Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2074
Peer-review started: December 4, 2023
First decision: January 23, 2024
Revised: February 4, 2024
Accepted: March 8, 2024
Article in press: March 8, 2024
Published online: May 15, 2024
Processing time: 157 Days and 10 Hours
Colon cancer is acknowledged as one of the most common malignancies worldwide, ranking third in United States regarding incidence and mortality. Notably, approximately 40% of colon cancer cases harbor oncogenic KRAS mutations, resulting in the continuous activation of epidermal growth factor receptor signaling.
To investigate the key pathogenic genes in KRAS mutant colon cancer holds considerable importance.
Weighted gene co-expression network analysis, in combination with additional bioinformatics analysis, were conducted to screen the key factors driving the progression of KRAS mutant colon cancer. Meanwhile, various in vitro experi
Integrated analysis demonstrated that TGM2 acted as an independent prognostic factor for progression-free survival. Immunohistochemical analysis on tissue microarrays revealed that TGM2 was associated with an elevated probability of perineural invasion in patients with KRAS mutant colon cancer. Additionally, biological roles of the key gene TGM2 was also assessed, suggesting that the downregulation of TGM2 attenuated the proliferation, invasion, and migration of the KRAS mutant colon cancer cell line.
This study underscores the potential significance of TGM2 in the progression of KRAS mutant colon cancer. This insight not only offers a theoretical foundation for therapeutic approaches but also highlights the need for additional clinical trials and fundamental research to support our preliminary findings.
Core Tip: In the present study, we aim to mine the pathogenic hub genes of KRAS mutant colon cancer via the multi-omics resources of public databases. We use a variety of bioinformatics methods to reveal core genes related to KRAS mutant colon cancer, and subsequently identify the core gene through colon cancer tissue array. Besides, our in vitro results demonstrate that small interfering RNA-induced transglutaminase 2 (TGM2) downregulation suppress the proliferation, migration and invasion of KRAS-mutated colon cell line, suggesting that TGM2 might be a therapeutic target for the treatment of colon cancer.
- Citation: Peng WB, Li YP, Zeng Y, Chen K. Transglutaminase 2 serves as a pathogenic hub gene of KRAS mutant colon cancer based on integrated analysis. World J Gastrointest Oncol 2024; 16(5): 2074-2090
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2074.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2074
Based on statistics from the International Association of Cancer Registries, colorectal cancer (CRC) ranks as the third most prevalent neoplasm and remains a primary contributor to cancer-related mortality, leading to approximately 1 million global deaths annually[1,2]. The primary conventional therapeutic modalities for CRC include surgery, chemotherapy, and targeted therapy. Among these modalities, surgery is especially suitable for early-stage patients with lesions confined to the colon. Unfortunately, at the time of diagnosis, approximately 20% of patients with CRC have already developed distant metastases, thereby rendering surgical intervention unfeasible. Chemotherapy has become the predominant therapeutic recommendation for patients with metastatic CRC. Furthermore, considering the mutation profile Ras genes and the tumor site, a combination of specific targeted agents, such as bevacizumab or cetuximab, is used to potentiate the anti-tumor effects of chemotherapeutic agents. Despite these efforts, the prognosis for patients remains unfavorable.
The KRAS oncogene has been extensively investigated in human malignancies since its identification in the 1960s[3,4]. KRAS mutations have been identified in over 20% of human malignancies, resulting in the persistent activation of downstream signaling pathways, including the Ras/Raf/Mek and PI3K/Akt pathways. This activation, in turn, instigates a series of molecular interactions affecting various cellular activities, such as proliferation, differentiation, survival, growth, and apoptosis, which are malignant biological characteristics associated with cancer[5].
Fearon and Vogelstein[6] initially highlighted the association between KRAS mutation status and CRC in 1990. Since then, research has identified nearly 3000 different types of KRAS point mutations[7]. Among these mutations, the most prevalent mutation is observed at codon 12 (exon 2), representing 83% of cases. Additionally, mutations at codon 13 (exon 2), codon 146 (exon 4), and codon 61 (exon 3) account for 17%, 4%, and 2% of cases, respectively[8,9].
A notable breakthrough in the therapeutic management of patients with metastatic CRC involves the application of monoclonal antibodies targeting epidermal growth factor receptor (EGFR), including panitumumab and cetuximab. These therapeutic agents have been proven to considerably enhance the survival rates of individuals in advanced stages. Importantly, they provide an advantageous alternative to chemotherapy, minimizing adverse effects and ultimately enhancing the overall quality of life for patients. However, the presence of a mutant KRAS status in these patients has been consistently correlated with an inadequate response to EGFR-targeted therapy[10,11]. Therefore, the clinical efficacy of single-agent panitumumab and cetuximab is specifically limited to patients harboring wild-type KRAS.
This investigation was undertaken with the objective of identifying the pivotal molecules in patients with KRAS mutant colon cancer using The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets. The weighted gene co-expression network analysis (WGCNA), in combination with additional bioinformatics analysis, was conducted to screen the key factors driving the progression of this cancer. Subsequently, we proceeded to substantiate this finding through immunohistochemistry (IHC) on tissue microarrays and in vitro assays. In summary, a comprehensive investigation into the core pathogenic genes associated with KRAS mutant CRC bears significant importance. Such an in-depth analysis facilitates the identification of pivotal drivers in the progression of colon cancer, offering potential avenues for the development of novel targeted therapeutic strategies. These advancements hold promise for enhancing the long-term prognosis of patients with chemorefractory metastatic colon cancer in the future.
Data encompassing transcriptome profiling (n = 458), simple nucleotide variation (n = 398), and comprehensive clinical profiles (n = 427) of patients with colon adenocarcinoma (COAD) were acquired from the TCGA database (https://portal.gdc.cancer.gov/), with the exclusion criteria applied to individuals with survival time < 30 d and incomplete disease-related information. Ultimately, a total of 367 samples were incorporated into the study from the TCGA-COAD dataset subsequent to the intersection. This cohort included 158 patients with KRAS mutant colon cancer and 209 with KRAS wild-type colon cancer. Concurrently, an independent external dataset was constructed by utilizing a cohort of 193 patients with colon cancer from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE87211. This dataset was established following the exclusion of 10 patients for whom the KRAS mutation status remained undetermined. Hence, 84 patients with KRAS mutant colon cancer and 109 patients with KRAS wild-type colon cancer were included in this study. Data normalization and processing were conducted using the R software (version 4.0.2) and the “limma[12]” package.
Scale-free networks, derived from gene expression profiles, were established independently for the TCGA-COAD and GSE87211 datasets. These networks were constructed using the WGCNA package (version 1.71) in the R software environment following established methodologies[13]. Gene expression matrices were transformed into matrices encapsulating the pairwise similarities among mRNA molecules. Subsequently, these matrices were further converted into adjacency matrices using the Pearson correlation coefficient test. An optimal soft-threshold power value was selected to ensure that the adjacency matrix conformed to the criteria of a scale-free topology. Following this selection, topological overlap matrix (TOM) and dissimilarity TOM (1-TOM) were created using TOM similarity and dissimilarity modules based on the adjacency matrix. Finally, the identification of the module was achieved through the dynamic tree-cutting method, with the minimum module size specified as 50. Modules exhibiting significant similarity scores were subsequently merged based on a predetermined threshold value. To ascertain the modules with robust associations with the examined clinical features, the expression profiles of each module were condensed into the eigenvector correlated with the foremost principal component in the expression matrix using module eigengene. The association between specific genes and the clinical characteristics of interest was evaluated through the estimation of gene significance. Based on these two metrics, modules demonstrating a notable correlation with KRAS mutant status were identified as KRAS mutation-related modules.
The Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were carried out on common genes across modules using the R package “clusterProfiler”[14] (version 4.4.4). An adjusted P-value < 0.05 was established as the criterion for a significant result in the enrichment analysis.
The protein-protein interaction (PPI) network of genes from KRAS mutation-related modules was constructed using the STRING website (http://string-db.org, version 11.0) and Cytoscape software (http://www.cytoscape.org/, Version 3.8.2). The CytoHubba plugin in the Cytoscape software was employed to calculate the degree of connectivity of each node. Typically, nodes with greater connectivity indicate a more substantial role in the network overall[15]. Following a comprehensive screening, a total of ten candidate hub genes closely linked to KRAS mutations in colon cancer were obtained for further analyses.
Paraffin-embedded tissue microarrays containing colon cancer samples (HColA180Su21) were purchased from Outdo Biotech (Shanghai, China). Comprehensive clinicopathological data were collected for further analysis. The tissue microarray comprised 94 specimens of colon cancer paired with their corresponding 86 adjacent normal tissue samples. This array encompassed patients with colon cancer who were 29-86 years old, with a median age of 65 years. These individuals had undergone surgical procedures from February 2012 to September 2014 and were enrolled in the microarray analysis. The follow-up period ended in July 2018. In the subsequent statistical analysis, a total of 90 patients were incorporated, with the exclusion of 4 cases characterized by exceptionally brief survival durations. The characteristics of patients with colon cancer represented in the selected tissue microarray are summarized in Table 1.
| Characteristic | All patients (n = 90) | |
| Age, mean ± SD | 64.11 ± 13.17 | |
| Gender, n (%) | Female | 46 (51.11) |
| Male | 44 (48.89) | |
| Histologic grade, n (%) | Grade II | 57 (63.33) |
| Grade III | 33 (36.67) | |
| Maximum tumor diameter, median (Q1, Q3) | 5.40 (4.00, 7.00) | |
| Tumor location, n (%) | Right colon | 42 (46.67) |
| Left colon | 41 (45.56) | |
| Unknown | 7 (7.78) | |
| Lympho-vascular invasion, n (%) | No | 60 (66.67) |
| Yes | 30 (33.33) | |
| Perineural invasion, n (%) | No | 74 (82.22) |
| Yes | 16 (17.78) | |
| Pathologic T stage, n (%) | T2 | 9 (10.00) |
| T3 | 31 (34.44) | |
| T4a | 41 (45.56) | |
| T4b | 9 (10.00) | |
| Pathologic N stage, n (%) | N0 | 56 (62.22) |
| N1 | 18 (20.00) | |
| N2 | 16 (17.78) | |
| Clinical M stage, n (%) | M0 | 86 (95.56) |
| M1 | 4 (4.44) | |
| Disease stage, n (%) | I | 9 (10.00) |
| II | 45 (50.00) | |
| III | 32 (35.56) | |
| IV | 4 (4.44) | |
| CD8 positive rate, n (%) | < 5% | 60 (66.67) |
| ≥ 5% | 30 (33.33) | |
| PD-L1 positive rate, n (%) | < 5% | 63 (70.00) |
| ≥ 5% | 27 (30.00) | |
| PD1 positive rate, n (%) | < 5% | 66 (73.33) |
| ≥ 5% | 24 (26.67) | |
| KRAS mutant status, n (%) | Mutant | 28 (31.11) |
| Wild type | 62 (68.89) | |
The paraffin sections were initially subjected to a treatment process aimed at eliminating paraffin and achieving rehydration. This procedure involved the use of xylene and alcohol. Subsequently, antigen retrieval was conducted using a citric acid antigen retrieval buffer. Following the blocking step with 5% bovine serum albumin, the sections underwent an overnight incubation at 4 °C with primary antibodies (anti-TGM2, Catlog#ab109200, Abcam, diluted at 1:100; anti-JAK3, Catlog#ab45141, Abcam, diluted at 1:200). To visualize the immunoreaction, 3,3’-diaminobenzidine was used as a chromogenic substrate. The H-score was calculated using the following formula: H-score = [1 × (% of cell with weak intensity) + 2 × (% of cell with intermediate intensity) + 3 × (% of cell with strong intensity)] × 100. The final H-score was automatically determined through the utilization of the PerkinElmer InForm software (version 2.4.0).
The KRAS mutant human colon cancer cell line HCT-116 was used in this study. The cell line was purchased from the Chinese Academy of Sciences, Shanghai Institutes for Biological Sciences (Shanghai, China). These cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, United States) supplemented with 10% fetal bovine serum (FBS) (Gibco, United States) and antibiotics (100 U/mL of penicillin and 100 μg/mL of streptomycin) in a 37 °C humidified incubator with 5% CO2.
HCT-116 cells were plated in 6-well plates and allowed to reach a confluence of 50%-60% before transfection. The transfection was carried out using a mixture of small interfering RNA (siRNA) (TGM2-siRNA or NC-siRNA) obtained from RiboBio (Guangzhou, China) and lipofectamine 3000 reagents (Invitrogen, United States) following the guidelines provided by the manufacturer. Western blotting was performed 48 h after transfection to validate the efficiency of gene knockdown as previously reported[16]. Subsequent in vitro experiments were conducted 24 h post-transfection.
The proliferative potential of cancer cells was assessed using the Cell Counting Kit (CCK)-8 assay. In 96-well plates, 2000 cells were seeded per well and then incubated for 0-3 d. At the specified time points, the optical density at 450 nm was measured using a VERSA microplate reader (Molecular Devices, Sunnyvale, CA, United States) after the addition of CCK-8 solution (MedChemExpress, United States) and an additional incubation period of 2-4 h. For the colony formation assay, cells were seeded onto 6-well plates and allowed to incubate in a complete medium. After 7-10 d, the colonies were fixed with methanol and subsequently stained with 0.1% crystal violet. Following staining, the microscopic examination was conducted to count colonies, with each well requiring the presence of more than 50 cells in a colony for inclusion in the count.
In the migration assay, cells were resuspended in 200 μL serum-free DMEM and subsequently introduced into the upper chamber of the Transwell Boyden chamber system (Corning Life Sciences, NY, United States). In contrast, the upper chamber of the Transwell system was coated with Matrigel for the cell invasion assay. Subsequently, 600 μL of DMEM containing 10%-15% FBS was introduced into the lower chamber as a chemoattractant. Following 24 h, the number of migrated or invasive cells was counted under a microscope, with an examination of five randomly selected fields per well after fixation and crystal violet staining. Each assay was independently conducted three times.
Experiments were performed in triplicate when indicated, and the data were presented as the means ± SD. The student’s t-test was used to compare differences between two groups. Prism software v8.4 (GraphPad Software, La Jolla, CA, United States) and R software were used to conduct the statistical analyses. Differences with P < 0.05 were considered statistically significant.
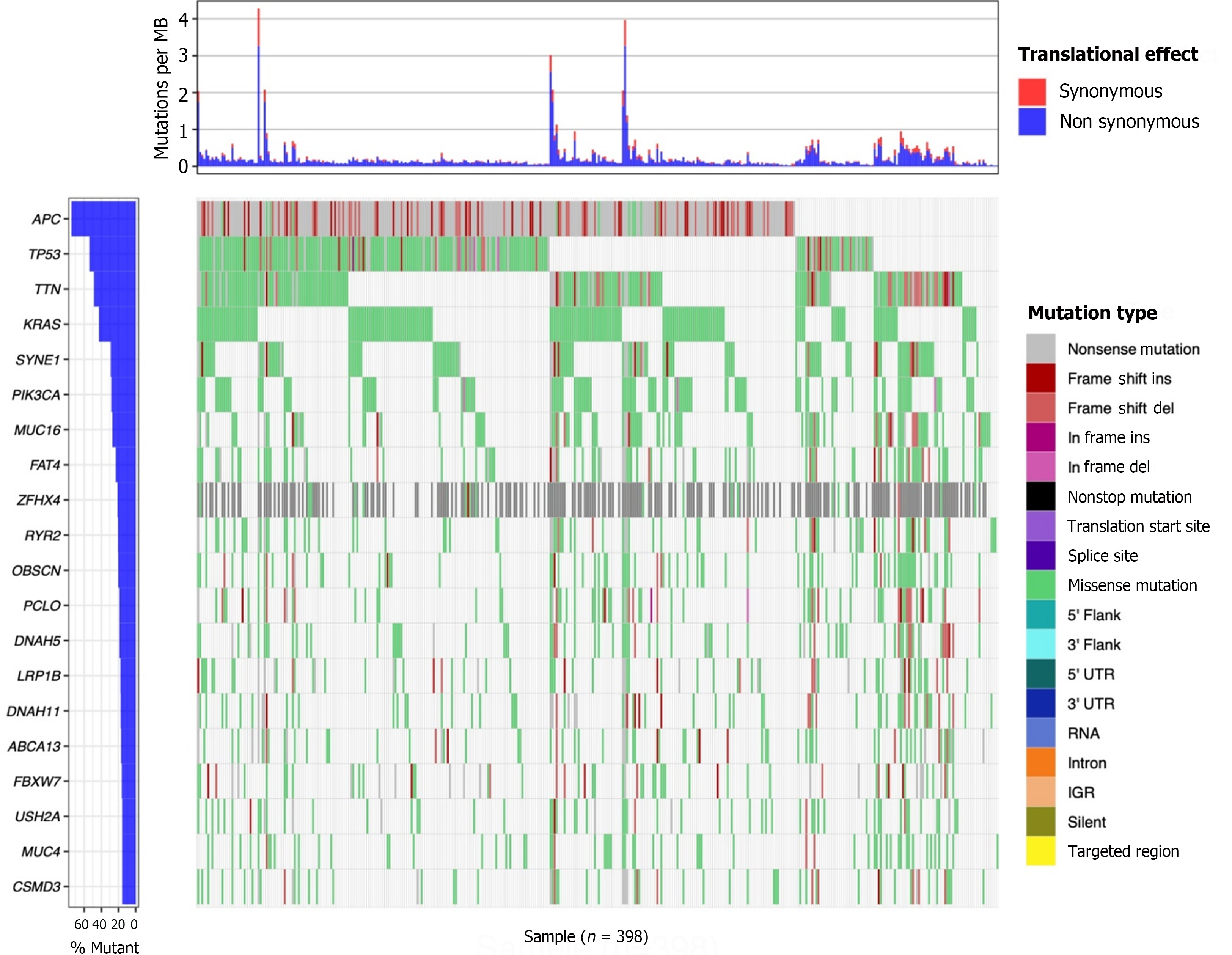
The landscape of mutation profiles in patients with colon cancer was graphically represented using the “maftools[17]” package. The top 20 mutated genes are displayed in Figure 1. Among 398 patients with colon cancer, the top five most frequently occurring genetic mutations were observed in APC, TP53, TTN, KRAS, and SYNE1. Notably, KRAS mutations accounted for 42.6% of these cases. Our subsequent research was directed towards an investigation into KRAS mutations in light of the significant contributions of KRAS to tumorigenesis, the widespread occurrence of KRAS mutations in colon cancer, and the unique significance of KRAS mutations in targeted therapy of colon cancer.
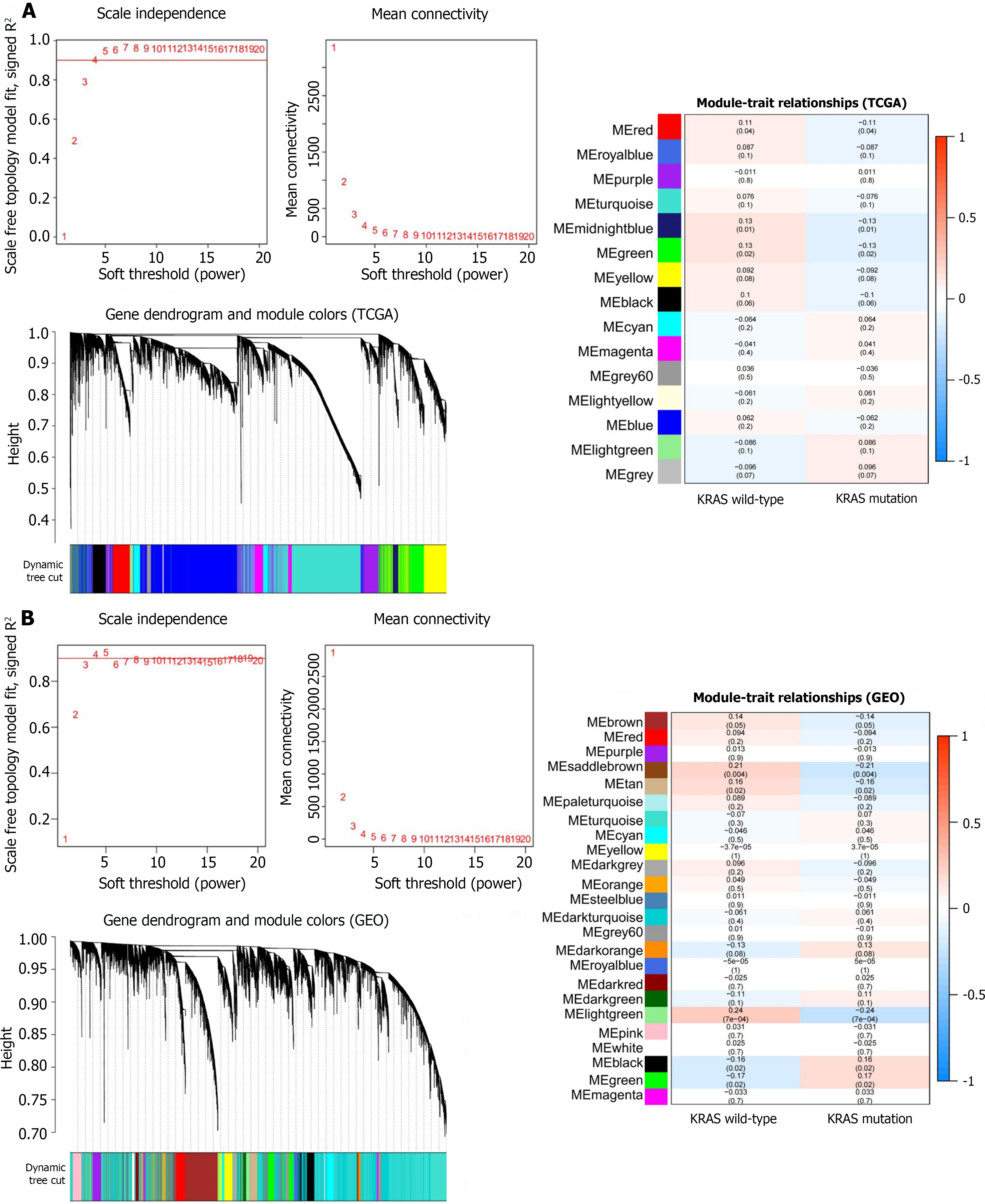
To unravel the pivotal molecules associated with KRAS mutations in colon cancer, the WGCNA method was employed to conduct a gene co-expression network on the selected TCGA and GEO cohorts using the “WGCNA” package. As presented in Figure 2, scale-free networks were successfully constructed for both the TCGA and GEO cohorts following the selection of the optimal soft threshold. In the TCGA-COAD dataset, we identified three different color modules related to KRAS mutations: The red module, the midnight-blue module, and the green module (P < 0.05 for all). Similarly, in the GSE87211 dataset, we identified five different color modules associated with KRAS mutation, namely, the saddle-brown module, tan module, light-green module, black module, and green module (P < 0.05 for all). The intersection of genes within the KRAS mutation-related modules from both the TCGA and GSE87211 datasets was determined using the R package “VennDiagram”[18]. Subsequently, a total of 36 genes were screened out (Supplementary Figure 1A). The KEGG enrichment analysis was performed to further explore the functional enrichment of these 36 KRAS mutation-related genes. As indicated in Supplementary Figure 1B, the most important KEGG pathway terms were primarily enriched in the regulation of viral infection- and immune response-related pathways.
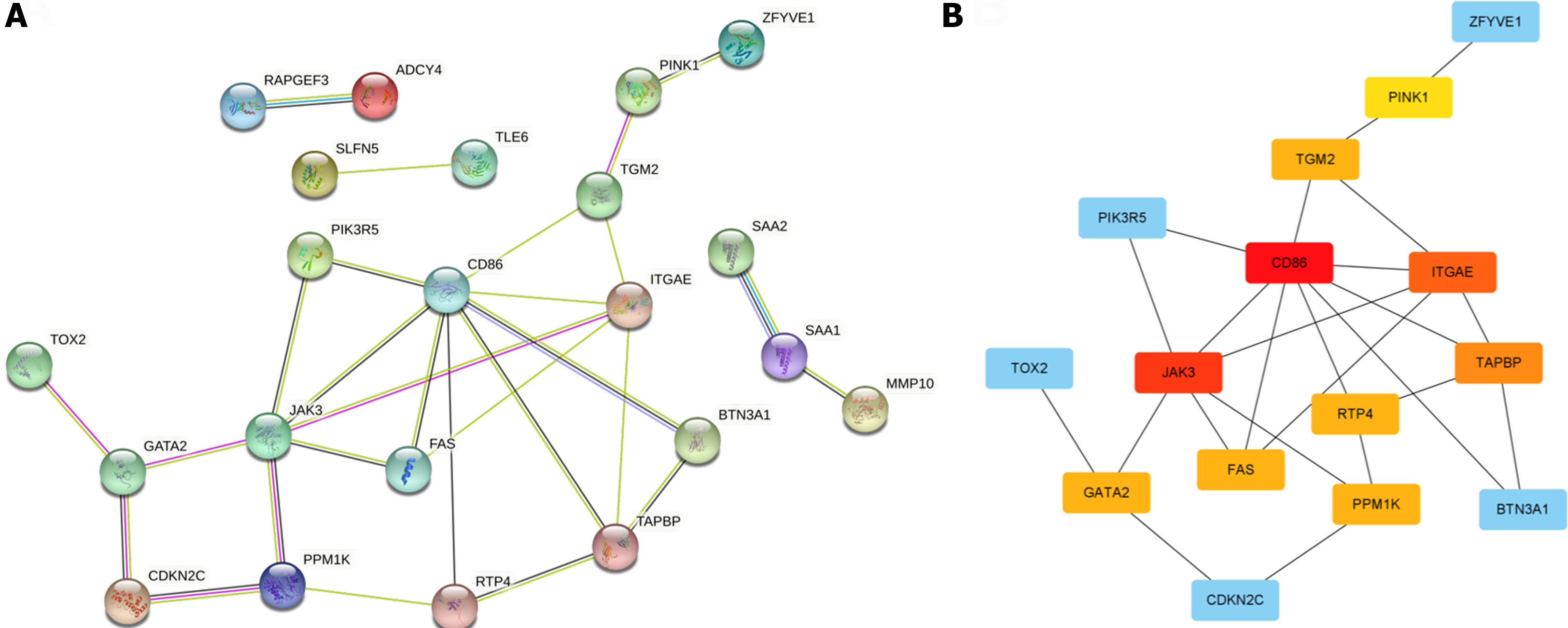
To investigate the most prominent clusters associated with KRAS mutations, the PPI network was constructed using the STRING database. We applied a confidence score threshold of 0.7 and removed protein nodes devoid of interactions with others. In total, 22 out of the 36 KRAS mutation-related genes were mapped into the PPI network, as displayed in Figure 3A. Subsequently, the CytoHubba plugin within the Cytoscape software was used to calculate the degree of connectivity for each node. Nodes with greater connectivity typically signify a more pivotal role in the network. Consequently, we pinpointed a set of 10 candidate hub genes closely associated with KRAS mutations in colon cancer for further analysis (Figure 3B).
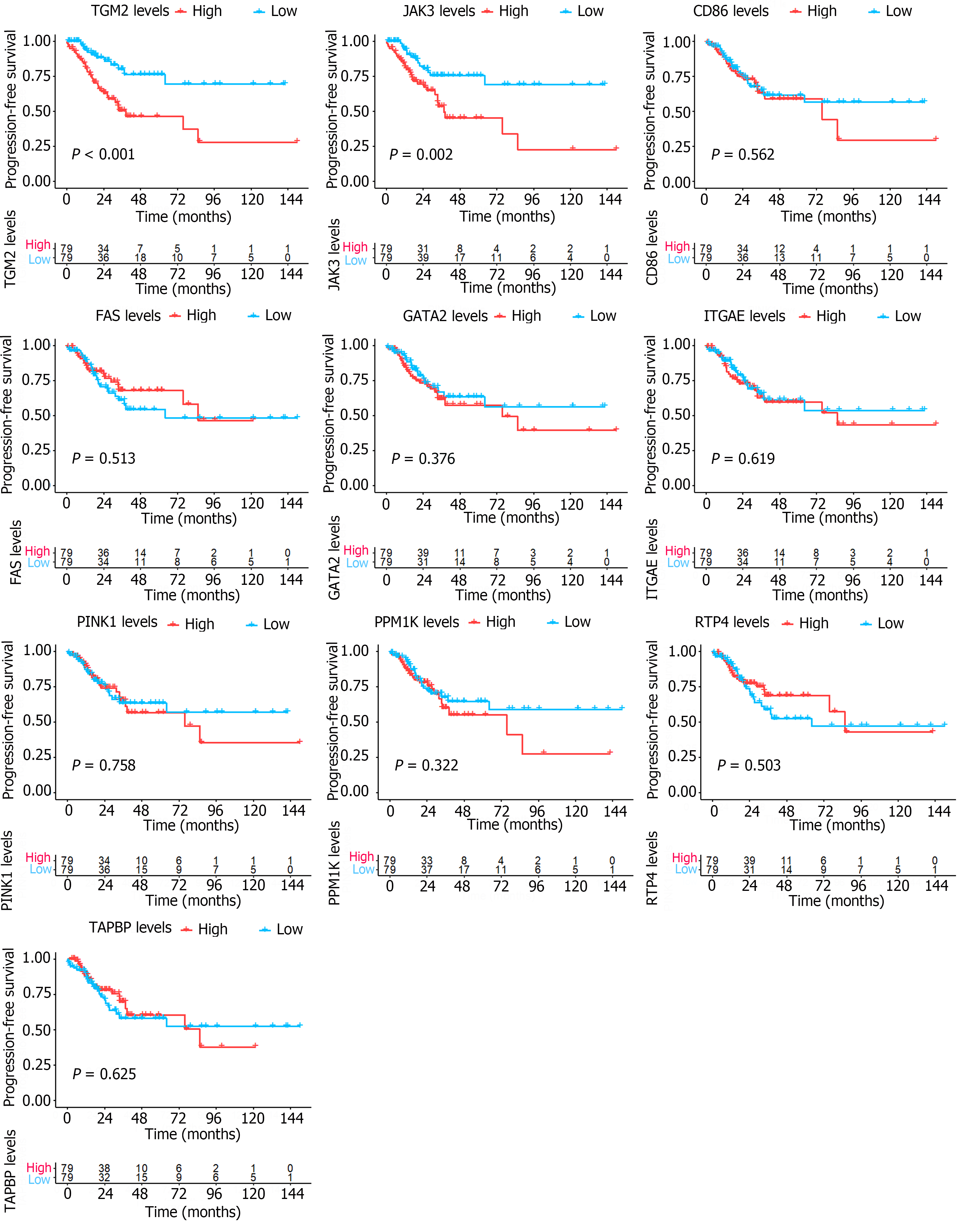
We conducted Kaplan-Meier survival analysis for both overall survival (OS) and progression-free survival (PFS) to further substantiate the prognostic relevance of these 10 candidate hub genes for patients afflicted with KRAS-mutant colon cancer. This analysis was carried out using a cohort of 158 individuals with KRAS mutant colon cancer and 209 individuals with KRAS wild-type colon cancer. Upon stratifying the gene expression into high- and low-expression groups using the median value, our analysis revealed that none of the ten candidate hub genes exhibited notable prognostic significance concerning OS among patients with KRAS mutant and KRAS wild-type colon cancer (Supplementary Figures 2 and 3, P > 0.05 for all). However, Kaplan-Meier survival analysis focusing on PFS unveiled that among the ten candidate hub genes, only elevated expression of JAK3 and TGM2 was associated with an unfavorable prognosis, as indicated in Figure 4. It should be noted that this phenotype was solely observed in patients harboring KRAS mutations and was not evident in patients with KRAS wild-type status (Supplementary Figure 4).
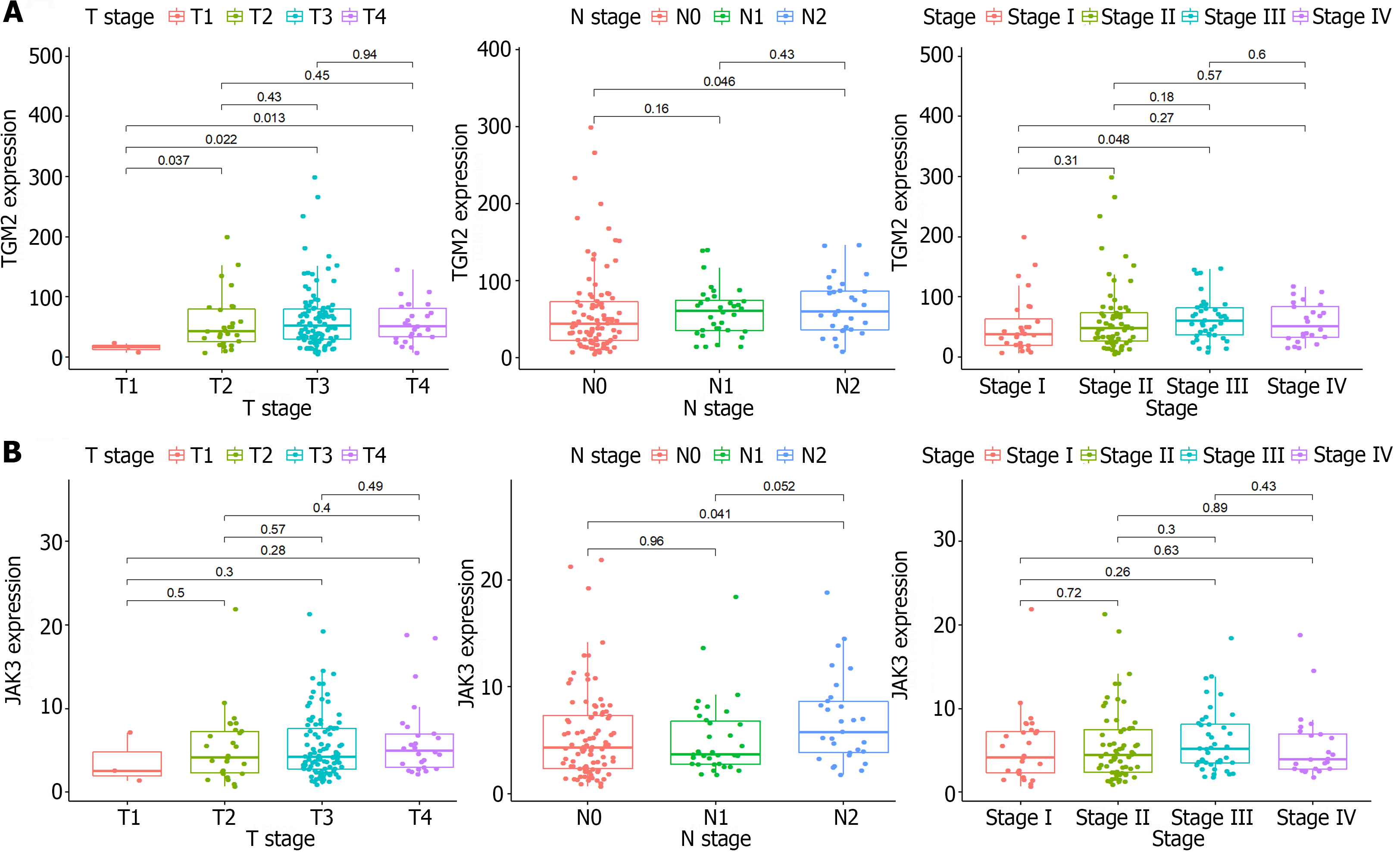
Subsequently, the study shifted to the examination of the correlation between the levels of TGM2 and JAK3 and the clinicopathological characteristics of patients with colon cancer. It was evident that the levels of TGM2 (Figure 5A, P = 0.046) and JAK3 (Figure 5B, P = 0.041) in N2 stage patients were significantly higher than those in N0 stage patients. This observation suggests that JAK3 and TGM2 may assume an important role in lymph node metastasis, consequently influencing the prognosis of patients with KRAS mutant colon cancer. Consistent with the aforementioned results, this phenotype was exclusive to patients with KRAS mutant colon cancer and not evident in those with KRAS wild-type status (Supplementary Figure 5).
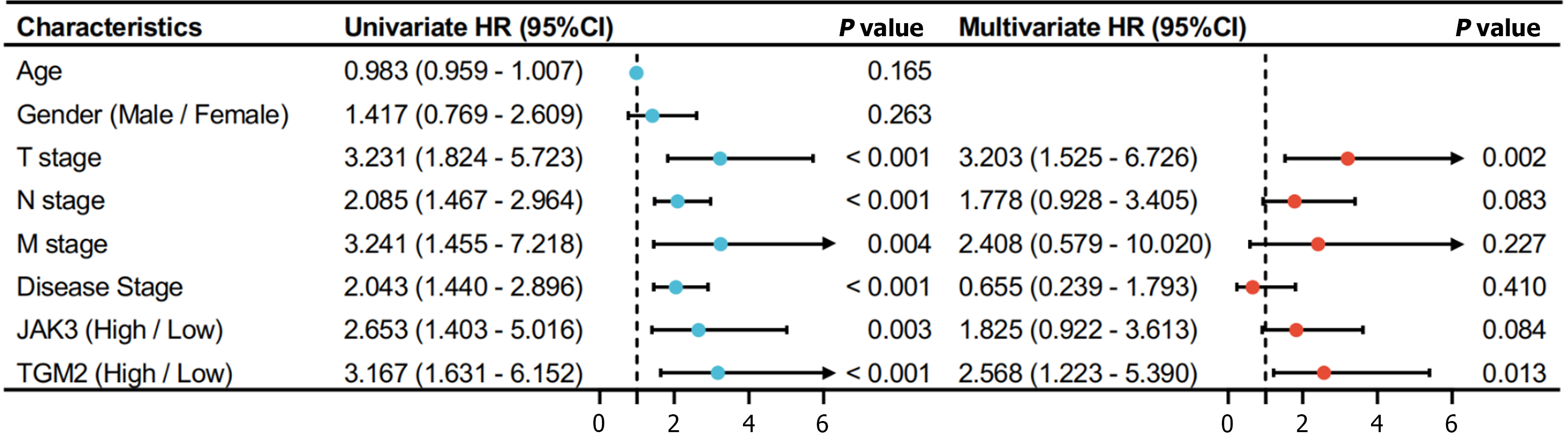
The univariate Cox regression analysis was further used to investigate PFS-related factors in KRAS mutant colon cancer samples. The analysis encompassed variables such as age, gender, tumor-node-metastasis (TNM) stages, and the levels of JAK3 and TGM2. As illustrated in Figure 6, the results of univariate Cox analysis indicated significant associations between PFS and TNM stages and the levels of JAK3 and TGM2 (P < 0.05 for all). However, only a high level of TGM2 [hazard ratio (HR) = 2.568, 95% confidence interval (CI): 1.223-5.390, P = 0.013] and the T stage (HR = 3.203, 95%CI: 1.525-6.726, P = 0.002) were identified as independent prognostic risk factors based on the multivariate cox analysis.
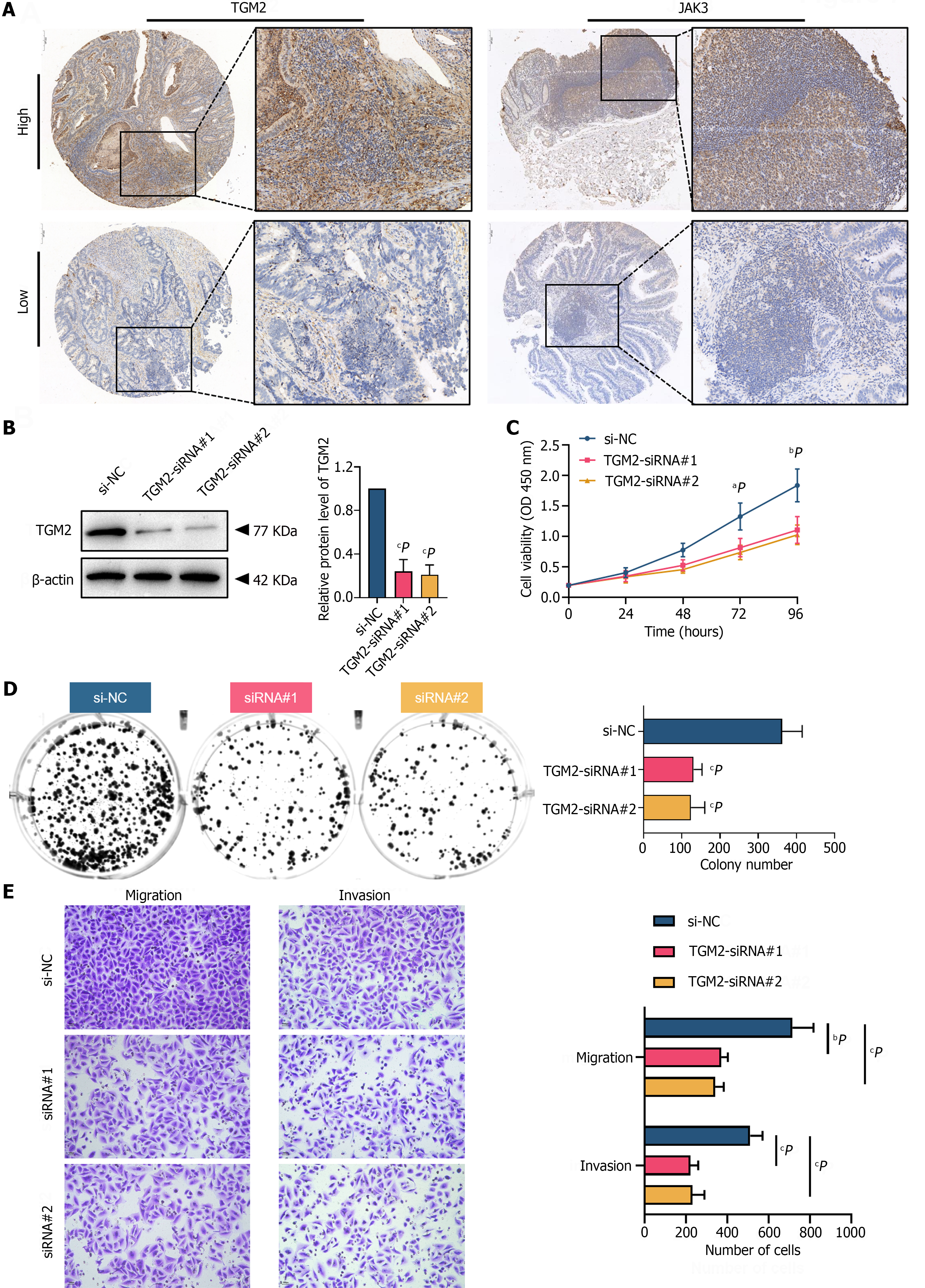
Paraffin-embedded colon cancer tissue microarrays were employed to corroborate the aforementioned findings observed in the TCGA-COAD cohort. The clinicopathological characteristics of the 90 patients are demonstrated in Table 1. The IHC results revealed that both TGM2 and JAK3 protein levels were predominantly localized in the nucleus and cytoplasm of colon cancer cells (Figure 7A). The median H-score for TGM2 was 10.25, with quartiles ranging from 2.56 to 68.27. Similarly, the median H-score for JAK3 was 60.49, with quartiles ranging from 41.78 to 93.79. The cut-off value for TGM2 H-score was established based on a threshold of 10. Patients with an H-score > 10 were classified as the high-expression group, and those with an H-score < 10 were classified as the low-expression group. Likewise, an H-score cut-off of 60 was designated for JAK3. χ2 tests were used to evaluate the associations between the expression of TGM2 and JAK3 and clinicopathological characteristics. The results demonstrated that the expression of TGM2 was associated with lympho-vascular invasion (P = 0.013) in the entire patient cohort (n = 90), as depicted in Table 2. In contrast, except for gender, there was no significant correlation between JAK3 expression and the clinicopathological features of patients (Supplementary Table 1).
| Clinical parameters | TGM2 expression level | χ2 | P value | |
| Low (n = 44) | High (n = 46) | |||
| Age | 0.148 | 0.701 | ||
| ≤ 60 | 18 (40.91) | 17 (36.96) | ||
| > 60 | 26 (59.09) | 29 (63.04) | ||
| Gender | 0.046 | 0.829 | ||
| Female | 23 (52.27) | 23 (50.00) | ||
| Male | 21 (47.73) | 23 (50.00) | ||
| Histologic grade | 0.667 | 0.414 | ||
| Grade II | 26 (59.09) | 31 (67.39) | ||
| Grade III | 18 (40.91) | 15 (32.61) | ||
| Tumor size | 0.525 | 0.469 | ||
| < 5 cm | 14 (31.82) | 18 (39.13) | ||
| ≥ 5 cm | 30 (68.18) | 28 (60.87) | ||
| Lympho-vascular invasion | 6.183 | 0.013a | ||
| No | 34 (77.27) | 24 (52.17) | ||
| Yes | 10 (22.73) | 22 (47.83) | ||
| Perineural invasion | 2.423 | 0.120 | ||
| No | 39 (88.64) | 35 (76.09) | ||
| Yes | 5 (11.36) | 11 (23.91) | ||
| Pathologic T stage | 1.076 | 0.300 | ||
| T2-T3 | 22 (50.00) | 18 (39.13) | ||
| T4a-T4b | 22 (50.00) | 28 (60.87) | ||
| Pathologic N stage | 0.073 | 0.787 | ||
| N0 | 28 (63.64) | 28 (60.87) | ||
| N1-2 | 16 (36.36) | 18 (39.13) | ||
| Clinical M stage | 0.000 | 1.000 | ||
| M0 | 42 (95.45) | 44 (95.65) | ||
| M1 | 2 (4.55) | 2 (4.35) | ||
| Disease stage | 0.067 | 0.796 | ||
| Stage I & II | 27 (61.36) | 27 (58.70) | ||
| Stage III & IV | 17 (38.64) | 19 (41.30) | ||
| CD8 positive rate | 0.022 | 0.881 | ||
| < 5% | 29 (65.91) | 31 (67.39) | ||
| ≥ 5% | 15 (34.09) | 15 (32.61) | ||
| PD-L1 positive rate | 0.686 | 0.408 | ||
| < 5% | 29 (65.91) | 34 (73.91) | ||
| ≥ 5% | 15 (34.09) | 12 (26.09) | ||
| PD-1 positive rate | 1.699 | 0.192 | ||
| < 5% | 35 (79.55) | 31 (67.39) | ||
| ≥ 5% | 9 (20.45) | 15 (32.61) | ||
| KRAS mutant status | 0.098 | 0.754 | ||
| Wild type | 31 (70.45) | 31 (67.39) | ||
| Mutation | 13 (29.55) | 15 (32.61) | ||
Additionally, χ2 tests were performed in a subset of 28 patients with KRAS mutant colon cancer isolated from the entire patient cohort. As indicated in Table 3 and Supplementary Table 2, elevated TGM2 expression was indicative of an increased probability of perineural invasion (P = 0.044) in the KRAS mutant subgroup (n = 28). Despite the affirmative finding, it is imperative to exercise caution in interpreting this conclusion due to the limited sample size in the population. Together, these results reveal that TGM2 may hold a significant role in tumor progression and invasion, underscoring the need for further investigation into the molecular functions of TGM2 in colon cancer.
| Clinical parameters | TGM2 expression level | P value | |
| Low (n = 13) | High (n = 15) | ||
| Age | 0.476 | ||
| ≤ 60 | 5 (38.46) | 8 (53.33) | |
| > 60 | 8 (61.54) | 7 (46.67) | |
| Gender | 1.000 | ||
| Female | 7 (53.85) | 7 (46.67) | |
| Male | 6 (46.15) | 8 (53.33) | |
| Histologic grade | 1.000 | ||
| Grade II | 8 (61.54) | 10 (66.67) | |
| Grade III | 5 (38.46) | 5 (33.33) | |
| Tumor size | 0.700 | ||
| < 5 cm | 6 (46.15) | 5 (33.33) | |
| ≥ 5 cm | 7 (53.85) | 10 (66.67) | |
| Lympho-vascular invasion | 0.670 | ||
| No | 9 (69.23) | 12 (80.00) | |
| Yes | 4 (30.77) | 3 (20.00) | |
| Perineural invasion | 0.044a | ||
| No | 13 (100.00) | 10 (66.67) | |
| Yes | 0 (0.00) | 5 (33.33) | |
| Pathologic T stage | 0.445 | ||
| T2-T3 | 7 (53.85) | 5 (33.33) | |
| T4a-T4b | 6 (46.15) | 10 (66.67) | |
| Pathologic N stage | 0.460 | ||
| N0 | 9 (69.23) | 8 (53.33) | |
| N1-2 | 4 (30.77) | 7 (46.67) | |
| Clinical M stage | 1.000 | ||
| M0 | 13 (100.00) | 14 (93.33) | |
| M1 | 0 (0.00) | 1 (6.67) | |
| Disease stage | 0.276 | ||
| Stage I & II | 9 (69.23) | 7 (46.67) | |
| Stage III & IV | 4 (30.77) | 8 (53.33) | |
| CD8 positive rate | 1.000 | ||
| < 5% | 9 (69.23) | 11 (73.33) | |
| ≥ 5% | 4 (30.77) | 4 (26.67) | |
| PD-L1 positive rate | 1.000 | ||
| < 5% | 9 (69.23) | 10 (66.67) | |
| ≥ 5% | 4 (30.77) | 5 (33.33) | |
| PD-1 positive rate | 1.000 | ||
| < 5% | 9 (69.23) | 10 (66.67) | |
| ≥ 5% | 4 (30.77) | 5 (33.33) | |
Next, a KRAS mutant colon cancer cell line HCT-116 was used in this study to explore the molecular function of TGM2. TGM2-siRNAs were transfected into HCT-116 cells, and western blotting was used to confirm the efficiency of TGM2 expression knockdown (Figure 7B). Cell proliferation dynamics were assessed through CCK-8 assays, demonstrating a substantial inhibition of cell proliferation in the KRAS mutant colon cancer cell line HCT-116 upon TGM2-siRNA transfection (Figure 7C). Furthermore, this observation was corroborated by colony formation assays (Figure 7D). In light of our prior findings, which established a close association between TGM2 and phenotypes such as lymph node metastasis and perineural invasion, we proceeded to investigate its impact on migration and invasion, two of the most important malignant characteristics. The Boyden chamber Transwell assay was employed in the presence or absence of Matrigel coating to assess invasion and migration, respectively. As depicted in Figure 7E, the number of invaded and migrated cells was visibly reduced in response to the siRNA-induced knockdown of TGM1 compared to the control group. In summary, these results provide clear evidence of the pivotal role played by TGM2 in KRAS mutant colon cancer cells.
Colon cancer is a highly prevalent malignancy globally, ranking third regarding incidence and mortality rates in the United States for both women and men[19]. Extensive genomic data reveal that KRAS mutations represent one of the most frequent factors leading to the aberrant activation of the EGFR signaling pathway in patients with colon cancer. Approximately 40% of CRC cases carry oncogenic KRAS mutations, particularly G12D, G12V, and G13D mutations, which persistently activate the EGFR signaling pathway[20,21]. These KRAS mutations can induce the synthesis of cell cycle protein Cyclin D1, which can automatically activate the EGFR pathway and initiate downstream signal transduction independent of EGFR signal reception. Consequently, this acceleration in cell proliferation and malignant transformation contributes to unfavorable survival outcomes[22]. Considering the well-established correlation between KRAS mutations and adverse prognosis in patients with colon cancer[23], the endeavor to develop therapeutic strategies focused on the targeting of KRAS or its signaling pathways is considered a paramount priority in clinical practice[24].
However, results from clinical trials indicate that the effectiveness of these small molecule inhibitors targeting KRAS mutations is less promising in the treatment of colon cancer[25-27]. Thus, the task of identifying pivotal mediators and developing therapeutic strategies to inhibit the excessive activation of downstream molecules resulting from KRAS mutations remains a formidable challenge. In this study, we aimed to identify the key pathogenic hub genes of KRAS mutant colon cancer using the multi-omics resources available in public databases. We employed various bioinformatics techniques to reveal core genes associated with KRAS mutant colon cancer and subsequently validated the significance of core genes using a colon cancer tissue microarray. Furthermore, our in vitro findings demonstrated that siRNA-induced TGM2 downregulation inhibited the proliferation, migration, and invasion of KRAS mutant colon cell line, indicating that TGM2 may be a therapeutic target for the treatment of colon cancer.
Several studies have demonstrated that TGM2 is a novel biomarker for various cancer types. Gu et al[28] reported that the overexpression of TGM2 induced aggressive phenotypes and drug resistance in colon cancer, which may be attributed to the regulation through activation of Wnt/β-catenin signaling. Consistent with the aforementioned findings, research conducted by Yang et al[29] similarly indicated a pro-tumorigenic role of TGM2 in colon cancer. Additionally, Torres et al[30] also demonstrated that TGM2 acted as one of the molecular markers of endometrial cancer. TGM2 has been shown to maintain the aggressive phenotype of mesothelioma cancer through MET receptor signaling[31]. In addition to its involvement in tumorigenesis, studies have indicated that TGM2 is associated with treatment resistance in various cancers, including lung cancer[32], laryngeal cancer[33], osteosarcoma[34], and pancreatic cancer[35]. Therefore, our results align with previous studies[36-39], all of which substantiate the significance of TGM2 in colon cancer. Notably, our results indicated that TGM2 could serve as a pivotal factor contributing to the unfavorable prognosis of KRAS mutant colon cancer, warranting further in-depth research in this context.
Another intriguing component of TGM2 lies in its potential as a candidate for anti-cancer therapy, as previously documented. Results from Malkomes et al[38] suggested that TGM2 inhibitors exhibited a significant inhibitory effect on tumor progression, demonstrating their promising therapeutic prospects. However, current treatment guidelines exclusively recommend EGFR-targeted therapy for patients with KRAS wild-type colon cancer without extending the recommendation to those with KRAS mutant colon cancer. The exploration of whether a combined approach involving TGM inhibitors with EGFR-targeted therapy can yield favorable therapeutic outcomes in KRAS mutant colon cancer remains a subject for further investigation.
Our study has several noteworthy strengths. To the best of our knowledge, this study is the first to assess the pathogenic hub genes implicated in KRAS mutant colon cancer using the multi-omics data available from public databases. Furthermore, we verified the biological function of the key gene TGM2 through loss-of-function experiments, conclusively establishing that the knockdown of TGM2 exerted inhibitory effects on the proliferation, invasion, and migration of the KRAS mutant colon cell line. Additionally, our IHC analysis of the tissue microarray revealed that TGM2 was associated with lympho-vascular invasion in the entire colon cancer population, while also serving as a predictive indicator for a higher probability of perineural invasion in the KRAS mutant colon cancer subgroup. This finding highlighted the clinical relevance of our study.
Nonetheless, it is imperative to acknowledge the limitations of this study. Firstly, the inclusion of only a single KRAS mutant colon cancer cell line represents a constraint. The functional roles and in-depth mechanisms of TGM2 were not completely examined. Secondly, the relatively modest sample size of the tissue microarray, particularly among those with KRAS mutations, imparts restrictions on the generalization of the conclusions. In the future, we intend to conduct comprehensive research, employing in vivo and in vitro experiments and more retrospective histological specimens, to elucidate the mechanism by which TGM2 augments the progression of KRAS mutant colon cancer.
In summary, our study delved into the pivotal molecules associated with KRAS mutant colon cancer via the multi-omics data available from public databases. Meanwhile, we unraveled the significant role played by TGM2 in driving the malignant phenotypes of the KRAS mutant colon cancer line, offering a theoretical basis for a comprehensive therapeutic approach. However, it is imperative that our findings be substantiated through extensive clinical investigations and further fundamental research.
Colon cancer stands as one of the prevalent malignancies worldwide, ranking third in terms of both incidence and mortality in United States. Approximately 40% of colon cancer carry oncogenic KRAS mutations, which persistently activate the epidermal growth factor receptor signaling pathway.
In-depth analysis of the core pathogenic genes of KRAS mutant colorectal cancer is of great significance.
This study aims to clarify the pivotal molecules in patients with KRAS mutant colon cancer.
The weighted gene co-expression network analysis and further bioinformatics analysis are conducted to screen the key KRAS mutation-driving factors using KRAS-mutated The Cancer Genome Atlas and Gene Expression Omnibus colon cancer cohort. Immunohistochemical assay on tissue array and lost-function experiments are used for further validation for the clinical relevance and biological function.
Integrated analysis demonstrated that transglutaminase 2 (TGM2) acted as an independent prognostic factor for progression-free survival of patients with KRAS mutant colon cancer. Immunohistochemical on tissue array reveals that TGM2 predicts a higher probability of perineural invasion in patients with KRAS mutant colon cancer. In vitro experiments demonstrated that the downregulation of TGM2 attenuated the proliferation, invasion, and migration of the KRAS mutant colon cancer cell line.
TGM2 might play an important role in the progression of KRAS mutant colon cancer.
This study underscores the potential significance of TGM2 in the progression of KRAS mutant colon cancer, providing a theoretical foundation for therapeutic approaches. Extensive clinical trials and fundamental research are required to substantiate our preliminary findings.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Hashimoto N, Japan S-Editor: Wang JJ L-Editor: A P-Editor: Cai YX
| 1. | Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 1586] [Article Influence: 264.3] [Reference Citation Analysis (2)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64686] [Article Influence: 16171.5] [Reference Citation Analysis (177)] |
| 3. | McCormick F. K-Ras protein as a drug target. J Mol Med (Berl). 2016;94:253-258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 4. | Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol. 2018;15:709-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 255] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 5. | Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 664] [Cited by in RCA: 5032] [Article Influence: 1677.3] [Reference Citation Analysis (0)] |
| 6. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8006] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 7. | Negri F, Bottarelli L, de'Angelis GL, Gnetti L. KRAS: A Druggable Target in Colon Cancer Patients. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 8. | Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lövig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001;85:692-696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 632] [Cited by in RCA: 666] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 9. | Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365-16385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 331] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 10. | Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2724] [Cited by in RCA: 2763] [Article Influence: 162.5] [Reference Citation Analysis (0)] |
| 11. | Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-Type KRAS Is Required for Panitumumab Efficacy in Patients With Metastatic Colorectal Cancer. J Clin Oncol. 2023;41:3278-3286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | Smyth GK. Limma: Linear Models for Microarray Data. In: Gentleman R, Carey VJ, Huber W, Irozarry RA, Dudoit S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer, 2005: 397-420. |
| 13. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10254] [Cited by in RCA: 16472] [Article Influence: 968.9] [Reference Citation Analysis (0)] |
| 14. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 4791] [Article Influence: 1197.8] [Reference Citation Analysis (0)] |
| 15. | Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1658] [Cited by in RCA: 3766] [Article Influence: 342.4] [Reference Citation Analysis (0)] |
| 16. | Chen K, Yang Q, Zha J, Deng M, Zhou Y, Fu G, Bi S, Feng L, Xu-Monette ZY, Chen XL, Dai Y, Young KH, Xu B. Preclinical evaluation of a regimen combining chidamide and ABT-199 in acute myeloid leukemia. Cell Death Dis. 2020;11:778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747-1756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1228] [Cited by in RCA: 3114] [Article Influence: 444.9] [Reference Citation Analysis (0)] |
| 18. | Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1253] [Cited by in RCA: 1849] [Article Influence: 132.1] [Reference Citation Analysis (0)] |
| 19. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3272] [Article Influence: 654.4] [Reference Citation Analysis (2)] |
| 20. | Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 271] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 21. | Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17:11-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 231] [Article Influence: 46.2] [Reference Citation Analysis (0)] |
| 22. | Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6:519-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 349] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 23. | Fiala O, Buchler T, Mohelnikova-Duchonova B, Melichar B, Matejka VM, Holubec L, Kulhankova J, Bortlicek Z, Bartouskova M, Liska V, Topolcan O, Sedivcova M, Finek J. G12V and G12A KRAS mutations are associated with poor outcome in patients with metastatic colorectal cancer treated with bevacizumab. Tumour Biol. 2016;37:6823-6830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 24. | Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov. 2020;19:533-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 709] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 25. | Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, Feng J, Chen JH, Li S, Long YO, Thach C, Liu Y, Zarieh A, Ely T, Kucharski JM, Kessler LV, Wu T, Yu K, Wang Y, Yao Y, Deng X, Zarrinkar PP, Brehmer D, Dhanak D, Lorenzi MV, Hu-Lowe D, Patricelli MP, Ren P. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell. 2018;172:578-589.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 589] [Cited by in RCA: 843] [Article Influence: 120.4] [Reference Citation Analysis (0)] |
| 26. | Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, Gaida K, Holt T, Knutson CG, Koppada N, Lanman BA, Werner J, Rapaport AS, San Miguel T, Ortiz R, Osgood T, Sun JR, Zhu X, McCarter JD, Volak LP, Houk BE, Fakih MG, O'Neil BH, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Hong DS, Ouyang W, Henary H, Arvedson T, Cee VJ, Lipford JR. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 843] [Cited by in RCA: 1498] [Article Influence: 249.7] [Reference Citation Analysis (0)] |
| 27. | Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, Sudhakar N, Bowcut V, Baer BR, Ballard JA, Burkard MR, Fell JB, Fischer JP, Vigers GP, Xue Y, Gatto S, Fernandez-Banet J, Pavlicek A, Velastagui K, Chao RC, Barton J, Pierobon M, Baldelli E, Patricoin EF 3rd, Cassidy DP, Marx MA, Rybkin II, Johnson ML, Ou SI, Lito P, Papadopoulos KP, Jänne PA, Olson P, Christensen JG. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020;10:54-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 899] [Article Influence: 149.8] [Reference Citation Analysis (0)] |
| 28. | Gu C, Cai J, Xu Z, Zhou S, Ye L, Yan Q, Zhang Y, Fang Y, Liu Y, Tu C, Wang X, He J, Li Q, Han L, Lin X, Li A, Liu S. MiR-532-3p suppresses colorectal cancer progression by disrupting the ETS1/TGM2 axis-mediated Wnt/β-catenin signaling. Cell Death Dis. 2019;10:739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 29. | Yang P, Yu D, Zhou J, Zhuang S, Jiang T. TGM2 interference regulates the angiogenesis and apoptosis of colorectal cancer via Wnt/β-catenin pathway. Cell Cycle. 2019;18:1122-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 30. | Torres A, Pac-Sosińska M, Wiktor K, Paszkowski T, Maciejewski R, Torres K. CD44, TGM2 and EpCAM as novel plasma markers in endometrial cancer diagnosis. BMC Cancer. 2019;19:401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 31. | Naselsky W, Adhikary G, Shrestha S, Chen X, Ezeka G, Xu W, Friedberg JS, Eckert RL. Transglutaminase 2 enhances hepatocyte growth factor signaling to drive the mesothelioma cancer cell phenotype. Mol Carcinog. 2022;61:537-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 32. | Park KS, Kim HK, Lee JH, Choi YB, Park SY, Yang SH, Kim SY, Hong KM. Transglutaminase 2 as a cisplatin resistance marker in non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:493-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | Jin T, Lin HX, Lin H, Guo LB, Ge N, Cai XY, Sun R, Chen WK, Li QL, Hu WH. Expression TGM2 and BNIP3 have prognostic significance in laryngeal cancer patients receiving surgery and postoperative radiotherapy: a retrospective study. J Transl Med. 2012;10:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Li C, Cai J, Ge F, Wang G. TGM2 knockdown reverses cisplatin chemoresistance in osteosarcoma. Int J Mol Med. 2018;42:1799-1808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 35. | Zhang S, Yao HF, Li H, Su T, Jiang SH, Wang H, Zhang ZG, Dong FY, Yang Q, Yang XM. Transglutaminases are oncogenic biomarkers in human cancers and therapeutic targeting of TGM2 blocks chemoresistance and macrophage infiltration in pancreatic cancer. Cell Oncol (Dordr). 2023;46:1473-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Kang S, Oh SC, Min BW, Lee DH. Transglutaminase 2 Regulates Self-renewal and Stem Cell Marker of Human Colorectal Cancer Stem Cells. Anticancer Res. 2018;38:787-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | Malkomes P, Lunger I, Oppermann E, Abou-El-Ardat K, Oellerich T, Günther S, Canbulat C, Bothur S, Schnütgen F, Yu W, Wingert S, Haetscher N, Catapano C, Dietz MS, Heilemann M, Kvasnicka HM, Holzer K, Serve H, Bechstein WO, Rieger MA. Transglutaminase 2 promotes tumorigenicity of colon cancer cells by inactivation of the tumor suppressor p53. Oncogene. 2021;40:4352-4367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 38. | Malkomes P, Lunger I, Oppermann E, Lorenz J, Faqar-Uz-Zaman SF, Han J, Bothur S, Ziegler P, Bankov K, Wild P, Bechstein WO, Rieger MA. Transglutaminase 2 is associated with adverse colorectal cancer survival and represents a therapeutic target. Cancer Gene Ther. 2023;30:1346-1354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 39. | Ayinde O, Wang Z, Pinton G, Moro L, Griffin M. Transglutaminase 2 maintains a colorectal cancer stem phenotype by regulating epithelial-mesenchymal transition. Oncotarget. 2019;10:4556-4569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |