Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1295
Peer-review started: February 27, 2023
First decision: March 14, 2023
Revised: March 27, 2023
Accepted: May 8, 2023
Article in press: May 8, 2023
Published online: July 15, 2023
Processing time: 135 Days and 3.3 Hours
The carcinogenesis of stomach adenocarcinoma (STAD) involves many different molecules and multiple pathways, including the NOTCH signaling pathway. As a key factor that functions as a critical link in the NOTCH pathway, mind bomb 1 (MIB1) is upregulated in various tumors and has been reported to promote cell metastasis and invasion. However, studies on the role of MIB1 in STAD are limited. Here, we evaluated the prognostic value of MIB1 in STAD and its association with immune infiltration and copy number variation.
To elucidate the relationship between MIB1 gene and gastric cancer (GC) and provide a new idea for the treatment of GC.
We identified mutations in the MIB1 gene by searching the cBioPortal database and then analyzed their relationship with the overall survival rate and disease-free survival rate using the Kaplan-Meier method. The Cancer Genome Atlas (TCGA) database provided transcript levels for MIB1 in STADs and normal tissues. As a method of distinguishing the STAD tissues from adjacent normal tissues, a receiver operating characteristic (ROC) curve was generated. Kaplan-Meier plotter was used to determine the effect of MIB1 expression on survival. Based on the LinkedOmics database, we were able to identify the coexpressed genes of the MIB1 gene, the top 50 positively correlated genes, and the top 50 negatively correlated genes. STRING was used to construct protein-protein interaction networks related to the MIB1 gene. An analysis of functional enri
According to the cBioPortal database, the MIB1 mutation rate in 287 patients in the TCGA dataset was approximately 6%. Kaplan-Meier survival analysis showed that patients with STAD in the mutated group had a worse prognosis than those in the unmutated group (P = 0.0156). There was a significant upregulation of MIB1 expression in STAD tissues compared to adjacent normal tissues. A high T stage was associated with increased MIB1 mRNA expression. The ROC curve analysis revealed 59.4% sensitivity and 85.6% specificity of MIB1 for differentiating STAD tissues from adjacent normal tissues at a truncation level of 2.248. Kaplan-Meier plotter indicated that patients with higher MIB1 levels had a worse prognosis than those with lower levels (26.4 mo vs 56.2 mo, P = 0.0330). A correlation analysis demonstrated an association between immune infiltrates and MIB1 mRNA expression.
Upregulation of MIB1 expression is significantly associated with poor survival rate and immune infiltration in gastric adenocarcinoma. MIB1 may be a biomarker for the poor prognosis of STAD patients and a potential immunotherapeutic target.
Core Tip: NOTCH signaling pathway is involved in the occurrence and development of many tumors. Mind bomb 1 (MIB1) is one of many E3 ubiquitin ligases in ubiquitin proteasome system, which plays a key role in NOTCH signaling pathway. Several studies showed that MIB1 participated in the proliferation and metastasis of certain tumor cells, but its role in gastric cancer (GC) remained still unclear. The purpose of our study was to elucidate the relationship between MIB1 gene and GC and provide a new idea for the treatment of GC.
- Citation: Wang D, Wang QH, Luo T, Jia W, Wang J. Comprehensive bioinformatic analysis of mind bomb 1 gene in stomach adenocarcinoma. World J Gastrointest Oncol 2023; 15(7): 1295-1310
- URL: https://www.wjgnet.com/1948-5204/full/v15/i7/1295.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i7.1295
Gastric cancer (GC) was the world’s leading cause of cancer deaths until the 1980s, when it was surpassed by lung cancer. Currently, the incidence of GC ranks fifth in the world and the fatality rate ranks fourth[1]. Despite its worldwide decline in morbidity and mortality rates in the past five years, GC has maintained a high mortality rate of 75% in most parts of the world, which is also the main cause of the global DALY-adjusted life year burden[2], and it is the most burdensome gastrointestinal disease in China[3]. Despite worldwide advances in clinical diagnosis and treatment, GC is still characterized by a low early diagnosis rate, low radical resection rate, and low 5-year survival rate, and most patients are first diagnosed when the disease is in an advanced stage[4]. Although many therapeutic advances, including surgical treatment, targeted therapy and immunological therapy, have been made in GC[5,6], the 5-year survival rate of patients primarily diagnosed with advanced stage is still as low as 18%[7], and the peritoneal recurrence rate after surgery is as high as 60%[8]. These findings indicate that there is a huge demand for more precise diagnosis and treatment of GC. Therefore, there is an urgent need to find new molecular markers to judge the prognosis of patients with GC.
GC is a multifactorial disease, and the recognized risk factors include age, male sex, genetic predisposition, Helicobacter pylori (H. pylori) infection, gastroesophageal reflux disease, and lifestyle factors such as smoking, alcohol consumption, and dietary composition[9,10]. Among the different types, 95% of GC cases are stomach adenocarcinoma (STAD)[11]. The combination of several variables, including genetics, epigenetics and the external environment, that may collectively result in the unregulated signaling pathway of cancer pathogenesis can be characterized as the pathogenesis of GC[12,13]. In addition, it is widely believed that dysfunctional oncogenic pathways contribute to the pathogenesis of GC, which might include the epidermal growth factor receptor, Notch, Hedgehog, nuclear factor-κB, and Wnt/β-catenin pathways[14]. Among these pathways, the Notch signaling pathway is involved in direct cell-to-cell communication, cell differentiation, proliferation and apoptosis[15].
Notch signaling is a highly conserved pathway in multicellular animals that regulates the cell fates and upholds homeostasis in adult tissues. Numerous reports have confirmed the role of Notch signaling in both carcinogenesis and antitumor effects in different backgrounds[16,17]. Notch secretion signaling can modulate heterotypic interactions between the stroma and tumor and vice versa. These interactions have been shown to regulate many aspects of oncobiology, such as angiogenesis, cancer stem cell maintenance, immune infiltration, and resistance to therapy. These functions provide evidence for the environmental dependence of Notch-induced cellular responses[18].
Mind bomb 1 (MIB1), a large multidomain RING-type E3 ubiquitin-protein ligase[19], which activates Notch signaling by promoting ubiquitination, endocytosis and subsequent activation of Notch ligands, plays a central role in the conduction of Notch signaling pathway. Inhibition of MIB1 leaded to the decrease of Notch signal activation in mammalian cells, which was fatal to mouse embryos with Notch activation deficiency[20,21]. Vitro experiments confirmed that MIB1 can induce degradation of suppressor of tumorigenicity 7 protein (ST7) to upregulate the IQ motif containing GTPase activating protein 1 (IQGAP1) in pancreatic cancer cells to promote tumor growth and progression, and also regulate the resistance of pancreatic cancer cells to gemcitabine[22,23]. It has been reported that MIB1 was ubiquitous in breast cancer to mediate JAG1 ubiquitination and activate Notch signal[24]. Aside from ubiquitinating the NOTCH ligand, MIB1 also ubiquitinated Ctnnd1 to regulate the migration of cells[25]. However, it remains unclear that the influence of MIB1 gene on GC because of the limited research on MIB1. Our study aimed to determine whether MIB1 was associated with prognosis in STAD and whether MIB1 could be regarded as a potential therapeutic target.
Briefly, the design of this study was as follows: First, the mutation of MIB1 in The Cancer Genome Atlas (TCGA)-STAD data was investigated, and the differences in overall survival (OS) and disease-free survival (DFS) between the patient group with mutations and the group without mutations in MIB1 were obtained. Second, data on the expression of MIB1 in pan-cancer and STAD were acquired. Survival analysis was performed to study the prognostic value of MIB1 in STAD from the aspect of receiver operating characteristic (ROC) curve and OS. Then, we obtained coexpressed genes from LinkedOmics, conducted Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis on the top 200 related genes, and subsequently displayed the top 50 positively and negatively correlated genes. Finally, enrichment analysis of differentially expressed genes was conducted to determine the biological function of MIB1 significantly differentially enriched genes (DEGs). In addition, the role of MIB1 in STAD was explored by studying the correlation between MIB1 and immune infiltrating cells.
The cBioPortal for Cancer Genomics (https://www.cbioportal.org/) was used to study the relationship between the mutation of the MIB1 gene in STAD and the OS or DFS of patients, and visual analysis was performed. In this database, STAD (TCGA, Nature 2014) was selected for analysis.
The official website of the TCGA (https://portal.gdc.cancer.gov/) was used to download the RNA-seq expression data of MIB1 for STAD. Thirty-two examples of neighboring normal tissues and a total of 375 cases of gastric adenocarcinoma were preserved. The chosen samples included data on MIB1 gene expression as well as pertinent clinical data, such as age, sex, HP, T stage, N stage, and M stage. The mean and standard deviation were used to describe the mRNA expression data. No permission from the ethical committee was needed for this investigation because all of the data were downloaded from the public database.
Kaplan-Meier curves were drawn using the Kaplan-Meier Plotter Web tool (https://kmplot.com/). Based on median gene expression, patients were split into two groups, and the log-rank test was used to compare the survival rates between the “high” expression group (red line) and the “low” expression group (blue line). We evaluated predictive factors, specifically OS.
With the use of LinkedOmics database (http://www.linkedomics.org/), a volcano plot showing the relationship between MIB1 members and 200 co-expressed genes in GC was created and the top 50 positively and negatively correlated genes were analyzed. The Metascape database (https://metascape.org/) was used to provide GO enrichment analysis and KEGG pathway keywords for these top 200 genes. STRING (https://string-db.org/) was used to find the genes having the strongest interactions with MIB1, and generated the associated protein-protein interaction network with an interaction score > 0.4.
The median MIB1 expression level was used to categorize expression data (HTseq-Counts) into high and low expression groups, which were then further examined using the DESeq2 R package (3.6.3). Adjusted P < 0.05 and |log2(FC)| > 1.5 were considered the thresholds to obtain DEGs, and GO enrichment analysis and KEGG pathway analysis of DEGs were performed by the “Cluster Profiler” package and visualized by the “ggplot2” package.
Tumor Immune Evaluation Resource (TIMER, https://cistrome.shinyapps.io/timer/) is a comprehensive online resource for systematically analyzing immune infiltration in various cancer types. The connection between MIB1 expression and six different immune infiltrating cells in gastric adenocarcinomas, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, was examined in our study by using TIMER. We also obtained the correlation between MIB1 and 24 tumor-infiltrating lymphocytes (TILs) by single-sample gene set enrichment analysis (ssGSEA), which was realized by the GSVA package. The Spearman test was used to measure the correlation between MIB1 and TILs.
All statistical analyses were performed using R (V 3.6.3). The differences between the gastric adenocarcinoma tissues and surrounding normal tissues were assessed using paired t tests and Mann-Whitney tests. The pROC software program was used to create a ROC curve in order to determine the MIB1 cutoff value. The impact of MIB1 on survival was assessed using Kaplan-Meier and log-rank testing.
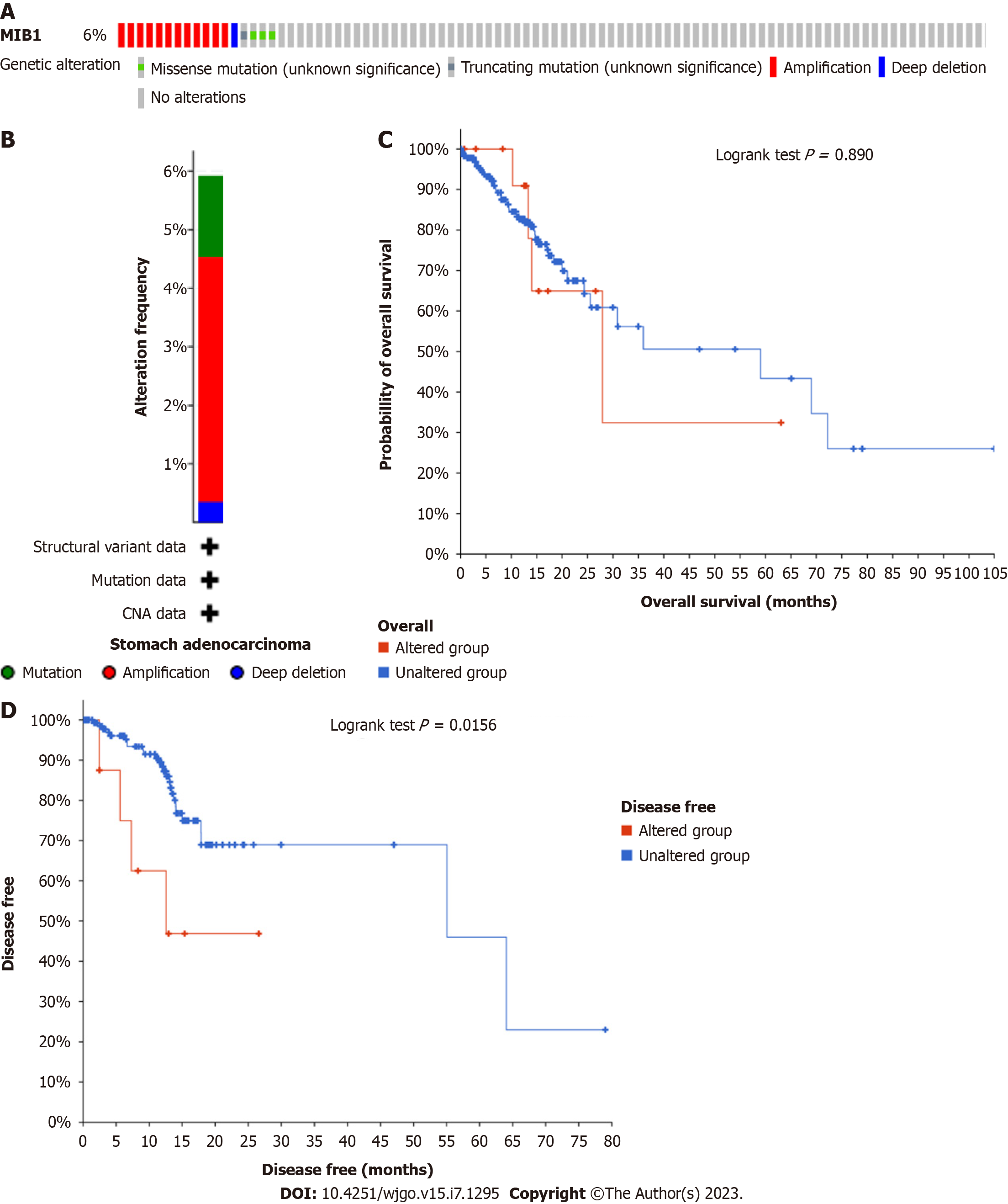
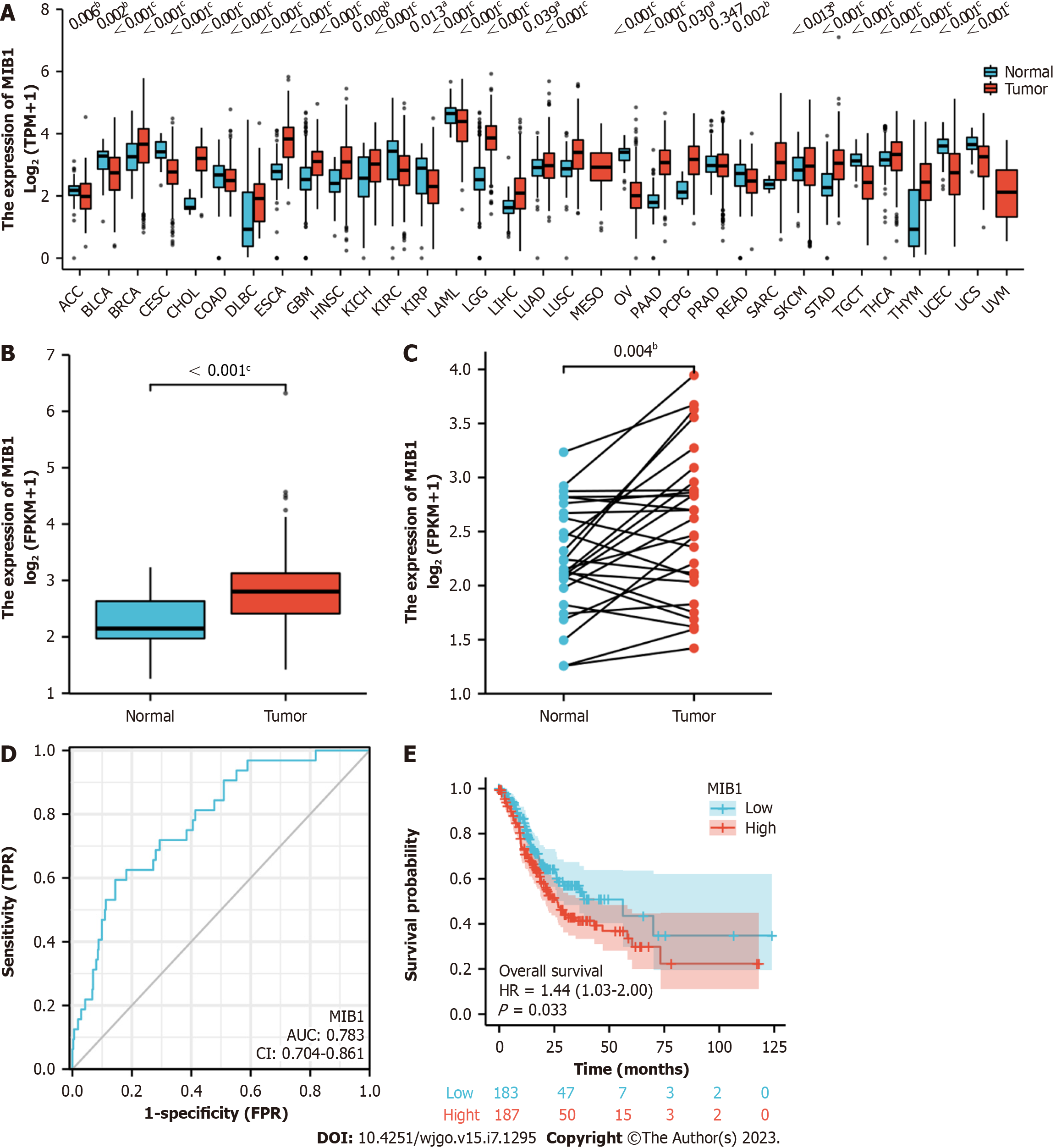
Mutation of the MIB1 gene was analyzed in STAD patients using the online cBioPortal database, and the genetic alterations of MIB1 in STAD were 6% (Figure 1A). Mutation data and copy number alteration (CNA) data were shown in Figure 1B. Patients with and without mutations did not have a significantly different OS rate (P = 0.8900). Nevertheless, the DFS rate of the group with mutations was much lower than that of the group without mutations (P = 0.0156) (Figure 1C and 1D). As shown in Figure 2A, MIB1 was considerably upregulated in a range of tumor tissues when compared to adjacent normal tissues, demonstrating that mRNA expression of MIB1 was abnormally expressed in several cancer types. Analysis of unpaired data showed that the MIB1 mRNA in STAD tissues (n = 375) was significantly higher than that in adjacent normal tissues (n = 32) (Figure 2B; 2.807 ± 0.584 vs 2.218 ± 0.495, Mann-Whitney U test, P < 0.001). Paired data analysis also showed that the mRNA expression level of MIB1 in gastric adenocarcinoma tissues was significantly higher than that in adjacent normal tissues (n = 27) (Figure 2C; 2.562 ± 0.696 vs 2.239 ± 0.506, P < 0.01).
The Mann-Whitney U test and logistic regression analysis were performed to evaluate the relationship between the mRNA expression of MIB1 and the clinicopathological characteristics of gastric adenocarcinoma samples. As shown in Table 1, the expression level of MIB1 was higher in patients with a high T stage (P = 0.017) and pathological stage (P = 0.032). However, the expression level of MIB1 was associated with other clinicopathological features, such as age (P = 0.423), sex (P = 0.884), N stage (P = 0.433), M stage (P = 1.000), tissue type (P = 0.448), and H. pylori infection (P = 0.470). In conclusion, MIB1 was associated with high T stage and pathological stage, which further suggested that MIB1 might be used as a biomarker for the poor prognosis of gastric adenocarcinoma.
| Characteristics | Total | Low expression of MIB1 | High expression of MIB1 | P value |
| T stage | 0.017 | |||
| T1 | 19 (5.2) | 11 (3.0) | 8 (2.2) | |
| T2 | 80 (21.8) | 37 (10.1) | 43 (11.7) | |
| T3 | 168 (45.8) | 99 (27.0) | 69 (18.8) | |
| T4 | 100 (27.2) | 40 (10.9) | 60 (16.3) | |
| N stage | 0.433 | |||
| N0 | 111 (31.1) | 61 (17.1) | 50 (14.0) | |
| N1 | 97 (27.2) | 49 (13.7) | 48 (13.4) | |
| N2 | 75 (21.0) | 32 (9.0) | 43 (12.0) | |
| N3 | 74 (20.7) | 38 (10.6) | 36 (10.1) | |
| M stage | 1.000 | |||
| M0 | 330 (93.0) | 164 (46.2) | 166 (46.8) | |
| M1 | 25 (7.0) | 12 (3.4) | 13 (3.7) | |
| Pathologic stage | 0.032 | |||
| Stage I | 53 (15.1) | 24 (6.8) | 29 (8.2) | |
| Stage II | 111 (31.5) | 69 (19.6) | 42 (11.9) | |
| Stage III | 150 (42.6) | 72 (20.5) | 78 (22.2) | |
| Stage IV | 38 (10.8) | 15 (4.3) | 23 (6.5) | |
| Gender | 0.884 | |||
| Female | 134 (35.7) | 68 (18.1) | 66 (17.6) | |
| Male | 241 (64.3) | 119 (31.7) | 122 (32.5) | |
| Histological type | 0.448 | |||
| Diffuse type | 63 (16.8) | 36 (9.6) | 27 (7.2) | |
| Mucinous type | 19 (5.1) | 11 (2.9) | 8 (2.1) | |
| Not otherwise specified | 207 (55.3) | 103 (27.5) | 104 (27.8) | |
| Papillary type | 5 (1.3) | 1 (0.3) | 4 (1.1) | |
| Signet ring type | 11 (2.9) | 4 (1.1) | 7 (1.9) | |
| Tubular type | 69 (18.4) | 32 (8.6) | 37 (9.9) | |
| Histologic grade | 0.305 | |||
| G1 | 10 (2.7) | 4 (1.1) | 6 (1.6) | |
| G2 | 137 (37.4) | 63 (17.2) | 74 (20.2) | |
| G3 | 219 (59.8) | 117 (32.0) | 102 (27.9) | |
| H. pylori infection | 0.470 | |||
| No | 145 (89.0) | 63 (38.7) | 82 (50.3) | |
| Yes | 18 (11.0) | 10 (6.1) | 8 (4.9) | |
| OS event | 0.022 | |||
| Alive | 228 (60.8) | 125 (33.3) | 103 (27.5) | |
| Dead | 147 (39.2) | 62 (16.5) | 85 (22.7) | |
| Age, mean ± SD | 65.39 ± 10.80 | 66.28 ± 10.51 | 0.423 |
The usefulness of MIB1 in separating GC samples from normal samples was investigated using ROC curve analysis. As shown in Figure 2D, the ROC curve showed that the area under the curve (AUC) value of MIB1 was 0.783 (95%CI: 0.704-0.861). The sensitivity and specificity of MIB1 were respectively 59.4% and 85.6% at the cutoff value of 2.248. Positive and negative predictive values were 26.0% and 96.1%, respectively. These findings suggested that MIB1 might be a potential biomarker to differentiate between normal tissues and stomach cancer tissues.
As shown in Figure 2E, patients with gastric adenocarcinoma who had high MIB1 Levels compared to those who had low MIB1 Levels had significantly worse OS (26.4 mo vs 56.2 mo, P = 0.033). This suggested that high mRNA expression of MIB1 was a biomarker for poor prognosis in gastric adenocarcinoma.
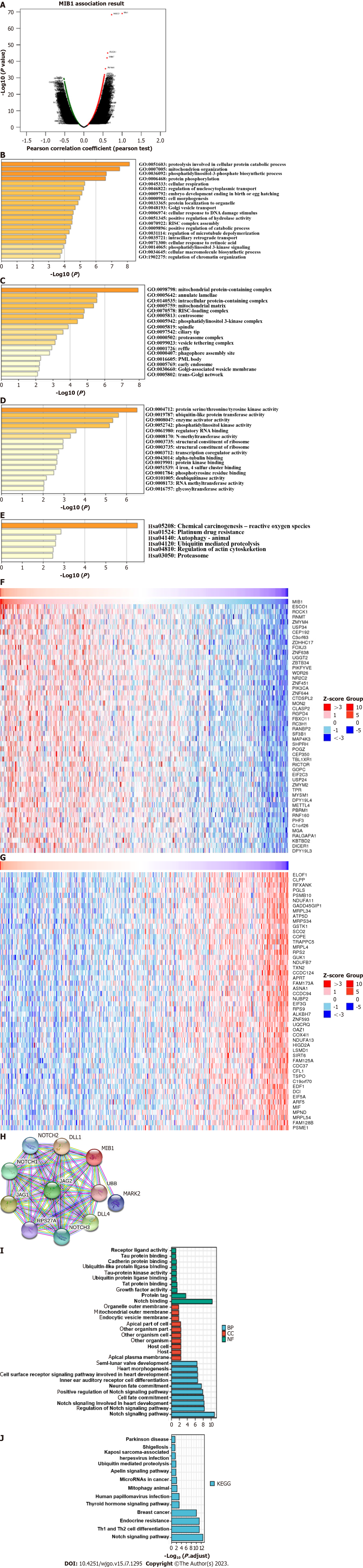
A volcano plot of MIB1 and coexpressed genes in GC was generated in the LinkedOmics database (Figure 3A). The Metascape database was then used to examine the GO and KEGG pathway terms of these top 200 genes. GO analysis was used to investigate the functional mechanism of MIB1 in GC. The BP terms “proteolysis involved in cellular protein catabolic process”, “mitochondrion organization”, “phosphatidylinositol-3-phosphate biosynthetic process”, and “protein phosphorylation” were significantly enriched (Figure 3B). Among the enriched CC terms was “mitochondrial protein-containing complex” (Figure 3C). “Protein serine/threonine/tyrosine kinase activity”, “ubiquitin-like protein transferase activity”, and “enzyme activator activity” were the most commonly enriched MF phrases (Figure 3D). The target genes were primarily linked to the phrases “chemical carcinogenesis-reactive oxygen species”, “platinum drug resistance”, and “ubiquitin mediated proteolysis”, according to KEGG pathway analysis (Figure 3E). The top 50 genes with positive relationship (Figure 3F) and the top 50 genes with negative relationship (Figure 3G) with MIB1 were displayed in a heatmap to further investigate the processes of MIB1 and its coexpressed genes. The ten coexpressed genes of MIB1 in the STRING database were NOTCH1, NOTCH2, NOTCH3, DLL1, DLL4, UBB, MARK2, JAG1, JAG2, and RPS27A (Figure 3H). These genes were analyzed by GO and KEGG, most of which were related to NOTCH pathway (Figure 3I and J).
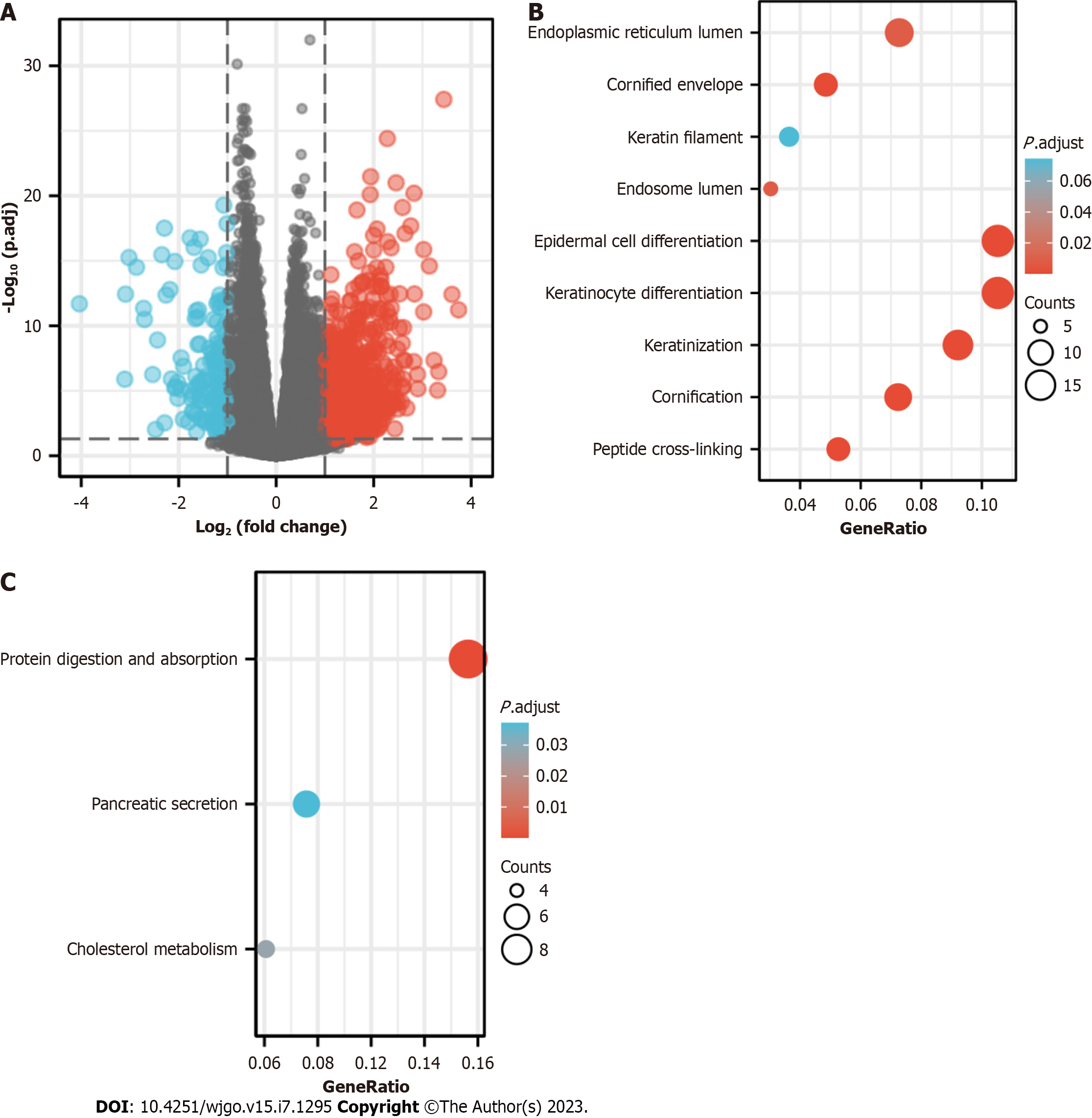
With a threshold of |logFC|< 1.5 and adjusted P < 0.05, 506 DEGs in total were discovered, of which 454 showed upregulation and 52 showed downregulation. The DEG expression was visualized in a volcano diagram (Figure 4A). The DEG-related MIB1 had strongly regulatory effects on the endoplasmic reticulum lumen, cornified envelope, keratin filament, endosome lumen, epidermal cell differentiation, and keratinocyte differentiation, keratinization, cornification and peptide cross-linking processes, according to GO enrichment analysis (Figure 4B). KEGG analysis showed that DEG-related MIB1 was associated with protein digestion and absorption, pancreatic secretion, and cholesterol metabolism pathways (Figure 4C).
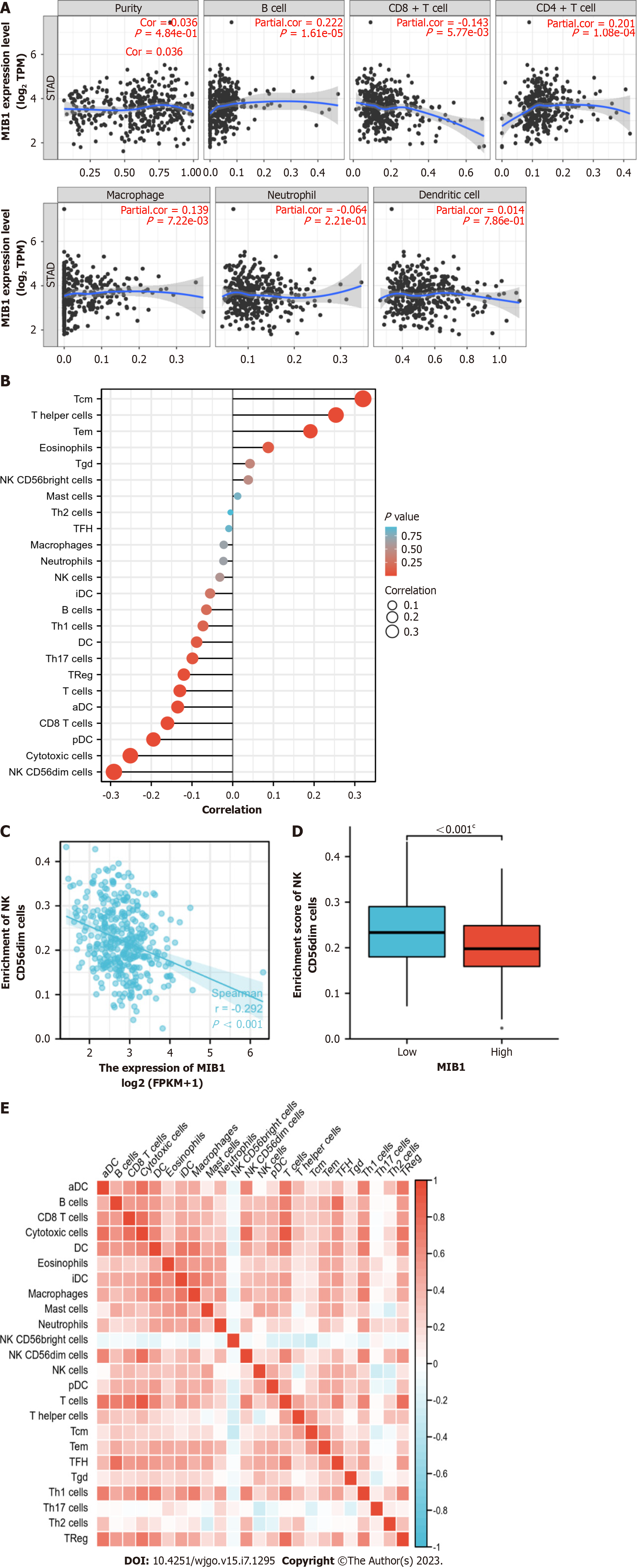
We analyzed the correlation between the expression of MIB1 and six types of tumor invasive immune cells in the TIMER database. As demonstrated in Figure 5A, MIB1 expression was favorably linked with B cells (r = 0.222, P = 1.61E-05), CD4+ T cells (r = 0.201, P = 1.08E-04), and macrophages (r = 0.139, P = 7.22E-03) and negatively connected with CD8+ T cells (r = -0.143, P = 5.77E-03). Figure 5B showed the relationship between MIB1 and 24 kinds of tumor immune infiltrating cells. Tcm, helper T cells, Tem, Tgd and NK CD56 bright cells were positively correlated with MIB1. MIB1 was inversely linked with cytotoxic cells, NK CD56dim cells, pDCs, aDCs, CD8 T cells, and Th17 cells. The relationship between the level of MIB1 expression and the degree of immune cell infiltration measured by the ssGSEA score was investigated using Spearman correlation. The extent of NK CD56dim cell infiltration was negatively correlated with MIB1 expression (r = -0.292, P < 0.001) (Figure 5C) and considerably decreased in the MIB1 high expression group (P < 0.001) (Figured 5D). These results demonstrated that MIB1 played an important role in the immune infiltration of GC. The proportions of 24 different subsets of tumor-infiltrating immune cells were compared using a heatmap to ascertain the degrees of association (Figure 5E).
Notch signaling is an evolutionarily conserved pathway that controls cell fate, determines cell differentiation, proliferation, tumor angiogenesis, stem cell maintenance, apoptosis and other cellular processes, and promotes the occurrence of GC through crosstalk with different signaling pathways, such as the Wnt, Ras, and NF-κB pathways[26,27]. Studies have indicated that endocytosis of Notch ligands is required to activate the receptor in the Notch pathway[28]. However, ubiquitination of the intracellular tail of Notch ligands is a critical event in the subsequent endocytosis and signal transduction of these molecules[29]. Initial genetic studies in flies and zebrafish identified two E3 ubiquitin ligase families capable of ligand ubiquitination: Mind Bomb (Mib) and Neuralized (Neur) proteins[30]. Then a series of studies concluded that MIB1, a member of the E3 ubiquitin-protein ligase family, played a major, possibly exclusive role in Notch ligand ubiquitination and transport in mammals[31].
The roles of some Notch family members in GC have not yet been fully understood, despite the fact that the impact of Notch signaling on GC has been extensively established[32,33]. In this work, we performed a thorough examination of MIB1, a NOTCH family member, in patients with GC to determine its mutation, expression, prognostic value, and immune infiltration.
The mRNA expression of MIB1 was observed to be increased in STAD tissues in our research. And positive correlation was found between the increase of mRNA expression and a high T stage. MIB1 might be a viable diagnostic biomarker for separating stomach cancer tissues from normal tissues, according to a ROC curve analysis. Kaplan-Meier curves and univariate analysis allowed us to demonstrate that increased mRNA expression of MIB1 was associated with short OS and might be used as a feasible biomarker for poor prognosis of GC. Additionally, MIB1 might have a unique function in the immunological infiltration of GC.
MIB1 is a ubiquitin-protein ligase. It was reported that overexpression of MIB1 significantly promoted cell proliferation, migration and invasion[25]. Recent studies have found that MIB1 played a carcinogenic function in a variety of human malignancies, including pancreatic, prostate, and lung cancer[23,34,35]. Studies have shown that MIB1 was an important biomarker leading to poor prognosis, and upregulation of MIB1 expression was associated with poor OS[22]. Furthermore, mutation of the MIB1 gene could lead to congenital heart disease by reducing Notch signaling activation[36]. Overexpression of E3 ubiquitin ligase MIB1 could reduce the apoptosis and inflammation of cardiac microvascular endothelial cells in coronary microvascular dysfunction[37].
In this study, analysis of the cBioportal database showed that the MIB1 mutation rate in 287 patients in the TCGA dataset was approximately 6%, and most of the changes were copy number amplification (CNA). In cancer, CNAs and deletions result in altered expression of tumor suppressor genes and oncogenes, respectively. It was reported that copy number variation of E3 ubiquitin ligase was associated with the occurrence and development of colorectal cancer[38]. We further analyzed the correlation between MIB1 gene changes and prognosis. We found no significant difference between MIB1 gene changes and OS, but the DFS of the patient group with mutations was much shorter than that of the group without mutations. Moreover, what we discovered were consistent with previous studies that MIB1 mRNA was abnormally expressed in many cancers, and we found that MIB1 was greatly increased in gastric adenocarcinoma via the TCGA database. According to this, gastric adenocarcinoma with a poor clinical prognosis might be identified using MIB1 as a possible biomarker for poor prognosis.
At present, the role of MIB1 in tumors and whether it acts through the NOTCH pathway have not been fully reported. There have been relatively many studies on MIB1 in pancreatic cancer. Some studies have shown that MIB1 can be used as a direct target of miRNA-198 and miRNA-195-5p. MIB1 has been considered as a new target of miRNA-198, which reduced the proliferation, migration and invasion of prostate cancer. However, this tumor inhibition role appeared to be independent of the Notch pathway[39]. MicroRNA-195-5p might regulate the proliferation and invasion of tumor cells by regulating MIB1, suggesting that miRNA-195-5p might be used to treat prostate cancer in the future[34]. Our results indicated that MIB1 may be an intriguing biomarker or an emerging target for cancer therapy. In addition, ectopic expression of MIB1 could induce epithelial-to-mesenchymal transition and stimulate cell migration through the Notch-dependent pathway, which might provide new insights into the treatment of MIB1-overexpressing cancer[35]. Other studies have shown that MIB1 promoted the progression of pancreatic cancer by inducing ST7 degradation and downregulating IQGAP1, suggesting that the MIB1/ST7/IQGAP1 axis was crucial in the advancement of pancreatic cancer, and inhibiting MIB1 might become a new therapeutic strategy for pancreatic cancer patients[22]. A study proved that MIB1 promoted pancreatic cancer proliferation by activating the β-catenin signaling pathway[23]. Therefore, whether MIB1 affects the progression of GC through the NOTCH pathway needs further in vivo and in vitro experiments.
A ROC curve analysis was performed to verify the clinical value of MIB1 in diagnosing gastric adenocarcinoma. With a sensitivity of 59.4% and a specificity of 85.6%, our findings demonstrated that MIB1 had a relatively higher AUC value to discover the patients with GC. Based on our research, we came to the conclusion that MIB1 might function as an applicable diagnostic biomarker to separate gastric adenocarcinoma from normal controls.
In addition, MIB1 was highly expressed in patients with gastric adenocarcinoma. MIB1 was correlated with multiple clinical features, such as pathological stage, T stage and OS, further suggesting that MIB1 was a prospective biomarker that merited additional clinical testing.
Cytotoxic cells, NK CD56dim cells, pDCs, aDCs, CD8 T cells and Th17 cells were negatively correlated with MIB1. Antitumor immunity was influenced by cytotoxic cells, including NK cells. The function of NK cells in innate immune surveillance is crucial in the fight against cancer[24]. In the course of transformation into toxic T cells, CD8+ T cells display cytotoxic abilities against tumor cells[25]. IFN-I produced by pDCs has antitumor activity[26], Th17 cells have a close connection to neutrophils and are essential for the immune response to tumors[27]. The decrease of these immune cells might contribute to the further development of GC. The results of ssGSEA further demonstrated that MIB1 was essential in controlling immune infiltration.
Our research revealed the complex role of MIB1 gene mutation and abnormal expression in the prognosis of GC. In addition, we also preliminarily discussed the relationship between the MIB1 gene and immune infiltration, as well as its mechanism and biological function in GC. However, our research also had some limitations. This study lacked in vivo or in vitro experiments to verify the role of the MIB1 gene, which will allow us to draw more general and accurate conclusions.
In brief, we found that MIB1 mRNA expression increased in STAD was positively attached with high T stage and pathological stage and negatively correlated with OS. According to our research, higher expression of MIB1 may be a useful predictive biomarker for identifying individuals with gastric adenocarcinomas who have a poor clinical prognosis and may have a special function in immune infiltration.
Gastric cancer (GC) is a disease with multi-etiology and multi-pathway involvement, and it is characterized by a low 5-year survival rate. NOTCH signaling pathway is also involved in the occurrence and development of GC. Mind bomb 1 (MIB1), an E3 ubiquitin ligase, plays a central role in activating the NOTCH pathway by mediating ubiquitination of NOTCH ligand. However, the effect of MIB1 on GC has not been reported.
To investigate the effect of MIB1 gene on the prognosis of GC.
To investigate the effect of expression and mutation of MIB1 gene on the prognosis of GC, the function of MIB1 in GC and its relationship with immune infiltration.
TCGA database, cBioPortal database, a receiver operating characteristic (ROC) curve, Kaplan-Meier plotter, LinkedOmics database, STRING database, The Gene Ontology enrichment , Kyoto Encyclopedia of Genes and Genomes pathway and TIMER database were used in this study.
The level of MIB1 expression had a certain impact on the survival rate of patients with GC. The prognosis of patients with high MIB1 was worse than that of patients with low MIB1. The increased expression of MIB1 gene was associated with high TNM staging, suggesting that MIB1 may play a role in the development of GC. The expression of MIB1 gene was associated with immune infiltration.
The up-regulation of MIB1 expression was significantly related to the low survival rate and immune infiltration in gastric adenocarcinoma.
MIB1 may be a biomarker for poor prognosis of gastric adenocarcinoma and a potential immunotherapeutic target.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ballini A, Italy; Inchingolo F, Italy; Santacroce L, Italy S-Editor: Chen YL L-Editor: A P-Editor: Ju JL
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64332] [Article Influence: 16083.0] [Reference Citation Analysis (174)] |
| 2. | Soerjomataram I, Lortet-Tieulent J, Parkin DM, Ferlay J, Mathers C, Forman D, Bray F. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet. 2012;380:1840-1850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 427] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 3. | Xie W, Yang T, Zuo J, Ma Z, Yu W, Hu Z, Song Z. Chinese and Global Burdens of Gastrointestinal Cancers From 1990 to 2019. Front Public Health. 2022;10:941284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 4. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2822] [Article Influence: 564.4] [Reference Citation Analysis (5)] |
| 5. | Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39:1010428317714626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 643] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 6. | Chen Z, Li Y, Tan B, Zhao Q, Fan L, Li F, Zhao X. Progress and current status of molecule-targeted therapy and drug resistance in gastric cancer. Drugs Today (Barc). 2020;56:469-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 7. | Asplund J, Kauppila JH, Mattsson F, Lagergren J. Survival Trends in Gastric Adenocarcinoma: A Population-Based Study in Sweden. Ann Surg Oncol. 2018;25:2693-2702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 8. | Nakagawa N, Kanda M, Ito S, Mochizuki Y, Teramoto H, Ishigure K, Murai T, Asada T, Ishiyama A, Matsushita H, Tanaka C, Kobayashi D, Fujiwara M, Murotani K, Kodera Y. Pathological tumor infiltrative pattern and sites of initial recurrence in stage II/III gastric cancer: Propensity score matching analysis of a multi-institutional dataset. Cancer Med. 2018;7:6020-6029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 897] [Cited by in RCA: 846] [Article Influence: 169.2] [Reference Citation Analysis (0)] |
| 10. | Yusefi AR, Bagheri Lankarani K, Bastani P, Radinmanesh M, Kavosi Z. Risk Factors for Gastric Cancer: A Systematic Review. Asian Pac J Cancer Prev. 2018;19:591-603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 161] [Reference Citation Analysis (0)] |
| 11. | Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 722] [Article Influence: 103.1] [Reference Citation Analysis (1)] |
| 12. | Houghton J, Wang TC. Helicobacter pylori and gastric cancer: a new paradigm for inflammation-associated epithelial cancers. Gastroenterology. 2005;128:1567-1578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 186] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 13. | Kankeu Fonkoua L, Yee NS. Molecular Characterization of Gastric Carcinoma: Therapeutic Implications for Biomarkers and Targets. Biomedicines. 2018;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 14. | Wu WK, Cho CH, Lee CW, Fan D, Wu K, Yu J, Sung JJ. Dysregulation of cellular signaling in gastric cancer. Cancer Lett. 2010;295:144-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 130] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 15. | Previs RA, Coleman RL, Harris AL, Sood AK. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clin Cancer Res. 2015;21:955-961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 16. | Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annu Rev Pathol. 2017;12:245-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 520] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 17. | Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25:318-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 298] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 18. | South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol. 2012;23:458-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 19. | Guo B, McMillan BJ, Blacklow SC. Structure and function of the Mind bomb E3 ligase in the context of Notch signal transduction. Curr Opin Struct Biol. 2016;41:38-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, Lee J, Chitnis AB, Kim CH, Kong YY. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459-3470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 200] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 21. | Yamamoto M, Morita R, Mizoguchi T, Matsuo H, Isoda M, Ishitani T, Chitnis AB, Matsumoto K, Crump JG, Hozumi K, Yonemura S, Kawakami K, Itoh M. Mib-Jag1-Notch signalling regulates patterning and structural roles of the notochord by controlling cell-fate decisions. Development. 2010;137:2527-2537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Zhang B, Cheng X, Zhan S, Jin X, Liu T. MIB1 upregulates IQGAP1 and promotes pancreatic cancer progression by inducing ST7 degradation. Mol Oncol. 2021;15:3062-3075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Fu X, Tang N, Xie W, Mao L, Qiu Y. Mind Bomb 1 Promotes Pancreatic Cancer Proliferation by Activating β-Catenin Signaling. J Nanosci Nanotechnol. 2020;20:7276-7282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2096] [Cited by in RCA: 2175] [Article Influence: 108.8] [Reference Citation Analysis (0)] |
| 25. | Iwahori K. Cytotoxic CD8(+) Lymphocytes in the Tumor Microenvironment. Adv Exp Med Biol. 2020;1224:53-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 26. | Saulep-Easton D, Vincent FB, Le Page M, Wei A, Ting SB, Croce CM, Tam C, Mackay F. Cytokine-driven loss of plasmacytoid dendritic cell function in chronic lymphocytic leukemia. Leukemia. 2014;28:2005-2015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 27. | Amicarella F, Muraro MG, Hirt C, Cremonesi E, Padovan E, Mele V, Governa V, Han J, Huber X, Droeser RA, Zuber M, Adamina M, Bolli M, Rosso R, Lugli A, Zlobec I, Terracciano L, Tornillo L, Zajac P, Eppenberger-Castori S, Trapani F, Oertli D, Iezzi G. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut. 2017;66:692-704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 143] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 28. | Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, Blacklow SC. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev Cell. 2015;33:729-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 271] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 29. | Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 638] [Cited by in RCA: 637] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 30. | Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development. 1998;125:4637-4644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 207] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 31. | Koo BK, Yoon MJ, Yoon KJ, Im SK, Kim YY, Kim CH, Suh PG, Jan YN, Kong YY. An obligatory role of mind bomb-1 in notch signaling of mammalian development. PLoS One. 2007;2:e1221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 32. | Brzozowa M, Mielańczyk L, Michalski M, Malinowski L, Kowalczyk-Ziomek G, Helewski K, Harabin-Słowińska M, Wojnicz R. Role of Notch signaling pathway in gastric cancer pathogenesis. Contemp Oncol (Pozn). 2013;17:1-5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Du X, Cheng Z, Wang YH, Guo ZH, Zhang SQ, Hu JK, Zhou ZG. Role of Notch signaling pathway in gastric cancer: a meta-analysis of the literature. World J Gastroenterol. 2014;20:9191-9199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 43] [Reference Citation Analysis (0)] |
| 34. | Chen B, Bai G, Ma X, Tan L, Xu H. MicroRNA1955p is associated with cell proliferation, migration and invasion in prostate cancer and targets MIB1. Oncol Rep. 2021;46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 35. | Wang H, Huang Q, Xia J, Cheng S, Pei D, Zhang X, Shu X. The E3 Ligase MIB1 Promotes Proteasomal Degradation of NRF2 and Sensitizes Lung Cancer Cells to Ferroptosis. Mol Cancer Res. 2022;20:253-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 36. | Li B, Yu L, Liu D, Yang X, Zheng Y, Gui Y, Wang H. MIB1 mutations reduce Notch signaling activation and contribute to congenital heart disease. Clin Sci (Lond). 2018;132:2483-2491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Qin XF, Shan YG, Gao JH, Li FX, Guo YX. E3 ubiquitin ligase mind bomb 1 overexpression reduces apoptosis and inflammation of cardiac microvascular endothelial cells in coronary microvascular dysfunction. Cell Signal. 2022;91:110223. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 38. | Bi H, Tian T, Zhu L, Zhou H, Hu H, Liu Y, Li X, Hu F, Zhao Y, Wang G. Copy number variation of E3 ubiquitin ligase genes in peripheral blood leukocyte and colorectal cancer. Sci Rep. 2016;6:29869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Ray J, Hoey C, Huang X, Jeon J, Taeb S, Downes MR, Boutros PC, Liu SK. MicroRNA198 suppresses prostate tumorigenesis by targeting MIB1. Oncol Rep. 2019;42:1047-1056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |