker gene, etc., respectively. The TCGA samples were divided into high and low risk groups depending on the risk score of the model. The log-rank test was utilized to discuss the survival rates of the two groups (P < 0.05). Receiver operational characteristic curves (ROC) were produced as well as area under the curve (AUC) values were acquired using the “survivalROC” software package to evaluate the reliability of the prognostic model. To further optimize the prognostic value of risk scores, we performed a univariate and multivariate Cox assay combining risk scores with clinical factors (gender, age, grading, T-stage, N-stage, M-stage, and staging) to estimate whether risk scores could be used as an independent prognostic element. Finally, the risk of immune evasion and the effectiveness of immunotherapy were estimated between the high- and low-risk groups depending on the model using TIDE (https://tide.dfci.harvard.edu/Login/).
All data are presented as mean ± SD. Statistical compared of data were tested by two-tailed Student’s test for multiple compares. The correlation between PLXDC1 expression and clinicopathologic features was examined by the chi-square check. OS curves were computed using the Kaplan-Meier method and comparison was made using the log-rank exam. Stepwise univariate and multi-variate analyses were performed using Cox proportional hazards regression models. Data were characterized using SPSS 23.0 software, GSEA (v 4.2.3), Perl (v 5.32.1.1), and R software (v 4.1.2). The methods component includes the statistical methods for each experiment typology and the respective R software packages. The critical standard for significant was P < 0.05.
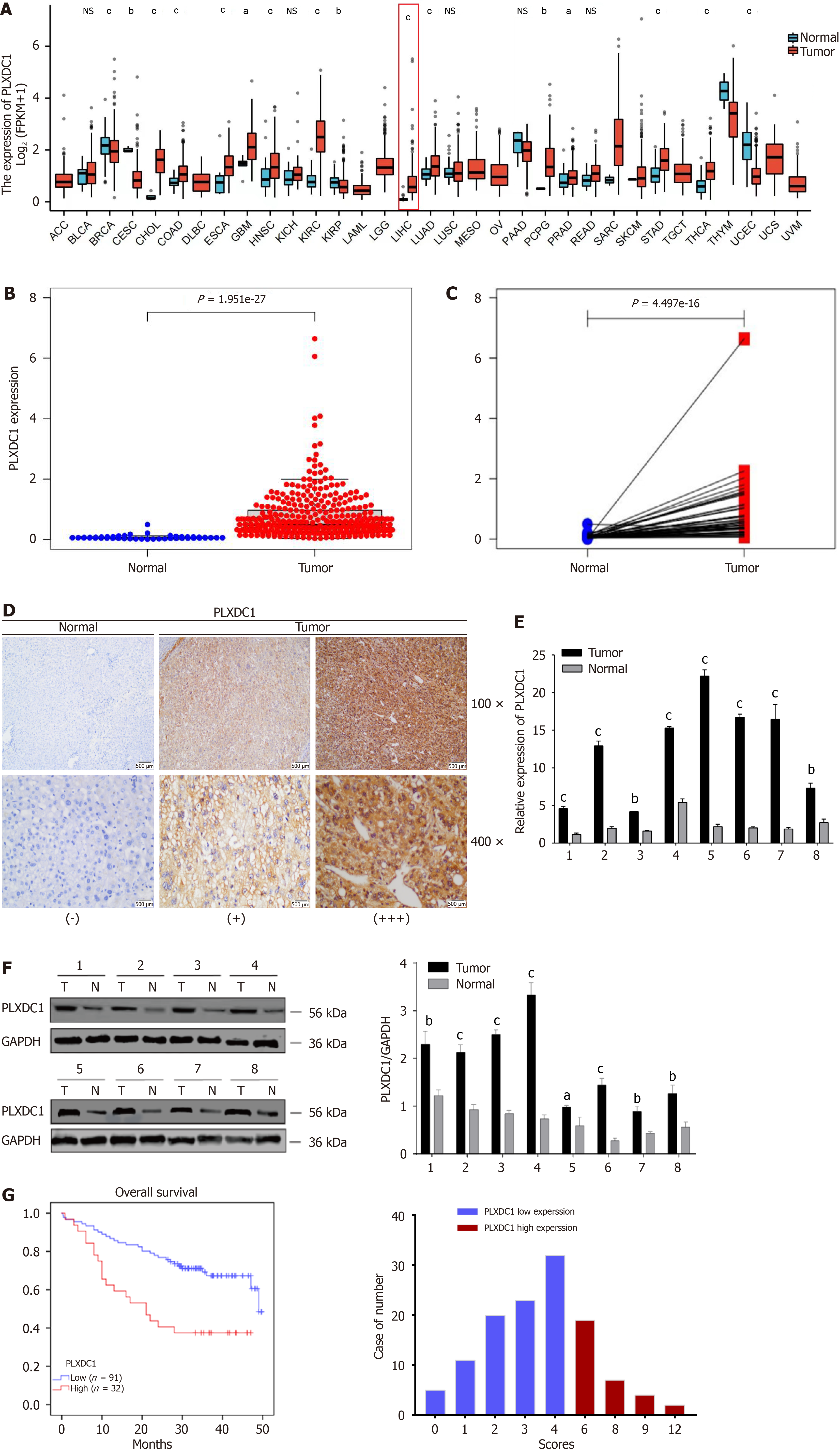
Pan-cancer varial expression profiling showed that PLXDC1 was clearly shown to be expressed in various cancerous tissues and normal tissues, with significantly decreased expression in endometrial, renal clear cell, and breast invasive carcinomas and significantly increased expression in the colon, bile duct, and HCC (Figure 1A). Unpaired difference analysis revealed that PLXDC1 expression was significantly greater in HCC tissues than in paraneoplastic tissues (Figure 1B), and paired difference analysis confirmed these findings (Figure 1C). At the tissue level, IHC staining analysis revealed that PLXDC1 expression was greater in HCC tissues than in paracancerous tissues (Figure 1D). To further assessment on whether PLXDC1 was overexpressed in HCC, we assessed the expression of PLXDC1 mRNA in the eight freshly frozen matched HCC tissues by qRT-PCR, and its relative expression and PLXDC1 protein expression were assessed by Western blotting analysis. In agreement with the above results, PLXDC1 mRNA levels were increased in HCC tissues compared with neighboring noncancerous tissues (Figure 1E). This was also observed at the protein level by Wb experiments.The expression of PLXDC1 was significantly greater in tumor tissues than in normal tissues (Figure 1F). In conclusion, the expression of PLXDC1 was significantly greater in HCC tissues than in normal tissues.
To investigate the role of PLXDC1 in HCC, we detected the PLXDC1 protein expression level in 123 HCC patients by immunohistochemistry and analyzed the correlation between the PLXDC1 protein level and clinicopathological features. As shown in Table 1, the PLXDC1 protein expression level was associated with the severity of cirrhosis (positive vs negative), albumin-bilirubin (ALBI) score (high vs low), aspartate aminotransferase to platelet ratio (APRI) score (high vs low), and the AFP concentration (high vs low). No significant relevance of other clinicopathological traits, like patient’s age, gender, tumor size, hepatitis B status and distant metastasis, was noted in connection with PLXDC1 protein levels. In addition, univariate Cox regression analysis revealed that the degree of cirrhosis (positive vs negative), ALBI score (high vs low), APRI score (high vs low), and the PLXDC1 protein level (low vs high) were four prognostic factors for OS in HCC patients. Further multifactorialmultivariate Cox regression analysis revealed that the PLXDC1 protein level (low vs high) and degree of cirrhosis (positive vs negative) were independent prognostic factors for OS in HCC patients (Table 2). According to Kaplan-Meier survival analysis, patients in the high-PLXDC1 expression subgroup had poorer OS than patients in the low-PLXDC1 expression subgroup did (Figure 1G). These findings suggested that the PLXDC1 level could be used as an indicator of HCC prognosis.
| Clinicopathological features | Number | Low expression, n (%) | High expression, n (%) | P value |
| Age | ||||
| < 60 | 68 | 51 (75.0) | 17 (25.0) | 0.467 |
| ≥ 60 | 55 | 40 (72.7) | 15 (27.3) | |
| Gender | ||||
| Male | 102 | 76 (74.5) | 26 (25.5) | 0.480 |
| Female | 21 | 15 (71.4) | 6 (28.6) | |
| Tumor size | ||||
| ≤ 5 cm | 65 | 49 (75.4) | 16 (24.6) | 0.432 |
| > 5 cm | 58 | 42 (72.4) | 16 (27.6) | |
| Cirrhosis | ||||
| Negative | 63 | 53 (84.1) | 10 (15.9) | 0.007a |
| Positive | 60 | 38 (63.3) | 22 (36.7) | |
| ALBI score | ||||
| ≤ -2.60 | 55 | 40 (72.7) | 15 (27.3) | 0.001a |
| > -2.60 | 68 | 51 (75.0) | 17 (25.0) | |
| APRI score | ||||
| > 2 | 38 | 26 (68.4) | 12 (31.6) | 0.010a |
| ≤ 2 | 85 | 65 (76.5) | 20 (23.5) | |
| AFP (ng/mL) | ||||
| ≤ 20 | 42 | 36 (85.7) | 6 (14.3) | 0.025a |
| > 20 | 81 | 55 (67.9) | 26 (32.1) | |
| HBV | ||||
| Negative | 12 | 8 (66.7) | 4 (33.3) | 0.381 |
| Positive | 111 | 83 (74.8) | 28 (25.2) | |
| Intrahepatic metastasis | ||||
| Negative | 111 | 82 (73.9) | 29 (26.1) | 0.619 |
| Positive | 12 | 9 (75.0) | 3 (25.0) |
| Variables | Overall survival | P value | |
| HR | 95%CI | ||
| Univariate analysis | |||
| Age | 1.320 | 0.753-2.315 | 0.332 |
| Gender | 0.972 | 0.678-1.395 | 0.878 |
| Tumor size | 1.168 | 0.666-2.049 | 0.588 |
| AFP (ng/mL, ≤ 20/> 20) | 1.397 | 0.771-2.532 | 0.270 |
| Cirrhosis | 2.474 | 1.376-4.449 | 0.002b |
| ALBI score | 2.254 | 1.275-3.975 | 0.005b |
| APRI score | 1.787 | 1.014-3.149 | 0.045a |
| HBV | 0.487 | 0.217-1.089 | 0.080 |
| PLXDC1 expression | 2.714 | 1.524-4.832 | 0.001b |
| Multivariate analysis | |||
| Age | 1.273 | 0.719-2.256 | 0.408 |
| Gender | 1.101 | 0.753-1.611 | 0.619 |
| Tumor size | 1.128 | 0.633-1.208 | 0.683 |
| AFP (ng/mL, ≤ 20/> 20) | 0.645 | 0.324-1.284 | 0.212 |
| Cirrhosis | 0.513 | 0.219-1.204 | 0.010a |
| ALBI score | 2.384 | 1.384-4.217 | 0.003b |
| APRI score | 1.941 | 1.096-3.435 | 0.023a |
| HBV | 0.448 | 0.196-1.027 | 0.125 |
| PLXDC1 expression | 2.583 | 1.137-4.993 | 0.005b |
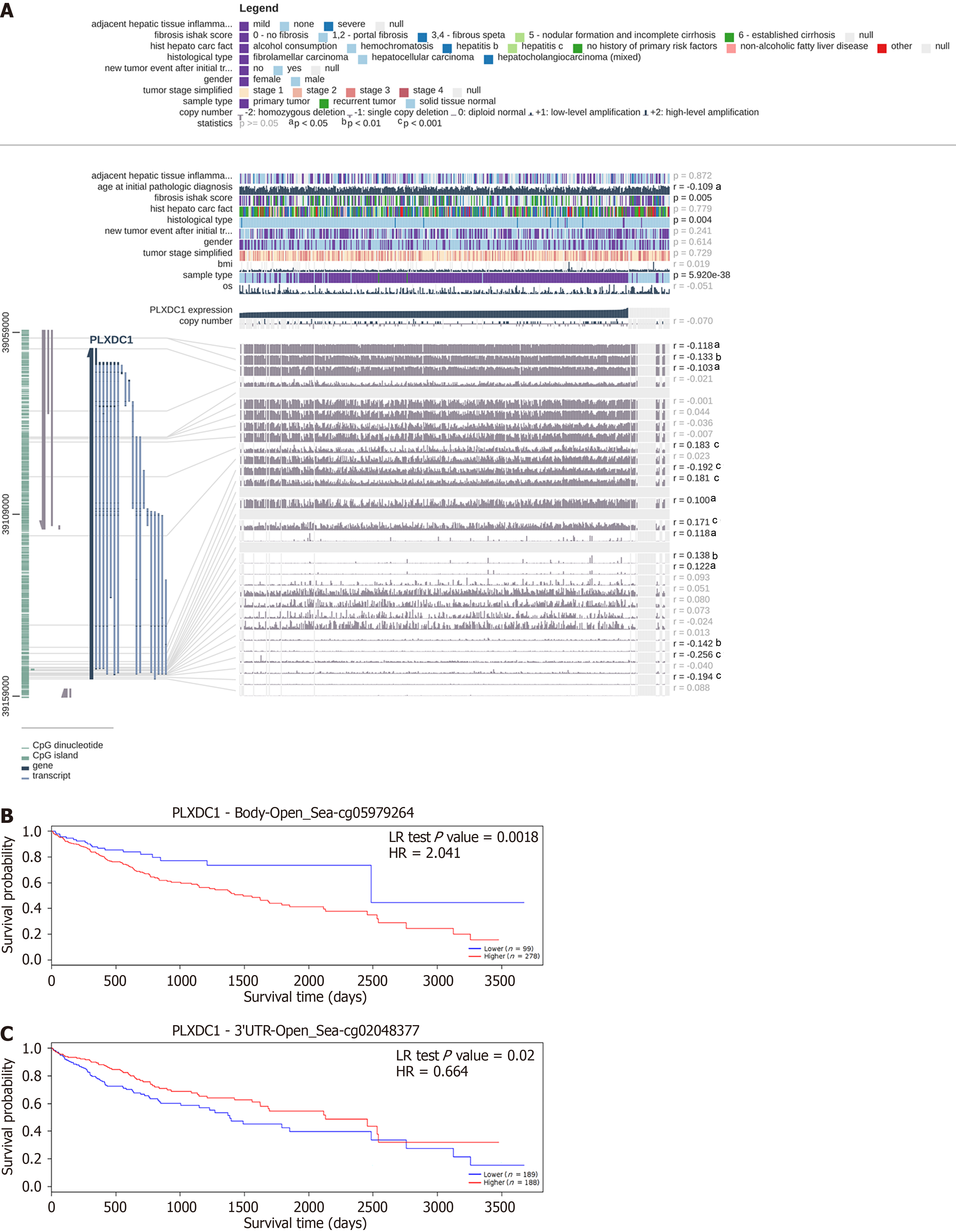
Methylation at 14 sites was shown to be associated with PLXDC1 expression in HCC according to EXPRESS. Among these PLXDC1 expression-associated methylation sites, methylation at cg00760203, cg05979264, cg06745656, cg24724583, cg06947852, cg04872113, and cg16178289 was significantly positively correlated with PLXDC1 expression (r > 0); methylation at cg08885306, cg02048377, cg05652255, cg02875558, cg10213328, and cg24670063 (r < 0) was significantly negatively correlated with PLXDC1 expression (Figure 2A). Survival analysis revealed that 2 PLXDC1 methylation sites were significantly related to patient prognosis. Methylation at cg05979264 was positively correlated with PLXDC1 expression, and the patients in the PLXDC1 expression group with a high level of methylation at these sites had a worse prognosis than did those in the low-expression group (Figure 2B). Methylation at cg02048377 had a negative relation with PLXDC1 expression, and the low-PLXDC1 expression group had a worse prognosis than the high-PLXDC1 expression group did (Figure 2C). Consistent with previous findings, the prognosis of HCC patients was worse in the group with high PLXDC1 and high methylation levels at PLXDC1-associated sites.
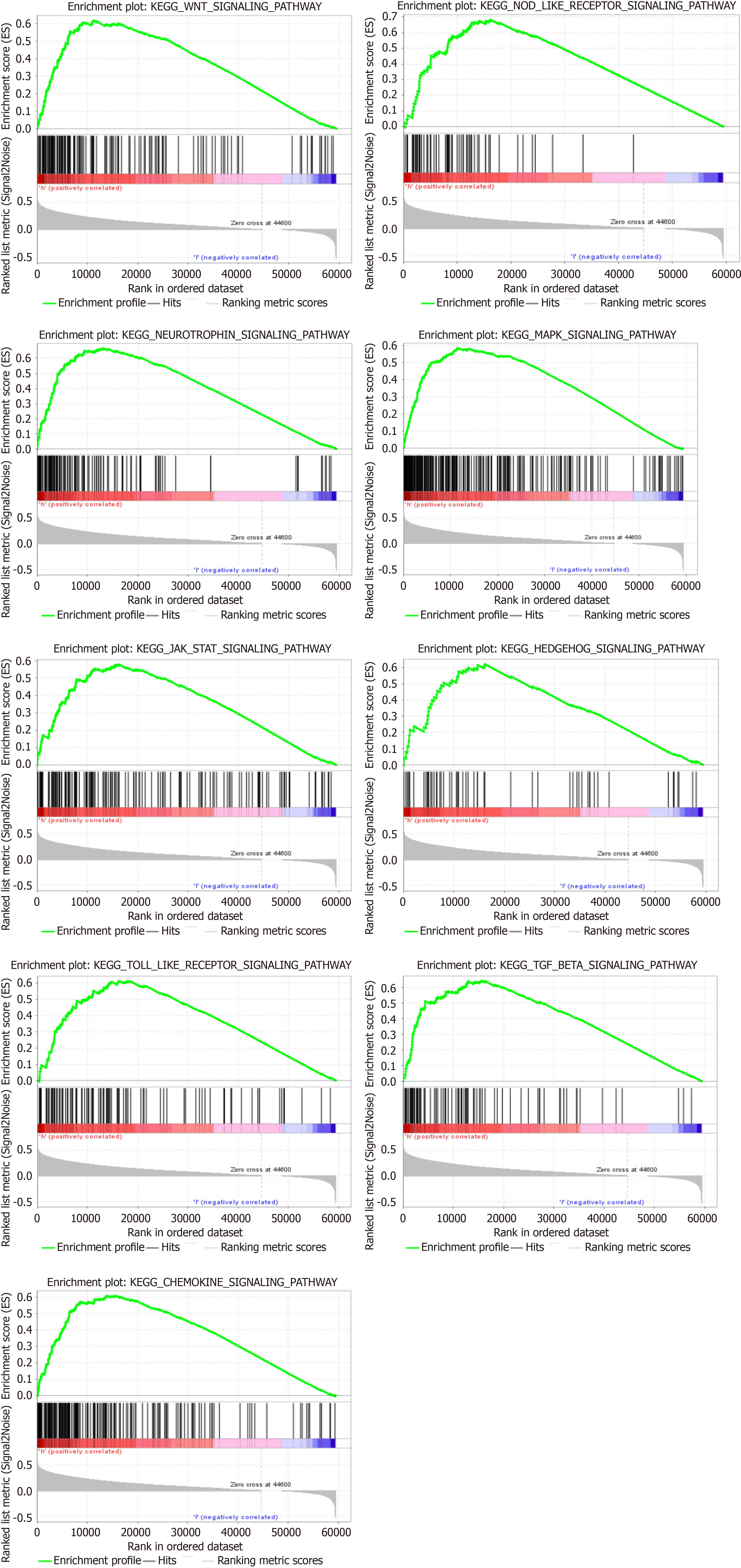
GSEA also revealed that PLXDC1 has participated in multiple signaling pathways in HCC. Table 3 shows topmost part 9 upregulated signaling pathways associated with PLXDC1 in HCC according to GSEA scores. We found that the Wnt and Hedgehog signaling pathways, which are associated with tumor-mesenchymal interactions, were upregulated. The chemokine, Toll-like receptor, and Nod-like receptor signaling pathways, which have participated in immune and inflammatory activation, were also upwarded (Figure 3). It was interesting to note that the high expression of PLXDC1 in HCC did not imply a favorable prognosis.
| GeneSet | NES | NOM P value | FDR q-value |
| WNT_SIGNALING_PATHWAY | 1.91 | 0 | 0.0054 |
| NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 1.89 | 0 | 0.0057 |
| NEUROTROPHIN_SIGNALING_PATHWAY | 1.90 | 0 | 0.0058 |
| MAPK_SIGNALING_PATHWAY | 1.91 | 0 | 0.0061 |
| JAK_STAT_SIGNALING_PATHWAY | 1.85 | 0 | 0.0069 |
| HEDGEHOG_SIGNALING_PATHWAY | 1.87 | 0 | 0.0069 |
| TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 1.84 | 0 | 0.0069 |
| TGF_BETA_SIGNALING_PATHWAY | 1.84 | 0 | 0.0070 |
| CHEMOKINE_SIGNALING_PATHWAY | 1.82 | 0 | 0.0079 |
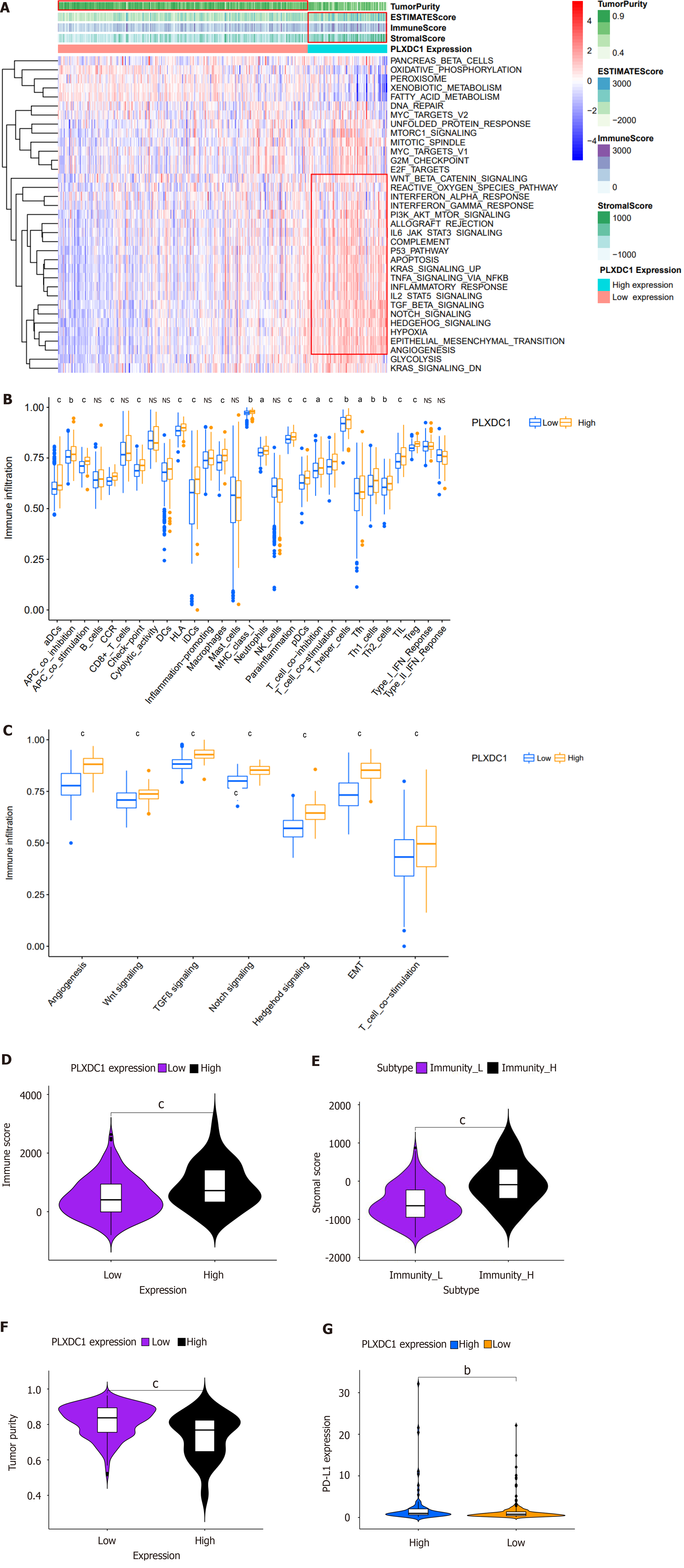
To investigate impacting PLXDC1 expression on HCC patients, the enrichment of hallmark channel-associated genes in the PLXDC1 higher- and lower-expression groups was estimated using GSEA. The results showed that the PLXDC1 high-expression group was enriched in the regulation of immune activation and inflammatory signaling pathways, like the TNFA signaling pathway via NF-κB, allograft rejection, complement, IL6-JAK-STAT3 signaling, IL2-STAT5 signaling, and inflammatory responses. In addition, the high-PLXDC1 expression group exhibited enrichment of mesenchymal signaling pathways, such as the TGF-β signaling pathway, angiogenesis pathway, the Wnt/beta-catenin signaling pathway, and the Notch signaling pathway, while the low-PLXDC1 expression group exhibited decreased enrichment of these pathways (Figure 4A). Heat maps and varial assays of immune cells and features in HCC performed using the ssGSEA approach indicated that immune cells are enriched to a greater extent in the high PLXDC1-expressing group than in the low PLXDC1-expressing set (Figure 4A and B). As shown in Figure 4C, signaling pathways related to EMT, angiogenesis, TGF-β, and Wnt were significantly more enriched in the high-PLXDC1-expressing group than in the low-PLXDC1-expressing group. ESTIMATE evaluation of HCC by TIME also indicated that the high-PLXDC1-expressing set had considerably higher immune cell scores (Figure 4A and D) and tumor mesenchymal scores (Figure 4A and E) than the low-PLXDC1-expressing set, whereas the tumor purity scores (Figure 4A and F) displayed the reverse tendency. These assays were consistent in suggesting that high PLXDC1 expression was linked to heavy immune cell infiltration. Nevertheless, this immune dominance did not indicate a survival benefit and thus became the main point of our attention. PD-L1 is currently a prominent goal of immunotherapy, and its expression level is an essential predictor of reaction to anti-PD-1/L1 treatment. We further evaluated the capacity of PD-L1 expression to forecast the response to immunotherapy and discovered that the high-PLXDC1-expressing set had higher PD-L1 expression than the low-PLXDC1-expressing set, indicating that the high-PLXDC1-expressing set might be immune evasive and have a more favorable immunotherapeutic efficacy (Figure 4G). Thus, high PLXDC1 expression has mediated TIME immune evasion in HCC.
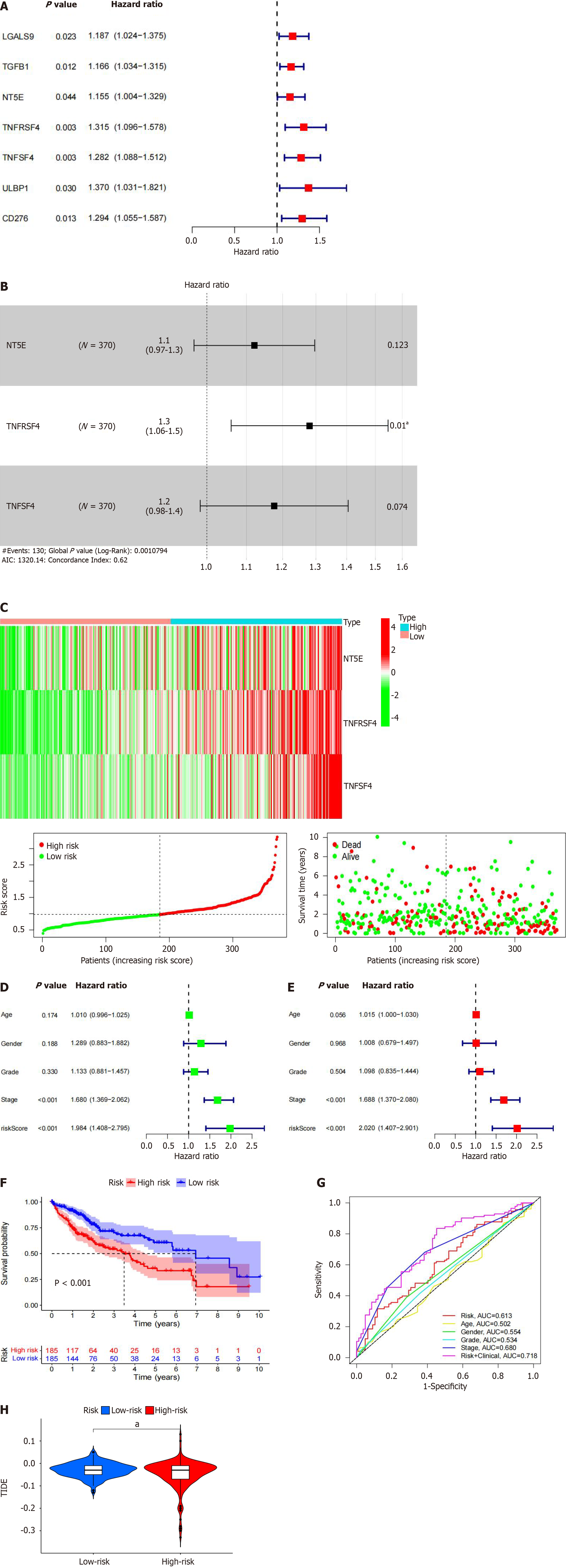
Choosing the database, we searched 66 marker genes localized to the surface of immune cells. These genes encode proteins, alternative name immunomodulators, which constitute immunostimulants or immunosuppressants, and research has indicated that immunomodulators can have a vital role in prognosis. According to our correlation analyses, 60 of the 66 immunomodulators were associated with PLXDC1 expression in HCC patients (Table 4). Cox analysis of the differentially expressed genes revealed that the NT5E (CD73), TGFB1, TNFSF4 (CD134), and TNFRSF4 genes were prognosis-related risk factors (Figure 5A). In addition, we constructed a PLXDC1-associated immune risk model based on prognostic immunomodulators. The risk score was calculated as follows: Risk score = (coefficient × TNFSF4 expression) + (coefficient × TNFRSF expression) + (coefficient × NT5E expression) (Figure 5B). Immune risk models were constructed to classify TCGA samples according to risk and prognosis (Figure 5C). Univariate Cox analysis revealed that age, stage, T stage, N stage, and risk score were risk factors associated with prognosis (Figure 5D), and multivariate Cox analysis revealed that age and risk score could be used as independent prognostic risk factors (Figure 5E). According to the model, survival rates were importantly lower in the high-risk group than in the low-risk group (Figure 5F). The ROC curve of the risk model showed the high accuracy of the model with the risk score alone (AUC = 0.613) and the model including the risk score and clinical factors (AUC = 0.718) (Figure 5G). These analyses indicated that enrichment of the TIME in PLXDC1-mediated HCC is associated with a poor prognosis. The TIDE score indicated that the risk of immune evasion was greater in the higher-risk group than in the lower-risk group (Figure 5H). This finding is in line with the results of previous research that PLXDC1 was mediating immune evasion in TIME of HCC.
| Gene | Cor | P value | Gene | Cor | P value |
| ADORA2A | -0.1481 | 0.0041 | ICOS | 0.3617 | 0 |
| BTLA | 0.2724 | 0 | ICOSLG | 0.0714 | 0.1682 |
| CD160 | 0.2320 | 0 | IL2RA | 0.4743 | 0 |
| CD244 | 0.1330 | 0.0100 | IL6 | 0.3618 | 0 |
| CD274 | 0.2110 | 0 | IL6R | -0.1707 | 0.0009 |
| CD96 | 0.3914 | 0 | KLRC1 | 0.2157 | 0 |
| CSF1R | 0.4258 | 0 | KLRK1 | 0.2004 | 0.0001 |
| CTLA4 | 0.3953 | 0 | LTA | 0.3886 | 0 |
| HAVCR2 | 0.5014 | 0 | MICB | 0.2998 | 0 |
| IL10 | 0.3530 | 0 | NT5E | -0.0069 | 0.8948 |
| IL10RB | 0.3147 | 0 | PVR | 0.1524 | 0.0031 |
| KDR | 0.2302 | 0 | RAET1E | 0.3120 | 0 |
| LAG3 | 0.1047 | 0.0430 | TMIGD2 | 0.2975 | 0 |
| LGALS9 | 0.5536 | 0 | TNFRSF13B | 0.2658 | 0 |
| PDCD1 | 0.3447 | 0 | TNFRSF13C | 0.3396 | 0 |
| PDCD1LG2 | 0.2444 | 0 | TNFRSF14 | 0.2982 | 0 |
| TGFB1 | 0.5562 | 0 | TNFRSF17 | 0.2448 | 0 |
| TGFBR1 | 0.4430 | 0 | TNFRSF18 | 0.4316 | 0 |
| TIGIT | 0.3372 | 0 | TNFRSF25 | 0.3400 | 0 |
| VTCN1 | 0.5346 | 0 | TNFRSF4 | 0.5544 | 0 |
| CD27 | 0.3269 | 0 | TNFRSF8 | 0.5020 | 0 |
| CD28 | 0.2856 | 0 | TNFRSF9 | 0.3496 | 0 |
| CD40 | 0.0859 | 0.0973 | TNFSF13 | 0.3648 | 0 |
| CD40LG | 0.3333 | 0 | TNFSF13B | 0.5252 | 0 |
| CD48 | 0.3965 | 0 | TNFSF14 | 0.1606 | 0.0018 |
| CD70 | 0.3658 | 0 | TNFSF15 | 0.5684 | 0 |
| CD80 | 0.4869 | 0 | TNFSF18 | 0.4378 | 0 |
| CD86 | 0.4568 | 0 | TNFSF4 | 0.3538 | 0 |
| CXCL12 | 0.3123 | 0 | TNFSF9 | 0.3690 | 0 |
| CXCR4 | 0.6607 | 0 | ULBP1 | 0.4382 | 0 |
| ENTPD1 | 0.7295 | 0 | KIR2DL1 | 0.0014 | 0.9782 |
| HHLA2 | 0.2502 | 0 | KIR2DL3 | 0.0596 | 0.2499 |
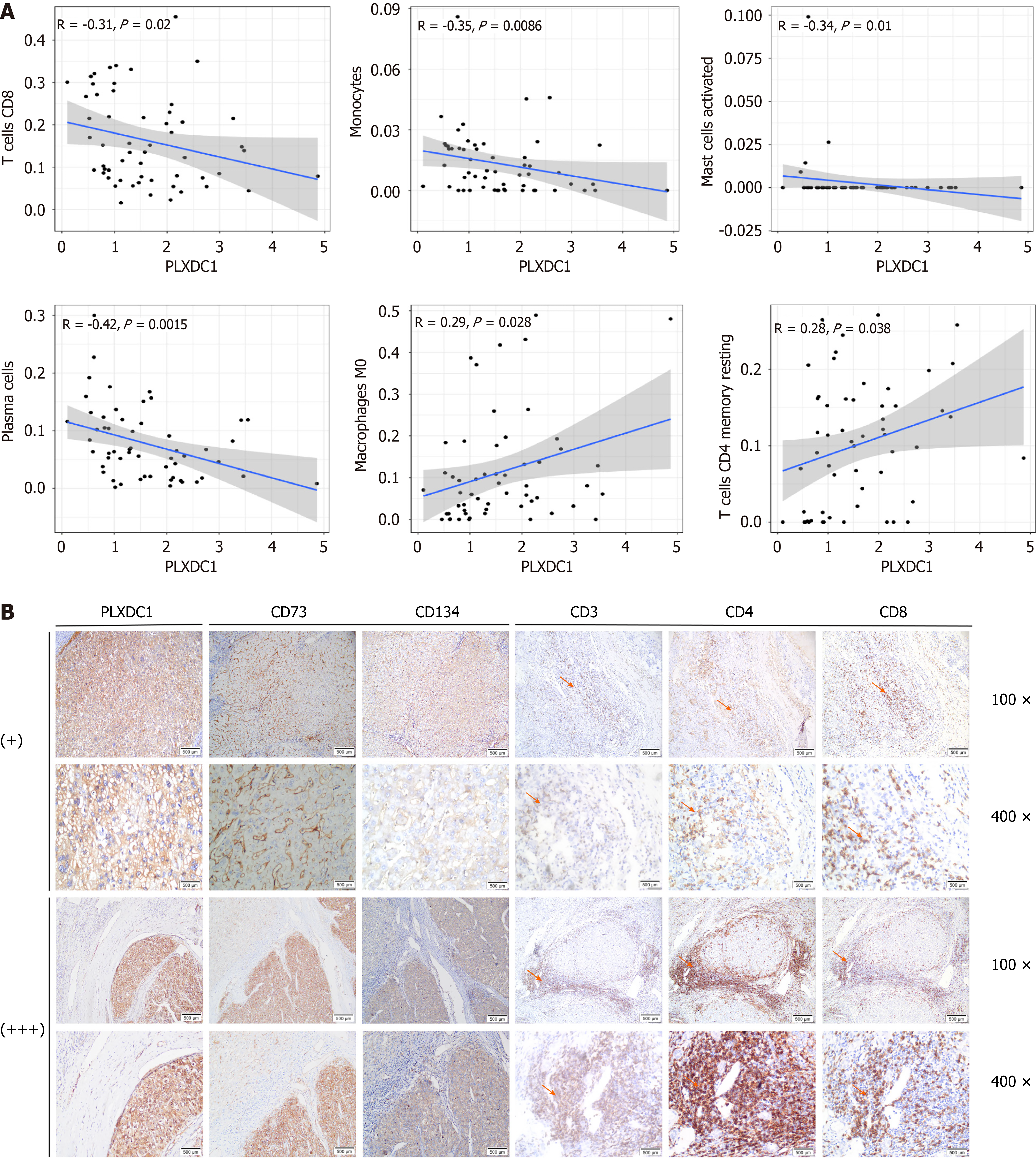
To investigate further the action of PLXDC1 in HCC in promoting TIME immune evasion, we used TIMER from the TCGA database and showed that PLXDC1 expression exhibit a negative relevance with the expression of markers of CD8+ T cells, monocytes, mast cells, and plasma cells and relevant with the expression of markers of macrophages and CD4+ T cells (Figure 6A). For immunohistochemistry, we chosed tissue samples with “+” or “++++” PLXDC1 status, as displayed in Figure 1D. An immunostaining experiment was performed to observe the infiltration of CD73 and CD134 cells on the basis of marker genes on the surface of HCC immune cells and on the surface of CD3+ T cells, CD4+ T cells, and CD8+ T cells in the surrounding tissues to decide whether the high expression of PLXDC1 in the TIME in line with the profile of immune evasion. The findings indicate that that tumor tissues with “++++” PLXDC1 expression had high expression of the CD73 and CD134 genes; additionally, CD3+ T cells, CD4+ T cells, and CD8+ T cells were mainly accumulated in the mesenchyme surrounding the tumor, with a few immune cells penetrating the mesenchyme into the tumor parenchyma. In contrast, in tumor tissues with “+” PLXDC1 expression, the CD73 and CD134 genes were expressed at low levels; CD3+ T cells, CD4+ T cells, and CD8+ T cells clustered less in the tumor periphery, while more immune cells penetrated the interstitium into the tumor parenchyma (Figure 6B). In summary, these findings further verified that high PLXDC1 expression mediates immune evasion in the TIME of HCC.
One of the characteristics of tumors is that they obtain a growth advantage by creating new blood vessels to provide nutrients. Angiogenesis plays an important role in tumorigenesis and progression; nutrients are predominantly supplied to HCC tumors via arteries, and these tumors undergo a high degree of angiogenesis[30-32]. Antiangiogenic drugs have been used successfully in the treatment of many types of cancer, such as lung, colorectal, esophageal, breast, and gastric cancer; HCC; and other malignancies[33-39]. Previous studies have revealed that PLXDC1 expression is closely associated with angiogenesis[40]. However, to our knowledge, we have demonstrated that PLXDC1 is overexpressed in HCC. In the present study, we proved the overexpression of PLXDC1 in HCC by utilizing publicly accessible databases in association with IHC staining, qRT-PCR, and Western blotting assays. Survival assays indicated that high expression of PLXDC1 was related to poor prognosis in HCC patients. In Furthermore, univariate, and multivariate Cox assays indicated that PLXDC1 was an isolated prognostic risk element for HCC. Survival analysis of HCC patients stratified according to the presence of PLXDC1-associated methylation sites consistently revealed that high methylation levels at these sites were correlated with poor prognosis in patients with HCC. These findings suggest that PLXDC1 may serve as a prognostic biomarker for HCC.
Notwithstanding the accomplishments of anti-angiogenic drugs in the therapy of HCC, some challenging issues have emerged. Suppression of angiogenesis via tumor flight mechanisms, involving upregulation of vicarious pathways, angiogenic mimicry, and recruitment of bone marrow-derived cells, offers only a short-lived survival benefits to patients[41-43]. Angiogenesis inhibitors can increase tumor aggressiveness and metastasis[14,44]. The use of antiangiogenic agents may lead to the development of an immunosuppressive phenotype in tumors. Previous studies have demon
However, there are several limitations to our research. We were able to see that the higher expression of PLXDC1 in HCC tissues mobilized immune evasion, but the precise mechanisms underlying this need to be further examined at both the cellular and animal levels. Furthermore, the unique angiogenic role of PLXDC1 may indicate its potential as a target for antiangiogenic therapy and immunotherapy for tumors; however, more subsequent research is needed.
In summary, we showed that PLXDC1 can be utilized as a bio-marker for poor prognosis and immune evasion in HCC patients from various biological angles.
For the first time, we investigated the oncological role of plexin domain-containing 1 (PLXDC1), also known as tumor endothelial marker 7, in hepatocellular carcinoma (HCC).
According to the latest literature reports and the previous research results of our research team, PLXDC1 participates in the regulation of the development mechanism of some common malignant tumors in the digestive system, such as gastric adenocellular carcinoma. However, whether the biological characteristics of PLXDC1 is related to HCC; HCC is one of the most common tumors of the digestive system, so we want to study whether PLXDC1 gene can affect the development of HCC and its possible mechanism.
To explore whether there is a correlation between the expression of PLXDC1 and the occurrence and development of HCC; explore the correlation between the expression of PLXDC1 and the clinical characteristics and clinical prognosis of HCC; analyze whether PLXDC1 can regulate the occurrence and development and related mechanisms of HCC in the tumor microenvironment, and determine whether this gene may be used as a potential biomarker for clinical prognosis evaluation and related immunotherapy.
Using the cancer genome map database, methylation level assessment, tumor immune microenvironment characteristics screening, tumor immune cell surface checkpoint expression, tumor immune dysfunction and exclusion (TIDE) scoring system and other bioinformatics methods to detect the expression of PLXDC1 in HCC, and evaluate the immune evasion potential of PLXDC1 in HCC;The survival time and other relevant clinical features of clinical HCC cases were collected. The mean ± SD of all clinical data were processed using SPSS 23.0 software, GSEA (v 4.2.3) algorithm, Perl (v 5.32.1.1), and the Kaplan-Meier method, and the correlation between PLXD 1 and HCC was analyzed by relevant statistical procedures such as chi-square test and log-rank test; Through immunohistochemical (IHC) staining analysis, real-time quantitative polymerase chain reaction (PCR) by qRT and Western, the expression level of PLXDC1 gene influences the occurrence and development in tissue metabolism, gene metabolism and protein metabolism of HCC.
Overexpression of PLXDC1 in HCC was associated with poor prognosis based on IHC staining, qRT-PCR, and Western blot assays. Univariate and multivariate Cox analyses suggested that PLXDC1 may be an independent prognostic risk factor. Methylation site analysis indicated that the prognosis of HCC patients with high methylation levels was poorer than that of those with low methylation levels. Pathway enrichment analysis of HCC tissues indicated that genes upregulated in the high-PLXDC1 subgroup were enriched in mesenchymal and immune activation signaling, and TIDE assessment revealed that the risk of immune evasion was significantly greater in the high-PLXDC1 subgroup than in the low-PLXDC1 subgroup. A high PLXDC1-related risk score also indicated a poor prognosis, and immune evasion was significantly greater in the high-risk group than in the low-risk group.
As a result of this research analyzing PLXDC1 from multiple biological perspectives, it was displayed that it is a biomarker of poor prognosis for HCC patients, and that it plays a role in determining immune evasion status.
Based on the results of our previous research in gastric cancer, we proposed the idea of whether PLXD1 affects HCC in the digestive system. To this end, we used the macroscopic bioinformatics perspectives and found that the expression of PLXD 1 affected HCC; By sorting out and analyzing the pathological characteristics of clinical cases, we again verified the correlation between the expression of PLXD 1 expression and clinical HCC prognosis from the individualized clinical perspectives; Finally, we used basic experimental methods such as immunohistochemistry staining analysis, real-time quantitative polymerase chain reaction and Western blotting to prove the above conjecture and results from the microscopic metabolic perspectives of tissue metabolism, gene metabolism, tissue metabolism, protein metabolism and so on.
We would like to express our sincere appreciation for the curators of the platforms and datasets from the open databases TCGA, Sento Academic, EXPRESS, SurvivalMeth, MSigDB, and TIDE.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Amin A, United Arab Emirates; Mizuguchi T, Japan S-Editor: Chen YL L-Editor: A P-Editor: Zhao S
| 1. | Sheng H, Huang Y, Xiao Y, Zhu Z, Shen M, Zhou P, Guo Z, Wang J, Wang H, Dai W, Zhang W, Sun J, Cao C. ATR inhibitor AZD6738 enhances the antitumor activity of radiotherapy and immune checkpoint inhibitors by potentiating the tumor immune microenvironment in hepatocellular carcinoma. J Immunother Cancer. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 156] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 2. | Nagaraju GP, Dariya B, Kasa P, Peela S, El-Rayes BF. Epigenetics in hepatocellular carcinoma. Semin Cancer Biol. 2022;86:622-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 175] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 3. | Wang Y, Zhang Y, Mi J, Jiang C, Wang Q, Li X, Zhao M, Geng Z, Song X, Li J, Zuo L, Ge S, Zhang Z, Wen H, Wang Z, Su F. ANKFN1 plays both protumorigenic and metastatic roles in hepatocellular carcinoma. Oncogene. 2022;41:3680-3693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 4. | Kwon JH, Song GW, Hwang S, Kim KH, Ahn CS, Moon DB, Ha TY, Jung DH, Park GC, Yoon YI, Shim JH, Kim KW, Lee SG. Surgical Outcomes of Spontaneously Ruptured Hepatocellular Carcinoma. J Gastrointest Surg. 2021;25:941-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15301] [Article Influence: 3060.2] [Reference Citation Analysis (4)] |
| 6. | Zhang ZZ, Hua R, Zhang JF, Zhao WY, Zhao EH, Tu L, Wang CJ, Cao H, Zhang ZG. TEM7 (PLXDC1), a key prognostic predictor for resectable gastric cancer, promotes cancer cell migration and invasion. Am J Cancer Res. 2015;5:772-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61:6649-6655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 162] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 8. | Li X, Fan Y, Tang M, Li H, Zhang Y, Mi J, Wang Y, Zhao M, Wang Z, Su F. PLXDC1 Can Be a Biomarker for Poor Prognosis and Immune Evasion in Gastric Cancer. J Inflamm Res. 2022;15:5439-5455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 9. | Nanda A, Buckhaults P, Seaman S, Agrawal N, Boutin P, Shankara S, Nacht M, Teicher B, Stampfl J, Singh S, Vogelstein B, Kinzler KW, St Croix B. Identification of a binding partner for the endothelial cell surface proteins TEM7 and TEM7R. Cancer Res. 2004;64:8507-8511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil Cytoskeleton. 2008;65:687-707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 218] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 11. | Weaver AM. Cortactin in tumor invasiveness. Cancer Lett. 2008;265:157-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 170] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 12. | Falchetti ML, D'Alessandris QG, Pacioni S, Buccarelli M, Morgante L, Giannetti S, Lulli V, Martini M, Larocca LM, Vakana E, Stancato L, Ricci-Vitiani L, Pallini R. Glioblastoma endothelium drives bevacizumab-induced infiltrative growth via modulation of PLXDC1. Int J Cancer. 2019;144:1331-1344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Fuchs B, Mahlum E, Halder C, Maran A, Yaszemski M, Bode B, Bolander M, Sarkar G. High expression of tumor endothelial marker 7 is associated with metastasis and poor survival of patients with osteogenic sarcoma. Gene. 2007;399:137-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15:325-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 745] [Cited by in RCA: 1391] [Article Influence: 198.7] [Reference Citation Analysis (0)] |
| 15. | Majidpoor J, Mortezaee K. Angiogenesis as a hallmark of solid tumors - clinical perspectives. Cell Oncol (Dordr). 2021;44:715-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 155] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 16. | Schito L. Bridging angiogenesis and immune evasion in the hypoxic tumor microenvironment. Am J Physiol Regul Integr Comp Physiol. 2018;315:R1072-R1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Lamplugh Z, Fan Y. Vascular Microenvironment, Tumor Immunity and Immunotherapy. Front Immunol. 2021;12:811485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 18. | Hamza AA, Heeba GH, Hassanin SO, Elwy HM, Bekhit AA, Amin A. Hibiscus-cisplatin combination treatment decreases liver toxicity in rats while increasing toxicity in lung cancer cells via oxidative stress- apoptosis pathway. Biomed Pharmacother. 2023;165:115148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 19. | Bouabdallah S, Al-Maktoum A, Amin A. Steroidal Saponins: Naturally Occurring Compounds as Inhibitors of the Hallmarks of Cancer. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 43] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 20. | Murali C, Mudgil P, Gan CY, Tarazi H, El-Awady R, Abdalla Y, Amin A, Maqsood S. Camel whey protein hydrolysates induced G2/M cellcycle arrest in human colorectal carcinoma. Sci Rep. 2021;11:7062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 21. | Hanna EM, Zaki N, Amin A. Detecting Protein Complexes in Protein Interaction Networks Modeled as Gene Expression Biclusters. PLoS One. 2015;10:e0144163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell. 2018;175:313-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 589] [Cited by in RCA: 985] [Article Influence: 164.2] [Reference Citation Analysis (0)] |
| 23. | Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 2949] [Article Influence: 245.8] [Reference Citation Analysis (0)] |
| 24. | Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3108] [Cited by in RCA: 3608] [Article Influence: 277.5] [Reference Citation Analysis (0)] |
| 25. | Kurebayashi Y, Ojima H, Tsujikawa H, Kubota N, Maehara J, Abe Y, Kitago M, Shinoda M, Kitagawa Y, Sakamoto M. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68:1025-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 333] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 26. | Lozon L, Saleh E, Menon V, Ramadan WS, Amin A, El-Awady R. Effect of safranal on the response of cancer cells to topoisomerase I inhibitors: Does sequence matter? Front Pharmacol. 2022;13:938471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 27. | Hamza AA, Mohamed MG, Lashin FM, Amin A. Dandelion prevents liver fibrosis, inflammatory response, and oxidative stress in rats. JoBAZ. 2020;81:43. [DOI] [Full Text] |
| 28. | Abdu S, Juaid N, Amin A, Moulay M, Miled N. Therapeutic Effects of Crocin Alone or in Combination with Sorafenib against Hepatocellular Carcinoma: In Vivo & In Vitro Insights. Antioxidants (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 51] [Reference Citation Analysis (0)] |
| 29. | Awad B, Hamza AA, Al-Maktoum A, Al-Salam S, Amin A. Combining Crocin and Sorafenib Improves Their Tumor-Inhibiting Effects in a Rat Model of Diethylnitrosamine-Induced Cirrhotic-Hepatocellular Carcinoma. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 37] [Reference Citation Analysis (0)] |
| 30. | Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339:58-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1390] [Cited by in RCA: 1358] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 31. | Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, Ferrara N. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2567] [Cited by in RCA: 2533] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 32. | Yang ZF, Poon RT. Vascular changes in hepatocellular carcinoma. Anat Rec (Hoboken). 2008;291:721-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 33. | Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, Shenkier T, Cella D, Davidson NE. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med. 2007;357:2666-2676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2393] [Cited by in RCA: 2301] [Article Influence: 127.8] [Reference Citation Analysis (0)] |
| 34. | Bear HD, Tang G, Rastogi P, Geyer CE Jr, Robidoux A, Atkins JN, Baez-Diaz L, Brufsky AM, Mehta RS, Fehrenbacher L, Young JA, Senecal FM, Gaur R, Margolese RG, Adams PT, Gross HM, Costantino JP, Swain SM, Mamounas EP, Wolmark N. Bevacizumab added to neoadjuvant chemotherapy for breast cancer. N Engl J Med. 2012;366:310-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 362] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 35. | Miles DW, Chan A, Dirix LY, Cortés J, Pivot X, Tomczak P, Delozier T, Sohn JH, Provencher L, Puglisi F, Harbeck N, Steger GG, Schneeweiss A, Wardley AM, Chlistalla A, Romieu G. Phase III study of bevacizumab plus docetaxel compared with placebo plus docetaxel for the first-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol. 2010;28:3239-3247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 664] [Cited by in RCA: 715] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 36. | Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542-2550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4457] [Cited by in RCA: 4446] [Article Influence: 234.0] [Reference Citation Analysis (0)] |
| 37. | Yang ZY, Liu L, Mao C, Wu XY, Huang YF, Hu XF, Tang JL. Chemotherapy with cetuximab versus chemotherapy alone for chemotherapy-naive advanced non-small cell lung cancer. Cochrane Database Syst Rev. 2014;2014:CD009948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335-2342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7832] [Cited by in RCA: 7727] [Article Influence: 368.0] [Reference Citation Analysis (1)] |
| 39. | Qin S, Li Q, Gu S, Chen X, Lin L, Wang Z, Xu A, Zhou C, Ren Z, Yang L, Xu L, Bai Y, Chen L, Li J, Pan H, Cao B, Fang W, Wu W, Wang G, Cheng Y, Yu Z, Zhu X, Jiang D, Lu Y, Wang H, Xu J, Bai L, Liu Y, Lin H, Wu C, Zhang Y, Yan P, Jin C, Zou J. Apatinib as second-line or later therapy in patients with advanced hepatocellular carcinoma (AHELP): a multicentre, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Gastroenterol Hepatol. 2021;6:559-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 154] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 40. | Bagley RG, Rouleau C, Weber W, Mehraein K, Smale R, Curiel M, Callahan M, Roy A, Boutin P, St Martin T, Nacht M, Teicher BA. Tumor endothelial marker 7 (TEM-7): a novel target for antiangiogenic therapy. Microvasc Res. 2011;82:253-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 41. | Presta M, Dell'Era P, Mitola S, Moroni E, Ronca R, Rusnati M. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 2005;16:159-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 910] [Cited by in RCA: 960] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 42. | Folberg R, Hendrix MJ, Maniotis AJ. Vasculogenic mimicry and tumor angiogenesis. Am J Pathol. 2000;156:361-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 489] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 43. | van Beijnum JR, Nowak-Sliwinska P, Huijbers EJ, Thijssen VL, Griffioen AW. The great escape; the hallmarks of resistance to antiangiogenic therapy. Pharmacol Rev. 2015;67:441-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 168] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 44. | Pàez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Viñals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1901] [Cited by in RCA: 1901] [Article Influence: 118.8] [Reference Citation Analysis (0)] |
| 45. | Tian L, Goldstein A, Wang H, Ching Lo H, Sun Kim I, Welte T, Sheng K, Dobrolecki LE, Zhang X, Putluri N, Phung TL, Mani SA, Stossi F, Sreekumar A, Mancini MA, Decker WK, Zong C, Lewis MT, Zhang XH. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature. 2017;544:250-254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 394] [Cited by in RCA: 601] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 46. | Paijens ST, Vledder A, de Bruyn M, Nijman HW. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell Mol Immunol. 2021;18:842-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 566] [Article Influence: 141.5] [Reference Citation Analysis (0)] |
| 47. | Zheng X, Fang Z, Liu X, Deng S, Zhou P, Wang X, Zhang C, Yin R, Hu H, Chen X, Han Y, Zhao Y, Lin SH, Qin S, Kim BY, Jiang W, Wu Q, Huang Y. Increased vessel perfusion predicts the efficacy of immune checkpoint blockade. J Clin Invest. 2018;128:2104-2115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 183] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 48. | Greene S, Robbins Y, Mydlarz WK, Huynh AP, Schmitt NC, Friedman J, Horn LA, Palena C, Schlom J, Maeda DY, Zebala JA, Clavijo PE, Allen C. Inhibition of MDSC Trafficking with SX-682, a CXCR1/2 Inhibitor, Enhances NK-Cell Immunotherapy in Head and Neck Cancer Models. Clin Cancer Res. 2020;26:1420-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 49. | Hendry SA, Farnsworth RH, Solomon B, Achen MG, Stacker SA, Fox SB. The Role of the Tumor Vasculature in the Host Immune Response: Implications for Therapeutic Strategies Targeting the Tumor Microenvironment. Front Immunol. 2016;7:621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 149] [Article Influence: 16.6] [Reference Citation Analysis (0)] |