Published online Dec 15, 2022. doi: 10.4251/wjgo.v14.i12.2353
Peer-review started: June 29, 2022
First decision: August 6, 2022
Revised: August 31, 2022
Accepted: November 4, 2022
Article in press: November 4, 2022
Published online: December 15, 2022
Processing time: 165 Days and 23.9 Hours
Mounting studies have highlighted the pivotal influence of anti-silencing function 1B (ASF1B) on the malignancy of cancers.
To explore the influence and mechanism of ASF1B in colorectal cancer (CRC).
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect mRNA expression of ASF1B. Immunohistochemical staining was performed to detect protein expression of ASF1B and Ki67 in tumor tissues. Western blot analysis was used to determine levels of ASF1B and proliferation/epithelial mesenchymal transition (EMT)/stemness-related proteins. In addition, the proliferation of CRC cells was assessed using Cell Counting Kit-8 and 5-Ethynyl-2’-Deoxyuridine assays. The migration and invasion of CRC cells were evaluated using transwell assays. Stemness of CRC cells was tested using the sphere formation assay. To construct a xenograft tumor model, HCT116 cells were introduced into mouse flanks via subcutaneous injection.
ASF1B expression was markedly increased in CRC tissues and cells, and it was inversely correlated with overall survival of CRC patients and was positively associated with the tumor node metastasis (TNM) stage of CRC patients. Silencing of ASF1B suppressed proliferation, migration, invasion, stemness and EMT of CRC cells as well as tumorigenesis of xenograft mice. Furthermore, protein levels of P-phosphatidylinositol 3-kinase (p-PI3K) and p-AKT were decreased after silencing of ASF1B in CRC cells. The inhibitory effects of ASF1B knockdown on cell proliferation, stemness and EMT were partly abolished by PI3K activator in CRC cells.
Silencing of ASF1B inactivated the PI3K/AKT pathway to suppress CRC malignancy in vitro.
Core Tip: Anti-silencing function 1B (ASF1B) expression was increased in colorectal cancer (CRC) tissues and cells, and was negatively associated with prognosis of CRC patients. Functionally, ASF1B knockdown repressed the malignant behaviors of CRC cells in vitro and tumorigenesis in vivo, therefore having potential for CRC treatment. Moreover, our findings showed that ASF1B down-regulation suppressed the malignant behaviors of CRC cells by inactivating the PI3K/AKT pathway.
- Citation: Yu GH, Gong XF, Peng YY, Qian J. Anti-silencing function 1B knockdown suppresses the malignant phenotype of colorectal cancer by inactivating the phosphatidylinositol 3-kinase/AKT pathway. World J Gastrointest Oncol 2022; 14(12): 2353-2366
- URL: https://www.wjgnet.com/1948-5204/full/v14/i12/2353.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i12.2353
Colorectal cancer (CRC) is a malignant gastrointestinal tumor, accounting for 8% of cancer-related deaths in 2018[1,2]. According to statistics, it leads to over 1 million deaths every year[3,4]. Despite great progress in CRC treatment including surgery, chemotherapy and combined therapy, CRC prognosis is still poor[5,6]. Therefore, deeper exploration of CRC pathogenesis and treatment targets is of importance to improve CRC therapy.
For the past few years, molecular targeting treatment has emerged as a research hotspot in CRC therapy[7,8]. Anti-silencing function 1 (ASF1), a conserved histone H3-H4 chaperone protein, is involved in the regulation of many processes such as transcription, DNA damage repair and DNA replication[9,10]. Of note, it includes two paralogous forms: ASF1A histone chaperone and ASF1B[11]. ASF1A is primarily implicated in regulation of DNA repair and cellular senescence, while ASF1B acts as a crucial regulator of cellular proliferation and cell cycle progression[10,12]. As a subtype of ASF1, up-regulated expression of ASF1B is reported to be associated with the poor prognosis of lung adenocarcinoma and breast cancer patients[13,14]. More importantly, ASF1B down-regulation is demonstrated to have anti-tumor ability in many cancers. For example, ASF1B knockdown suppresses cell proliferation, and promotes cell cycle arrest and apoptosis in cervical cancer[15]. ASF1B knockdown impairs proliferation, migration and invasion of lung cancer cells[16]. Silencing of ASF1B represses growth of hepatocellular carcinoma (HCC) cells, and induces cell cycle arrest[17]. Overall, ASF1B is gaining attention as an important player in the development of diverse cancers. However, the function and mechanistic understanding of ASF1B have rarely been reported in CRC.
The phosphatidylinositol 3-kinase (PI3K)/AKT pathway is widely found in tumors, and plays an important role in the growth and proliferation of tumor cells[18]. Notably, the PI3K/AKT pathway has been demonstrated to be involved in regulation of CRC progression and development[19,20]. For example, pleckstrin homology like domain family A member 2 down-regulation inactivates the PI3K/AKT pathway to inhibit proliferation, invasion and migration of CRC cells[19]. Pyrroline-5-carboxylate reductase 2 down-regulation suppresses the PI3K/AKT pathway to prevent proliferation, migration and invasion of CRC cells[20]. Inosine 5’-monophosphate dehydrogenase type II knockdown attenuates proliferation, invasion and migration abilities of CRC cells by inactivating the PI3K/AKT pathway[21]. In particular, ASF1B was revealed to affect the malignant behaviors of prostate cancer cells and pancreatic cancer cells by regulating the PI3K/AKT pathway[22,23]. Thus, we assumed that ASF1B may regulate the PI3K/AKT pathway to affect malignant progression of CRC. In this study, the expression pattern of ASF1B as well as the role of ASF1B was determined in CRC in vitro and in vivo. Furthermore, the molecular mechanism of ASF1B-mediated carcinogenesis was explored in CRC cells.
A total of 68 pairs of CRC tissues and adjacent normal tissues (3.0 cm away from the tumor margin) were obtained from CRC patients between June 2019 and January 2021, which were transported in liquid nitrogen to the laboratory and then stored at -80 °C until use. Pathological diagnosis of CRC patients (age range, 23-61 years; 35 males and 33 females) was conducted by three pathologists based on the eighth edition of the Union for International Cancer Control and the American Joint Committee on Cancer tumor node metastasis (TNM) classification[24,25]. The inclusion criteria were as follows: (1) Complete general information (including gender, age, ethnicity, past history and family history); and (2) Patients who underwent bidirectional endoscopy (colonoscopy performed immediately after gastroscopy). The exclusion criteria were as follows: (1) History of gastric cancer, peptic ulcer and other cancers; (2) Received antibiotics, proton pump inhibitors or glucocorticoids in the past month; (3) Patients who underwent chemotherapy, radiation therapy and other treatments for tumors; (4) Previous history of gastrointestinal surgery; (5) Presence of inflammatory bowel disease, Gardner’s syndrome (a disease that affects the incidence of CRC) or familial adenoma; and (6) A history of systemic diseases. The current study obtained permission from the Ethics Committee of Zhebei Mingzhou Hospital (ZBMZYYLL211028), and all subjects signed informed consents. In addition, CRC patients were divided into ASF1B high or low-expression groups based on the median value of ASF1B expression.
The survival of patients was followed for 90 mo through the records of reexamination or the telephone.
The online website GEPIA (http://gepia.cancer-pku.cn/detail.php) was used to compare the expression of ASF1B between tumor tissues and normal tissues in CRC.
A human normal colorectal mucosal cell line (FHC) and CRC cell lines (HT29, HCT116, LOVO, SW480 and SW620) were bought from the American Type Culture Collection (ATCC, Manassas, VA, United States). All cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, Carlsbad, CA, United States) containing 10% fetal bovine serum (FBS) (Gibco, Gaithersburg, MD, United States) and 1% penicillin/streptomycin (Gibco), which were incubated at 37 °C under 5% CO2. To eliminate mycoplasma contamination, all cells were routinely examined using MycAway (Yeasen, Shanghai, China).
Short hairpin RNAs against ASF1B (sh-ASF1B#1, sh-ASF1B#2 and sh-ASF1B#3) and sh-negative control (sh-NC) were bought from RiboBio (Guangzhou, China). Then HCT116 and SW620 cells were seeded in six-well plates to adjust the cell density to 5 × 105/mL. When the cell confluence reached 70%-80%, the above plasmids (final concentrations, 50 nM) were transiently transfected into cells through Lipofectamine 3000 (Invitrogen) for 48 h. To further explore the role of the PI3K/AKT pathway in CRC cells, transfected HCT116 cells were treated with PI3K activator (740 Y-P, 50 μg/mL, MedChemexpress, Shanghai, China) for 90 min[26].
Total RNAs in CRC tissues were isolated using TRIzol reagent (Invitrogen), and then used for synthesis of complementary DNAs (cDNAs) according to the instructions of a PrimeScript RT reagent kit (Takara, Dalian, China). Next, cDNAs and the SYBR Green polymerase chain reaction (PCR) kit (Takara) were used to perform quantitative real-time PCR (qRT-PCR) on a LightCycler 480 Real-Time PCR system (Roche, Shanghai, China). Reaction procedures for PCR were displayed as follows: 5 min at 94 °C, 38 cycles of 25 s at 94 °C, 1 min at 60 °C and 2 min at 72 °C. Primers bought from Sangon (Shanghai, China) were as follows: ASF1B-F, GATCAGCTTCGAGTGCAGTG; ASF1B-R, TGGTAGGTGCAGGTGATGAG; GAPDH-F, CCATCTTCCAGGAGCGAGAT; GAPDH-R, TGCTGATGATCTTGAGGCTG. Relative mRNA expression of ASF1B, which was normalized to GAPDH, was calculated using the 2-ΔΔct method.
Proliferation of HCT116 and SW620 cells was assessed through a Cell Counting Kit-8 (CCK-8) kit (Beyotime, Shanghai, China). Briefly, transfected cells (5 × 103 cells/well) were plated into 96-well plates, followed by incubation for 0 h, 24 h, 48 h, 72 h and 96 h. After that, CCK-8 solution (10 μL) was added to each well and the cells were incubated at 37 °C for 2 h. Finally, the optical density at 450 nm was measured using a microplate reader (MG LABTECH, Durham, NC, United States).
Proliferation of HCT116 and SW620 cells was assessed using the 5-Ethynyl-2’-Deoxyuridine (EDU) proliferation assay (RiboBio). In detail, HCT116 and SW620 cells (1 × 105 cells/well) were plated to 96-well plates. Then these cells were exposed to 50 μM EDU at 37 °C for 2 h, and fixed with 4% formaldehyde for 30 min and permeabilized with 0.1% Triton X-100 for 20 min. The EDU solution was then added to the culture, followed by staining of nuclei via Hoechst. EDU-positive cells were observed using a fluorescent microscope.
The invasion of HCT116 and SW620 cells was assessed using the transwell chamber (8.0 μm pore size; Millipore, Billerica, MA, United States) coated with Matrigel. In brief, cells (100000 cells/well) re-suspended in FBS-free medium were placed onto the upper compartment. Subsequently, the upper compartment was filled with 600 μL culture medium with 10% FBS. The chambers were then incubated for 24 h, and cells in the lower surface were fixed with 4% paraformaldehyde and dyed with 0.1% crystal violet. Invasive cells were counted under an optical microscope. In contrast, cell migration was assessed in the upper chamber without Matrigel, and the procedures were the same as the invasion evaluation.
Stemness of HCT116 and SW620 cells was evaluated by the sphere formation assay. Briefly, cells (250 cells/well) were placed into 24-well plates with ultra-low attachment, which were cultured in RPMI 1640 medium containing 10 ng/mL fibroblast growth factor, 10 μg/mL insulin, 10 ng/mL epidermal growth factor and 2% B-27. After 10 d of incubation, representative fields were photographed using a Nikon microscope.
Protein extraction of CRC tissues and cells was performed using RIPA lysis buffer (Beyotime). The lysates were then centrifuged (12000 r/min) for 10 min at 4 °C, and 5 × loading buffer was added to the supernatant to denature at 100 °C for 5 min. Determination of protein concentration was performed using a BCA Protein Assay Kit (Beyotime). Subsequently, the sodium dodecyl sulfate-polyacrylamide gel with 5% stacking gel and 10% separation gel was prepared, and these protein samples (10 μL) were separated by electrophoresis in 1 × buffer [500 mL ddH2O, 9.4 g glycine, 1.51 g Tris-base and 0.5 g sodium dodecyl sulfate (SDS)] at 80 V for 30 min and at 120 V for 1 h. Next, the proteins were subjected to 10% SDS polyacrylamide gel electrophoresis, after which the proteins were transferred to polyvinylidene difluoride membranes (Millipore, Darmstadt, Germany). After being blocked with 5% skim milk, primary antibodies (Abcam, Cambridge, CA, United States) including anti-ASF1B (1:1000, ab276071), anti-E-cadherin (1:10000, ab40772), anti-N-cadherin (1:1000, ab245117), anti-SOX2 (1:1000, ab92494), anti-OCT4 (1:10000, ab200834), anti-p-PI3K (1:500, ab278545), anti-PI3K (1:1000, ab32089), anti-p-AKT (1:500, ab38449), anti-AKT (1:500, ab8805) and anti-β-actin (1:200, ab115777) were added to immerse the membranes at 4 °C overnight. Then the secondary antibody (1:2000, ab6721) was added. Finally, the ECL chemiluminescent system was used to examine protein blots and Image J software (NIH, United States) was used to analyze the intensity of protein blots.
Mouse tumor tissues fixed with 4% paraformaldehyde were embedded using paraffin and cut into sections (4 μm), and then dried at 62 °C for 1 h. Next, these sections were deparaffinized using xylene for 20 min, and soaked in 100%, 95%, 90%, 80% and 70% ethyl alcohol for 5 min. Antigen repair was then performed at 120 °C in citrate buffer (pH 6.0), and the tissue sections were cooled at room temperature. After being washed with phosphate buffered saline (PBS), incubated in H2O2 and washed with PBS again, sections were blocked using bovine serum albumin (5%) for 1 h. Subsequently, primary antibodies including anti-ASF1B (1:200, ab235358, Abcam) and anti-Ki67 (1:200, ab16667, Abcam) were added and incubated overnight. The secondary antibody (1:500, ab6112, Abcam) was added, followed by addition of 3,3’-diaminobenzidine substrate solution. Finally, hematoxylin was used to stain the samples, and staining areas were observed by microscopy.
Animal experiments received permission from the Institutional Animal Care and Use Committee of Beijing Viewsolid Biotechnology Co., Ltd (VS212601454), which complied with the National Institutes of Health Guide for the Care and Use of Laboratory Animals as well as ARRIVE guidelines. Female BALB/C nude mice (5-wk old), were purchased from the Chinese Academy of Sciences (Shanghai, China), and were kept at 25 °C under a 12 h light/dark cycle and 50% humidity.
To establish a xenograft mouse model, 5 × 106 HCT116 cells transfected with sh-NC or sh-ASF1B#1 were introduced into the right flank of mice via subcutaneous injection[27]. The mice were randomly divided into two groups (n = 5/group): The sh-NC group and the sh-ASF1B#1 group. The tumor size was gauged every week after injection. Tumor volume was calculated using the formula V = (shortest diameter)2 × (longest diameter) × 0.5. On the 35th d after the injection, the mice were anesthetized by an intraperitoneal injection of sodium pentobarbital (50 mg/kg) and then euthanized by cervical dislocation. Finally, the tumors were collected for further analysis.
Data from three separate experiments are presented as mean ± SD. GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, United States) was used to perform statistical analysis. Student’s t-test was performed to analyze differences between two groups. Survival curves were plotted using the Kaplan-Meier method and differences between survival curves were assessed using the log-rank test. Clinicopathological characteristics between patients who demonstrated high ASF1B expression vs those with low expression were compared using the χ2 test. Differences of P < 0.05 were considered statistically significant.
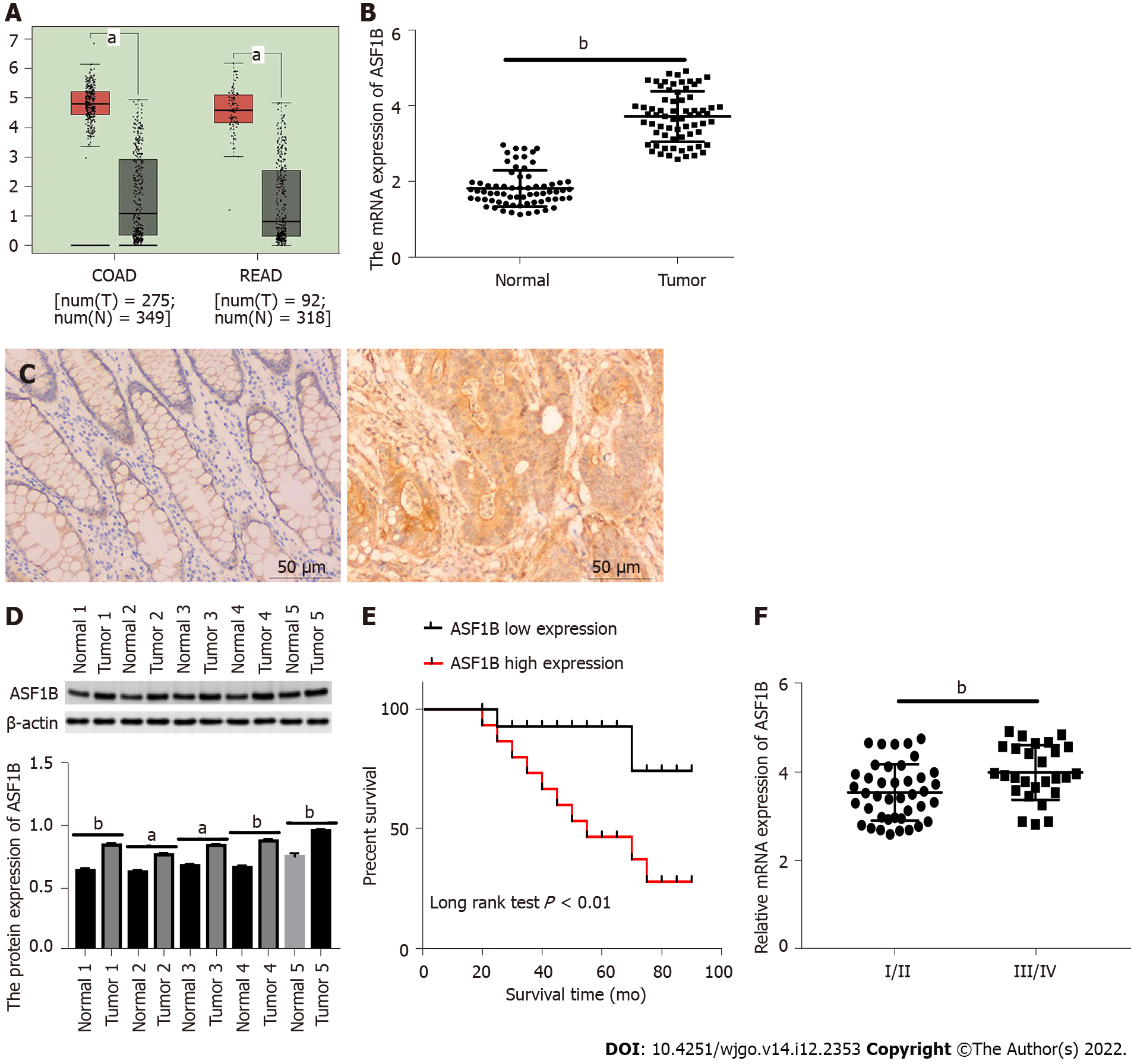
We firstly analyzed ASF1B expression in the TCGA database, and observed up-regulation of ASF1B expression in tissues of colon adenocarcinoma and rectum adenocarcinoma in contrast to corresponding noncancerous tissues (Figure 1A, P < 0.05). We validated mRNA expression of ASF1B using qRT-PCR. As expected, the mRNA expression of ASF1B was increased in CRC tissues relative to that in normal tissues (Figure 1B, P < 0.01). Moreover, the protein expression of ASF1B was detected by immunohistochemistry staining and western blot, and higher ASF1B expression was found in tumor tissues than in normal tissues (Figures 1C and 1D, P < 0.05). We further investigated the clinical significance of ASF1B in CRC and found that ASF1B expression was significantly associated with TNM stage, lymph node metastasis and distant metastasis (Table 1). The expression of ASF1B was markedly higher in CRC patients at TNM stage III/IV than in CRC patients at TNM stage I/II (Figure 1E, P < 0.01). ASF1B up-regulation resulted in poor survival of CRC patients (Figure 1F, P < 0.01).
| Variable | Total | ASF1B expression (n = 68) | P value | |
| Low (n = 34) | High (n = 34) | |||
| Age | 0.622 | |||
| < 45 yr | 28 (3.69 ± 0.74) | 15 (3.11 ± 0.33) | 13 (4.41 ± 0.37) | |
| 45 yr | 40 (3.73 ± 0.62) | 19 (3.20 ± 0.39) | 21 (4.21 ± 0.37) | |
| Gender | 0.4667 | |||
| Female | 33 (3.68 ± 0.68) | 18 (3.16 ± 0.35) | 15 (4.30 ± 0.41) | |
| Male | 35 (3.74 ± 0.66) | 16 (3.17 ± 0.39) | 19 (4.24 ± 0.37) | |
| Tumor size | 0.4921 | |||
| < 2 cm | 24 (3.77 ± 0.71) | 11 (3.14 ± 0.42) | 13 (4.31 ± 0.36) | |
| ≥ 2 cm | 44 (3.68 ± 0.65) | 24 (3.20 ± 0.35) | 20 (4.26 ± 0.40) | |
| TNM stage | 0.0035b | |||
| I/II | 41 (3.54 ± 0.64) | 27 (3.16 ± 0.38) | 14 (4.26 ± 0.35) | |
| III/IV | 27 (3.99 ± 0.62) | 8 (3.25 ± 0.35) | 19 (4.30 ± 0.40) | |
| Lymph node metastasis | 0.0158a | |||
| Negative | 36 (3.32 ± 0.68) | 29 (2.97 ± 0.35) | 7 (4.21 ± 0.42) | |
| Positive | 32 (3.95 ± 0.55) | 17 (3.40 ± 0.24) | 15 (4.30 ± 0.37) | |
| Distant metastasis | 0.0040b | |||
| Negative | 30 (3.39 ± 0.64) | 20 (3.02 ± 0.33) | 10 (4.12 ± 0.43) | |
| Positive | 38 (3.98 ± 0.57) | 12 (3.31 ± 0.28) | 26 (4.28 ± 0.38) | |
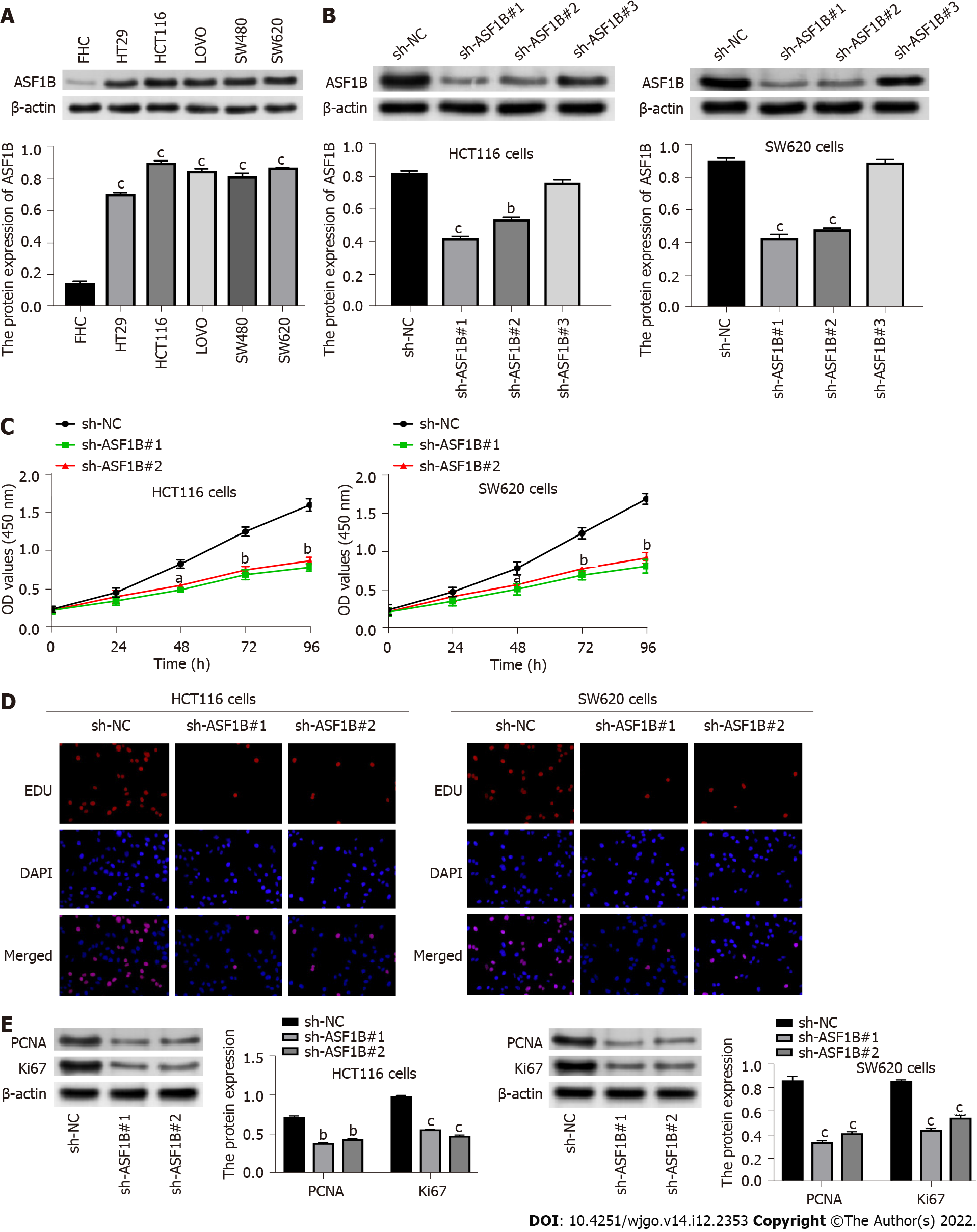
The protein expression of ASF1B was detected by western blot in a normal colorectal mucosal cell line (FHC) and CRC cell lines. Examination by western blot showed that ASF1B expression was increased in HT29, HCT116, LOVO, SW480 and SW620 cells compared to FHC cells (P < 0.001) and especially in SW620 and HCT116 cells. Therefore, the impact of ASF1B on CRC cell proliferation was investigated through loss-of-function assays in the HCT116 and SW620 cell lines.
Next, the impact of ASF1B on proliferation of CRC cells was probed through loss-of-function assays. Firstly, ASF1B was silenced by transfection of sh-ASF1B#1 and sh-ASF1B#2 in HCT116 and SW620 cells (Figure 2B, P < 0.01), and sh-ASF1B#1 and sh-ASF1B#2 were utilized for subsequent function experiments. The influence of ASF1B on proliferation of CRC cells was then determined by CCK-8 and EDU assays. It was found that the proliferation ability of HCT116 and SW620 cells was attenuated by ASF1B knockdown (Figures 2C and 2D, P < 0.05). In addition, we found that protein levels of proliferation markers (Ki67 and PCNA) were decreased after ASF1B knockdown in HCT116 and SW620 cells (Figure 2E, P < 0.01).
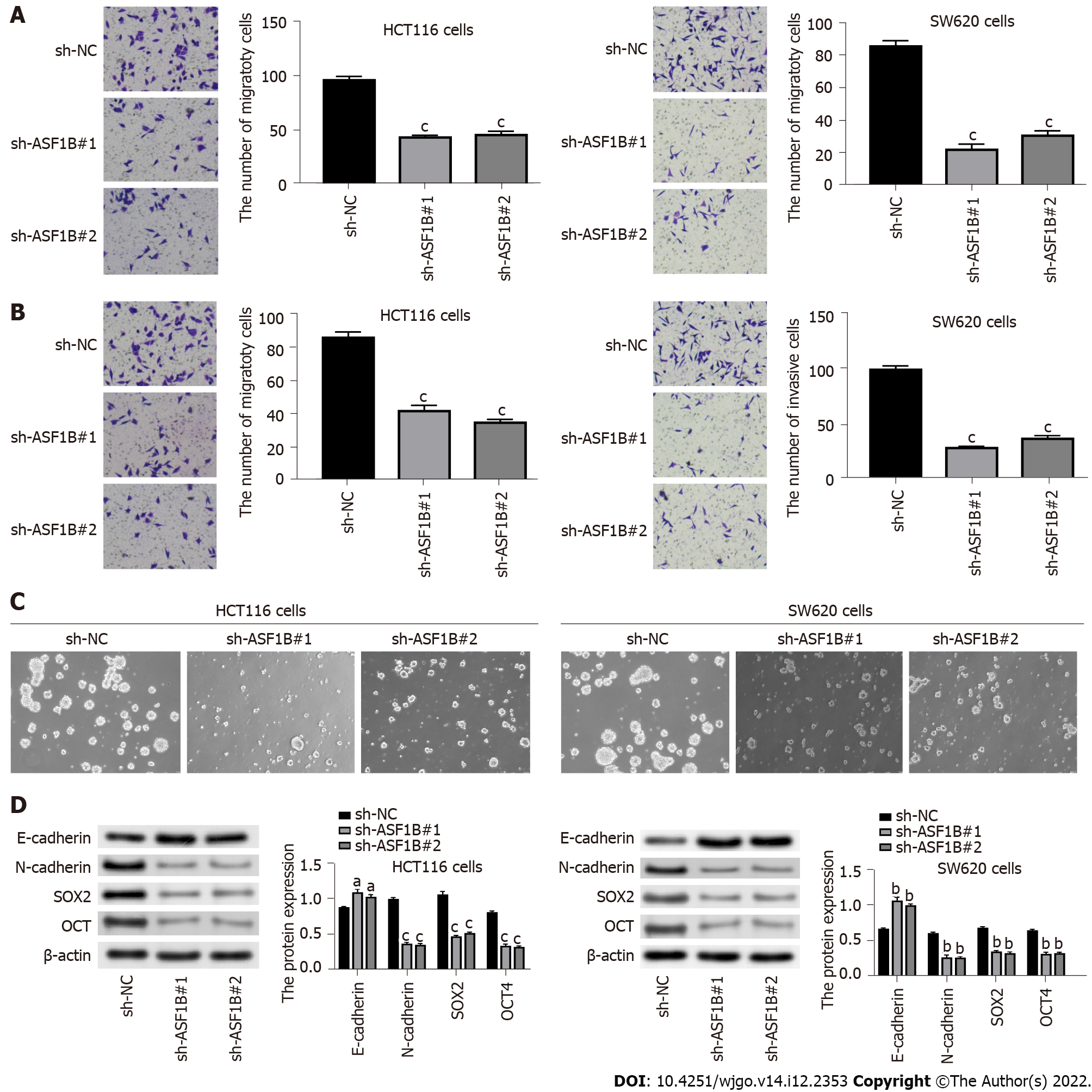
When we explored the influence of ASF1B on cell migration and invasion, and suppression of migration and invasion caused by ASF1B knockdown was found in HCT116 cells and SW620 cells (Figures 3A and 3B, P < 0.001). In addition, we detected the influence of ASF1B on cell stemness and EMT. It was demonstrated that ASF1B down-regulation led to an evident decrease in sphere formation of HCT116 and SW620 cells (Figure 3C), indicating that ASF1B down-regulation suppressed cell stemness. We also examined the levels of EMT-related proteins (E-cadherin and N-cadherin) and stemness marker proteins (SOX2 and OCT) using western blot. As expected, we observed an increased level of E-cadherin and a decreased level of N-cadherin as well as decreased levels of SOX2 and OCT after ASF1B down-regulation in HCT116 and SW620 cells (Figure 3D, P < 0.05).
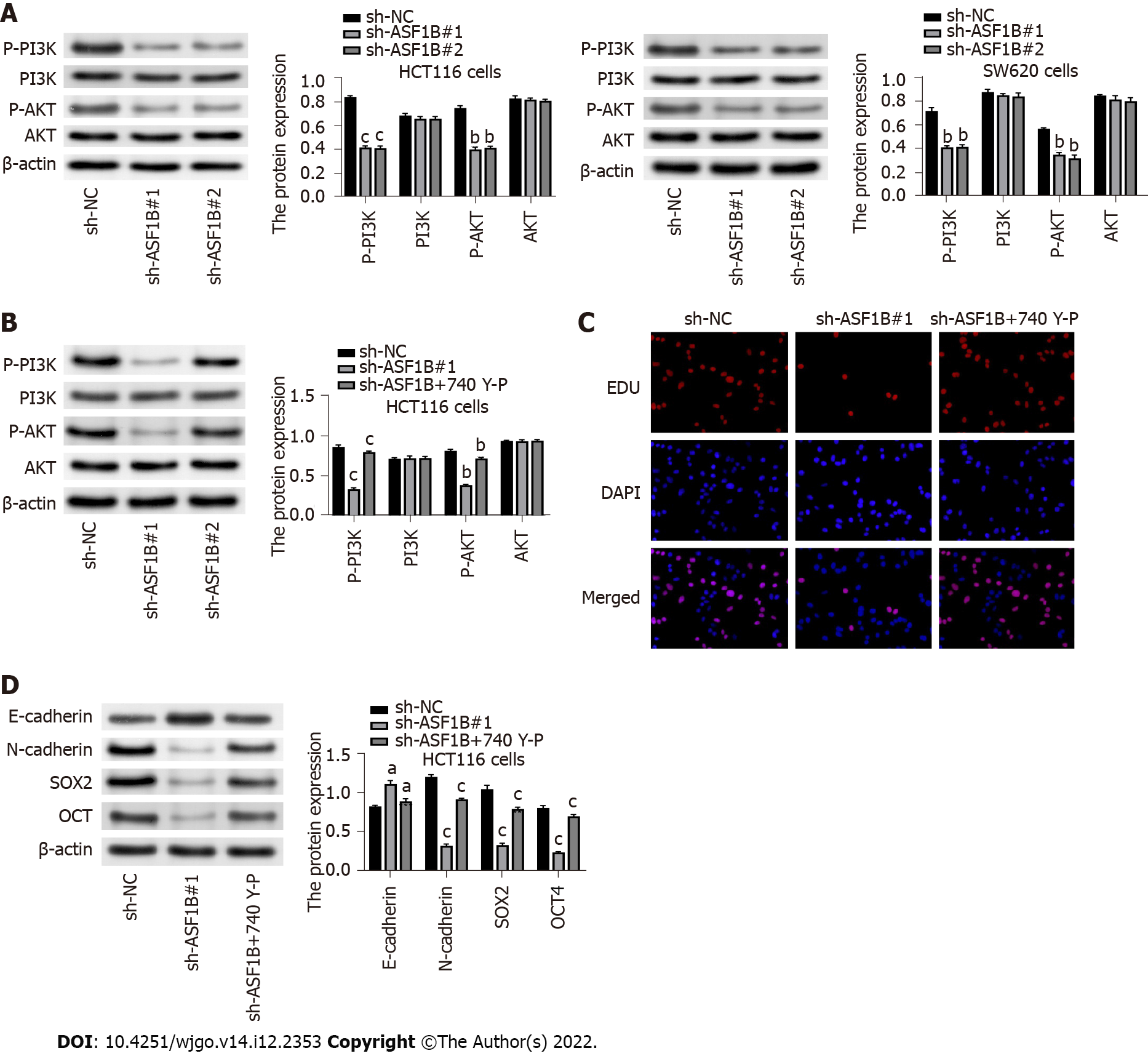
In order to examine the underlying mechanism involved in the regulatory impact of ASF1B on CRC malignancy, subsequent experiments were conducted. It was demonstrated that p-PI3K and p-AKT levels were reduced in response to ASF1B knockdown in HCT116 and SW620 cells (Figure 4A, P < 0.01). In addition, reduction of p-PI3K and p-AKT levels caused by ASF1B knockdown was reversed by addition of PI3K activator (740 Y-P) in HCT116 cells (Figure 4B, P < 0.05). More importantly, the inhibitory effect of ASF1B knockdown on proliferation was partly abolished by the addition of PI3K activator in HCT116 cells (Figure 4C). The inhibitory effects of ASF1B knockdown on protein levels of N-cadherin, SOX2 and OCT as well as the promoting effect of ASF1B knockdown on the level of E-cadherin were partly reversed by addition of PI3K activator in HCT116 cells (Figure 4D, P < 0.05). These results implied that ASF1B down-regulation had an impact on CRC malignancy by regulating the PI3K/AKT pathway.
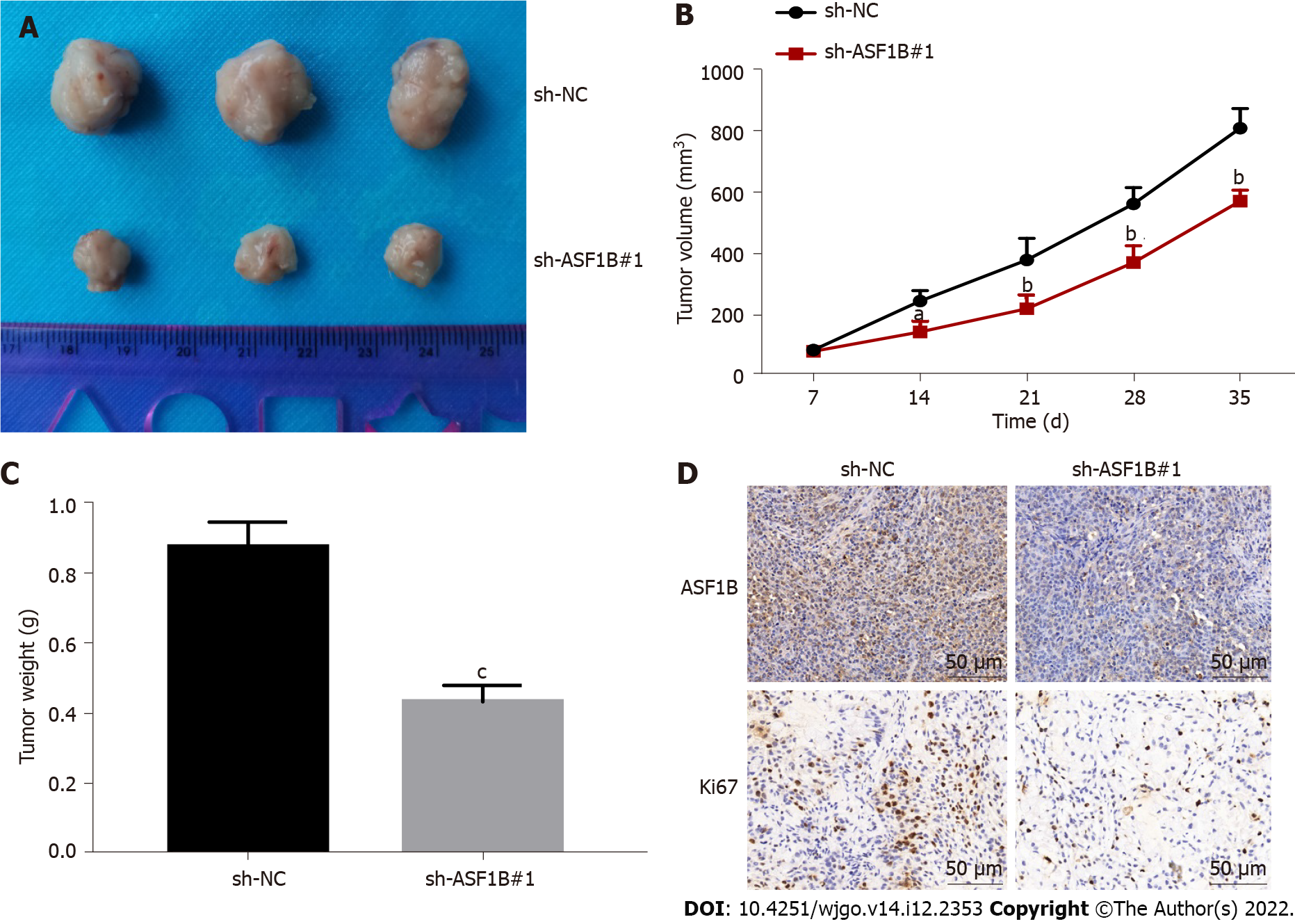
After detecting the function of ASF1B in CRC in vitro, we next verified its function in vivo. As illustrated in Figures 5A-C, silencing of ASF1B inhibited tumor growth, and reduced tumor volume and weight in xenograft mice (P < 0.05). Likewise, protein levels of Ki67 and ASF1B were reduced after ASF1B knockdown in tumor tissues of mice (Figure 5D).
Over the past years, the mortality rate of CRC patients is increasing and limited prognosis poses a severe threat to CRC patients[28-30]. Thus, developing new effective therapies for CRC patients is imperative. Emerging evidence has uncovered that ASF1B is highly expressed and shows potential clinical value for the prognosis of cancer patients[13,17]. For instance, ASF1B shows high expression in HCC tissues, and ASF1B up-regulation is correlated with poor survival of HCC patients[17]. ASF1B expression is increased in tissues of lung adenocarcinoma, and ASF1B up-regulation is correlated with worse overall survival in lung adenocarcinoma patients[13]. Here, we also found up-regulation of ASF1B expression in CRC tissues and cells, and our outcome was consistent with prior reports[14,31]. We also discovered that ASF1B expression was significantly associated with TNM stage, lymph node metastasis and distant metastasis. In addition, ASF1B expression was inversely relevant to survival data of CRC patients, and was markedly higher in CRC patients with advanced TNM stages than in patients with earlier stages. These results suggested that ASF1B may be a useful marker for prognosis and diagnosis assessment in CRC patients.
In previous studies, ASF1B was reported to be a key regulator in cancer progression[15,16]. For instance, ASF1B knockdown prevents cervical cancer cells from proliferating, migrating and invading, and suppresses tumor growth in mice[15]. ASF1B knockdown inhibits migration, invasion and EMT of lung cancer cells, and retards tumor growth in xenograft mice as well as the expression of Ki67[16]. Similarly, our outcomes in vitro displayed that ASF1B down-regulation distinctly attenuated proliferation, migration and invasion abilities of HCT116 and SW620 cells, and inhibited EMT and stemness of HCT116 and SW620 cells. As an important cancer hallmark in metastases and the “cadherin switch”, EMT was demonstrated to initiate CRC metastasis from the primary tumor to distant sites, especially to liver and lymph nodes[32,33]. It was reported that loss of E-cadherin can cause metastatic dissemination and activation of EMT transcription factors in cancer cells[34]. Many invasive and metastatic cancers are associated with high expression of E-cadherin, notably in prostate cancer[35], ovarian cancer[36], and glioblastoma[37], suggesting that E-cadherin facilitates metastasis in several tumors instead of inhibiting tumor progression. In addition, N-cadherin is reported to act as an indicator of ongoing EMT and N-cadherin down-regulation can cause metastatic dissemination[38]. In our study, we found that the silencing of ASF1B results in a reverse cadherin switch phenomenon, indicating a potentially important role of ASF1B silencing in reducing the metastatic potential of CRC cells in vitro. In addition, our in vivo results indicated that ASF1B down-regulation suppressed tumor growth in xenograft mice as well as Ki67 expression. These findings provided evidence of the anti-tumor role of ASF1B knockdown in CRC, and shed light on exploring promising therapeutic agents for CRC treatment.
Considering the important function of ASF1B in CRC, we attempted to examine how ASF1B affected CRC progression and development. It is noteworthy that ASF1B has been proved to affect progression and development of cancers by regulating the PI3K/AKT pathway[22,23]. For instance, suppression of the PI3K/AKT pathway is pertinent to the influence of ASF1B knockdown on prostate cancer[22]. Silencing of ASF1B impairs cell proliferation by inactivating the PI3K/AKT pathway in pancreatic cancer[23]. In this study, we also investigated whether the PI3K/AKT pathway participated in ASF1B knockdown-mediated tumor inhibition in CRC cells. It was found that protein levels of p-PI3K and p-AKT were decreased due to ASF1B knockdown in HCT116 and SW620 cells. Concurrently, suppressive influences of ASF1B knockdown on proliferation, stemness and EMT of HCT116 cells were reversed by PI3K activator. Combining the above findings, we concluded that ASF1B knockdown repressed the malignant behaviors of CRC cells by inactivation of the PI3K/AKT pathway.
However, there were three limitations in our study. Firstly, we only explored the role of ASF1B knockdown in CRC, and the overexpression experiment of ASF1B should be performed to further validate the influence of ASF1B on CRC. Secondly, only in vitro studies have been performed and therefore in vivo studies are recommended to strengthen the study’s hypothesis. Thirdly, we failed to perform a-priori sample calculation, and we determined the sample size through a literature search. Overall, the novelty of this study included two aspects. Firstly, we demonstrated the role of ASF1B in CRC in vitro and in vivo for the first time. Secondly, we uncovered a new mechanism by which ASF1B impacted CRC cells.
In summary, ASF1B expression was increased in CRC tissues and cells, and was negatively associated with the prognosis of CRC patients. Functionally, ASF1B knockdown repressed the malignant behaviors of CRC cells in vitro and tumorigenesis in vivo, therefore having potential for CRC treatment. Moreover, our findings revealed that ASF1B down-regulation repressed impaired behaviors of CRC cells by inactivating the PI3K/AKT pathway. Our study provides new insights into the functional importance of ASF1B in CRC, and indicates that ASF1B may be a promising prognostic marker and a target for the management of CRC.
ASF1B may be a potential diagnostic and prognostic biomarker for improving CRC patients’ outcome. More importantly, ASF1B may be a novel target for the treatment of CRC, showing promising prospects in clinical practice. In our future studies, we plan to perform in vivo studies to validate the role of ASF1B in CRC. Moreover, we expect more investigations into the other mechanisms of ASF1B in cancers.
Colorectal cancer (CRC) is identified as a malignant gastrointestinal tumor, with high prevalence and mortality. Abundant studies have proved the important role of anti-silencing function 1B (ASF1B) in cancers, but little is known about ASF1B in CRC.
In order to identify the prognosis biomarker and treatment target for CRC.
To evaluate the role and mechanism of ASF1B in CRC.
The mRNA expression of ASF1B was detected by quantitative real-time polymerase chain reaction. The clinical value of ASF1B for diagnosis and prognosis of CRC was assessed. The function of ASF1B was evaluated using in vitro assays and in vivo tumor formation experiments. The molecular mechanism of ASF1B on the phosphatidylinositol 3-kinase (PI3K)/AKT pathway was explored via the addition of PI3K activator.
The expression level of ASF1B was markedly increased in CRC tissues and cells, which was inversely associated with survival time of CRC patients and positively associated with tumor node metastasis stage of CRC patients. Biological functional analyses indicated that ASF1B knockdown may suppress the malignancy of CRC cells by regulating the PI3K/AKT pathway.
ASF1B is highly expressed in CRC tissues and cells, showing potential as a diagnostic and prognostic biomarker for CRC. Silencing of ASF1B inactivated the PI3K/AKT pathway to inhibit CRC malignancy in vitro.
Other mechanisms of ASF1B in CRC may be investigated in the future, and its application in anti-tumor therapy will be extended.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ekine-Afolabi B, United Kingdom; Martinou E, United Kingdom; Tanabe S, Japan S-Editor: Wang JJ L-Editor: Webster JR P-Editor: Wang JJ
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15468] [Article Influence: 2578.0] [Reference Citation Analysis (2)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55765] [Article Influence: 7966.4] [Reference Citation Analysis (132)] |
| 3. | Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2526] [Cited by in RCA: 2911] [Article Influence: 363.9] [Reference Citation Analysis (3)] |
| 4. | Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3058] [Cited by in RCA: 3293] [Article Influence: 411.6] [Reference Citation Analysis (3)] |
| 5. | Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 520] [Cited by in RCA: 903] [Article Influence: 112.9] [Reference Citation Analysis (2)] |
| 6. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3259] [Article Influence: 651.8] [Reference Citation Analysis (2)] |
| 7. | Pang SW, Awi NJ, Armon S, Lim WW, Low JS, Peh KB, Peh SC, Teow SY. Current Update of Laboratory Molecular Diagnostics Advancement in Management of Colorectal Cancer (CRC). Diagnostics (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Müller MF, Ibrahim AE, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469:125-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 263] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 9. | Peng H, Nogueira ML, Vogel JL, Kristie TM. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc Natl Acad Sci U S A. 2010;107:2461-2466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 10. | Messiaen S, Guiard J, Aigueperse C, Fliniaux I, Tourpin S, Barroca V, Allemand I, Fouchet P, Livera G, Vernet M. Loss of the histone chaperone ASF1B reduces female reproductive capacity in mice. Reproduction. 2016;151:477-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Soniat M, Cağatay T, Chook YM. Recognition Elements in the Histone H3 and H4 Tails for Seven Different Importins. J Biol Chem. 2016;291:21171-21183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Paul PK, Rabaglia ME, Wang CY, Stapleton DS, Leng N, Kendziorski C, Lewis PW, Keller MP, Attie AD. Histone chaperone ASF1B promotes human β-cell proliferation via recruitment of histone H3.3. Cell Cycle. 2016;15:3191-3202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Feng Z, Zhang J, Zheng Y, Wang Q, Min X, Tian T. Elevated expression of ASF1B correlates with poor prognosis in human lung adenocarcinoma. Per Med. 2021;18:115-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 14. | Corpet A, De Koning L, Toedling J, Savignoni A, Berger F, Lemaître C, O'Sullivan RJ, Karlseder J, Barillot E, Asselain B, Sastre-Garau X, Almouzni G. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30:480-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 15. | Liu X, Song J, Zhang Y, Wang H, Sun H, Feng X, Hou M, Chen G, Tang Q, Ji M. ASF1B promotes cervical cancer progression through stabilization of CDK9. Cell Death Dis. 2020;11:705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 16. | Wang W, Xiao L, Pan D, Hu L. ASF1B enhances migration and invasion of lung cancers cell via regulating the P53-mediated epithelial-mesenchymal transformation (EMT) signaling pathway. Neoplasma. 2022;69:361-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Ouyang X, Lv L, Zhao Y, Zhang F, Hu Q, Li Z, Zhu D, Li L. ASF1B Serves as a Potential Therapeutic Target by Influencing Cell Cycle and Proliferation in Hepatocellular Carcinoma. Front Oncol. 2021;11:801506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Davis WJ, Lehmann PZ, Li W. Nuclear PI3K signaling in cell growth and tumorigenesis. Front Cell Dev Biol. 2015;3:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 19. | Ma Z, Lou S, Jiang Z. PHLDA2 regulates EMT and autophagy in colorectal cancer via the PI3K/AKT signaling pathway. Aging (Albany NY). 2020;12:7985-8000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 20. | Yin F, Huang X, Xuan Y. Pyrroline-5-Carboxylate Reductase-2 Promotes Colorectal Cancer Progression via Activating PI3K/AKT/mTOR Pathway. Dis Markers. 2021;2021:9950663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Duan S, Huang W, Liu X, Chen N, Xu Q, Hu Y, Song W, Zhou J. IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. J Exp Clin Cancer Res. 2018;37:304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 22. | Han G, Zhang X, Liu P, Yu Q, Li Z, Wei X. Knockdown of anti-silencing function 1B histone chaperone induces cell apoptosis via repressing PI3K/Akt pathway in prostate cancer. Int J Oncol. 2018;53:2056-2066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Wang K, Hao Z, Fu X, Li W, Jiao A, Hua X. Involvement of elevated ASF1B in the poor prognosis and tumorigenesis in pancreatic cancer. Mol Cell Biochem. 2022;477:1947-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 24. | Bertero L, Massa F, Metovic J, Zanetti R, Castellano I, Ricardi U, Papotti M, Cassoni P. Eighth Edition of the UICC Classification of Malignant Tumours: an overview of the changes in the pathological TNM classification criteria-What has changed and why? Virchows Arch. 2018;472:519-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 25. | Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2341] [Cited by in RCA: 4392] [Article Influence: 549.0] [Reference Citation Analysis (4)] |
| 26. | Sun C, Zhang Z, He P, Zhou Y, Xie X. Involvement of PI3K/Akt pathway in the inhibition of hepatocarcinoma cell invasion and metastasis induced by SASH1 through downregulating Shh-Gli1 signaling. Int J Biochem Cell Biol. 2017;89:95-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 27. | Zheng Z, Li X, You H, Zheng X, Ruan X. LncRNA SOCS2-AS1 inhibits progression and metastasis of colorectal cancer through stabilizing SOCS2 and sponging miR-1264. Aging (Albany NY). 2020;12:10517-10526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 28. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21361] [Article Influence: 2136.1] [Reference Citation Analysis (3)] |
| 29. | Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M, Lhermitte B, Olshen AB, Wiedenmann B, Cantley LC, Gray JW, Hanahan D. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 760] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 30. | Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 968] [Cited by in RCA: 1153] [Article Influence: 96.1] [Reference Citation Analysis (0)] |
| 31. | Rosty C, Sheffer M, Tsafrir D, Stransky N, Tsafrir I, Peter M, de Crémoux P, de La Rochefordière A, Salmon R, Dorval T, Thiery JP, Couturier J, Radvanyi F, Domany E, Sastre-Garau X. Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene. 2005;24:7094-7104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 115] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 32. | Vu T, Datta PK. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 375] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 33. | Yang C, Dou R, Wei C, Liu K, Shi D, Zhang C, Liu Q, Wang S, Xiong B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther. 2021;29:2088-2107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 150] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 34. | Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645-3654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1038] [Cited by in RCA: 1157] [Article Influence: 68.1] [Reference Citation Analysis (0)] |
| 35. | Putzke AP, Ventura AP, Bailey AM, Akture C, Opoku-Ansah J, Celiktaş M, Hwang MS, Darling DS, Coleman IM, Nelson PS, Nguyen HM, Corey E, Tewari M, Morrissey C, Vessella RL, Knudsen BS. Metastatic progression of prostate cancer and e-cadherin regulation by zeb1 and SRC family kinases. Am J Pathol. 2011;179:400-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 36. | Reddy P, Liu L, Ren C, Lindgren P, Boman K, Shen Y, Lundin E, Ottander U, Rytinki M, Liu K. Formation of E-cadherin-mediated cell-cell adhesion activates AKT and mitogen activated protein kinase via phosphatidylinositol 3 kinase and ligand-independent activation of epidermal growth factor receptor in ovarian cancer cells. Mol Endocrinol. 2005;19:2564-2578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 104] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Lewis-Tuffin LJ, Rodriguez F, Giannini C, Scheithauer B, Necela BM, Sarkaria JN, Anastasiadis PZ. Misregulated E-cadherin expression associated with an aggressive brain tumor phenotype. PLoS One. 2010;5:e13665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 38. | Lammens T, Swerts K, Derycke L, De Craemer A, De Brouwer S, De Preter K, Van Roy N, Vandesompele J, Speleman F, Philippé J, Benoit Y, Beiske K, Bracke M, Laureys G. N-cadherin in neuroblastoma disease: expression and clinical significance. PLoS One. 2012;7:e31206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |