Published online Jan 15, 2019. doi: 10.4251/wjgo.v11.i1.9
Peer-review started: October 18, 2018
First decision: November 14, 2018
Revised: December 5, 2018
Accepted: December 17, 2018
Article in press: December 17, 2018
Published online: January 15, 2019
Processing time: 90 Days and 5.2 Hours
Colorectal cancer (CRC) is a common malignancy of the gastrointestinal tract. The worldwide mortality rate of CRC is about one half of its morbidity. Ubiquitin is a key regulatory factor in the cell cycle and widely exists in eukaryotes. Human leukocyte antigen F-associated transcript 10 (FAT10), known as diubiquitin, is an 18 kDa protein with 29% and 36% homology with the N and C termini of ubiquitin. The function of FAT10 has not been fully elucidated, and some studies have shown that it plays an important role in various cell processes.
To examine FAT10 expression and to analyze the relationship between FAT10 expression and the clinicopathological parameters of CRC.
FAT10 expression in 61 cases of CRC and para-cancer colorectal tissues was measured by immunohistochemistry and Western blotting. The relationship between FAT10 expression and clinicopathological parameters of CRC was statistically analyzed.
Immunohistochemical analysis showed that the positive rate of FAT10 expression in CRC (63.93%) was significantly higher than that in tumor-adjacent tissues (9.84%, P < 0.05) and normal colorectal mucosal tissue (1.64%, P < 0.05). Western blotting also indicated that FAT10 expression was significantly higher in CRC than in tumor-adjacent tissue (P < 0.05). FAT10 expression was closely associated with clinical stage and lymphatic spread of CRC. FAT10 expression also positively correlated with p53 expression.
FAT10 expression is highly upregulated in CRC. FAT10 expression is closely associated with clinical stage and lymphatic spread of CRC.
Core tip: Colorectal cancer (CRC) is a common malignancy of the gastrointestinal tract. Genetic studies have demonstrated that the development of CRC is a complex process involving the activation of proto-oncogenes, inactivation of tumor suppressor genes, gene mutations, and dysregulation of apoptosis-related genes. Human leukocyte antigen F-associated transcript 10 (FAT10) is a regulatory protein of the ubiquitin-like modifier family that regulates various cell processes including mitosis, chromosome stability, apoptosis, immune control, and 26S-proteasome-mediated protein degradation. Our study investigated FAT10 expression in tumor and tumor-adjacent tissues of CRC patients and analyzed the relationship between FAT10 expression and the clinicopathological parameters of CRC.
- Citation: Zhang CY, Sun J, Wang X, Wang CF, Zeng XD. Clinicopathological significance of human leukocyte antigen F-associated transcript 10 expression in colorectal cancer. World J Gastrointest Oncol 2019; 11(1): 9-16
- URL: https://www.wjgnet.com/1948-5204/full/v11/i1/9.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i1.9
Colorectal cancer (CRC) is a common malignancy of the gastrointestinal tract. In China, the incidence of CRC ranks fifth in men and fourth in women among all malignant tumors, and its mortality ranks fifth[1-9]. The worldwide mortality rate of CRC is about one half of its morbidity. Although the treatment of this malignancy has improved, its prognosis is still not satisfactory. The etiology and pathogenesis of CRC are related to environmental (mainly diet, such as fat and animal protein) and genetic factors. Genetic factors contribute to the development of CRC in many ways. It is well known that CRC is commonly seen in patients with familial adenomatous polyposis and Lynch syndrome, although the incidence in patients with other polyp types is low. Genetic studies have demonstrated that the development of CRC is a complex process involving the activation of proto-oncogenes, inactivation of tumor suppressor genes, gene mutations, and dysregulation of apoptosis-related genes.
Ubiquitin, a polypeptide containing 76 amino acid residues, is a key regulatory factor in the cell cycle and is widely expressed in eukaryotes[10-13]. In recent years, a growing number of ubiquitin-related low-molecular-weight proteins, known as ubiquitin-like proteins, have been identified and are associated with a variety of cell processes[14-16]. So far, two ubiquitin-like protein families, ubiquitin-like modifiers (UBLs) and ubiquitin-domain proteins, have been identified[17-19].
Human leukocyte antigen F-associated transcript 10 (FAT10), also known as diubiquitin, belongs to the UBL family. FAT10 is an 18-kDa protein with 29% and 36% homology with the N and C termini of ubiquitin, respectively. It is located on chromosome 6 and was originally thought to be a gene of the major histocompatibility complex. Although the function of FAT10 has not been fully elucidated, some studies have shown that it plays an important role in various cell processes[20].
This study investigated FAT10 expression in tumor and tumor-adjacent tissues of CRC patients by immunohistochemistry and Western blotting and analyzed the relationship between FAT10 expression and the clinicopathological parameters of CRC.
Sixty-one surgical samples were collected from patients who underwent surgery for CRC at our hospital between March 2010 and March 2011. None of the patients underwent preoperative radiotherapy or chemotherapy. There were 38 men and 23 women, with a median age of 67 years (range, 39–97 years). The median tumor size was 50 mm (range, 25–110 mm). Of all patients included, 46 had highly differentiated tumors, 15 had lowly differentiated tumors, 22 had stage I/II disease, 39 had stage III/IV disease, 38 had lymph node metastasis, and 23 had no lymph node metastasis. Tumor-adjacent samples were collected 2 cm away from the tumor, and normal colorectal mucosal samples were collected from surgical margins (> 5 cm away from the tumor). All samples were immediately preserved in liquid nitrogen for further use. Institutional review board approval of Central Hospital Affiliated to Shenyang Medical College was obtained for this study.
Tissue samples were fixed in neutral buffered formalin solution, embedded in paraffin, and cut into 4-μm sections. The sections were dewaxed in xylene, hydrated in a graded ethanol series, and subjected to immunohistochemical staining for FAT10 using the streptavidin–peroxidase method. Mouse anti-FAT10 monoclonal antibody (dilution, 1:200; Santa Cruz Biotechnology, Santa Cruz, CA, United States) was used as a primary antibody. Following diaminobenzidine staining, yellowish-brown granules present in the nucleus were regarded as positive signals. Five high-power (400×) fields with the most strongly positive signal were selected to count the number of positive cells among 200 tumor cells. The percentage of immunoreactive cells was scored as follows: no staining, 0; 1%-10% staining, 1; 11%-50%, 2; 51%-80%, 3; and 81%-100%, 4. Staining intensity was scored on the following 0-3 scale: negative, 0; light yellowish-brown, 1; yellowish-brown, 2; and brown, 3. Immunoreactive score (IS; 0-7) was calculated as the sum of the score of the percentage of immunoreactive cells and the score of staining intensity. Samples with IS < 4 were considered negative, while those with IS ≥ 4 were considered positive.
Tissue samples were washed in pre-cooled phosphate-buffered saline three times and lysed in a nondenaturing tissue lysis buffer containing protease inhibitors to extract total proteins. Cell lysate proteins were resolved by polyacrylamide gel electrophoresis and electrophoretically transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% normal fetal bovine serum at room temperature for 2 h, incubated with mouse anti-FAT10 monoclonal antibody (dilution, 1:400; Santa Cruz Biotechnology) or anti-β-actin antibody (dilution, 1:200; Zhongshan Golden Bridge, Beijing, China) at 4°C overnight, washed with Tween Tris Base Buffer Solution (commonly known as TTBS) three times, and then incubated with a horseradish-peroxidase-labeled secondary antibody (dilution, 1:400; Zhongshan Golden Bridge) at room temperature for 2 h. After the membranes were washed three times with TTBS, the proteins were detected by enhanced chemiluminescence. Images were obtained using the EC3 Imaging System, and the bands were semiquantitatively analyzed using ImageJ software to calculate the relative expression of FAT10 to β-actin. The experiment was repeated at least three times, with mean values calculated for further analysis.
Statistical analyses were performed using SPSS version 21.0. Data are presented as mean ± SD. The relationship between FAT10 expression and clinicopathological parameters of CRC was analyzed by χ2 test. Comparisons between groups were evaluated by t test, and comparisons among three or more groups were analyzed by analysis of variance, followed by the least significant difference test or Tamhane’s test. P < 0.05 was considered statistically significant.
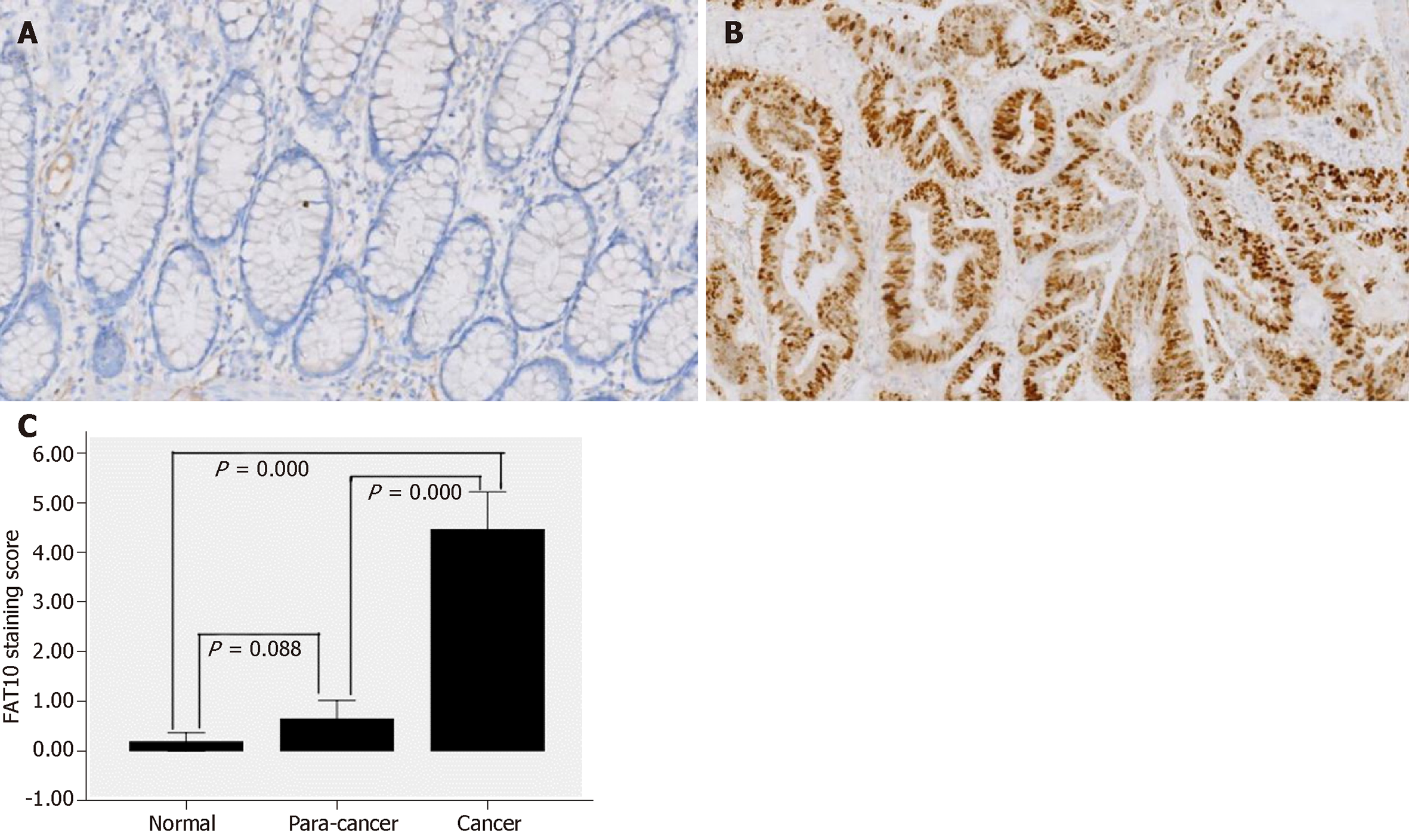
Immunohistochemical staining showed that positive signals, most of which were weak, were present only in four (6.56%) normal colorectal mucosal tissues and in 11 (18.03%) tumor-adjacent tissues (Figure 1A). According to IS, only one (1.64%) normal colorectal mucosal tissue and six (9.84%) tumor-adjacent tissues were positive for FAT10. In contrast, 46 (75.41%) CRC tissues were positive for FAT10, of which 39 showed moderately to strongly positive expression (Figure 1B). FAT10 expression was significantly higher in CRC than in normal colorectal mucosa and tumor-adjacent tissues (P < 0.05), although there was no significant difference between normal colorectal mucosa and tumor-adjacent tissues (P > 0.05; Table 1 and Figure 1C).
| Group | Staining score | P value | FAT10 | P value | |
| Negative | Positive | ||||
| (IS < 4) | (IS ≥ 4) | ||||
| Normal | 0.18 ± 0.72 | 0.000 | 60/61 (98.36) | 1/61 (1.64) | 0.000 |
| Para-cancer | 0.64 ± 1.46 | 55/61 (90.16) | 6/61 (9.84) | ||
| Cancer | 4.46 ± 2.97 | 22/61 (36.07) | 39/61 (63.93) | ||
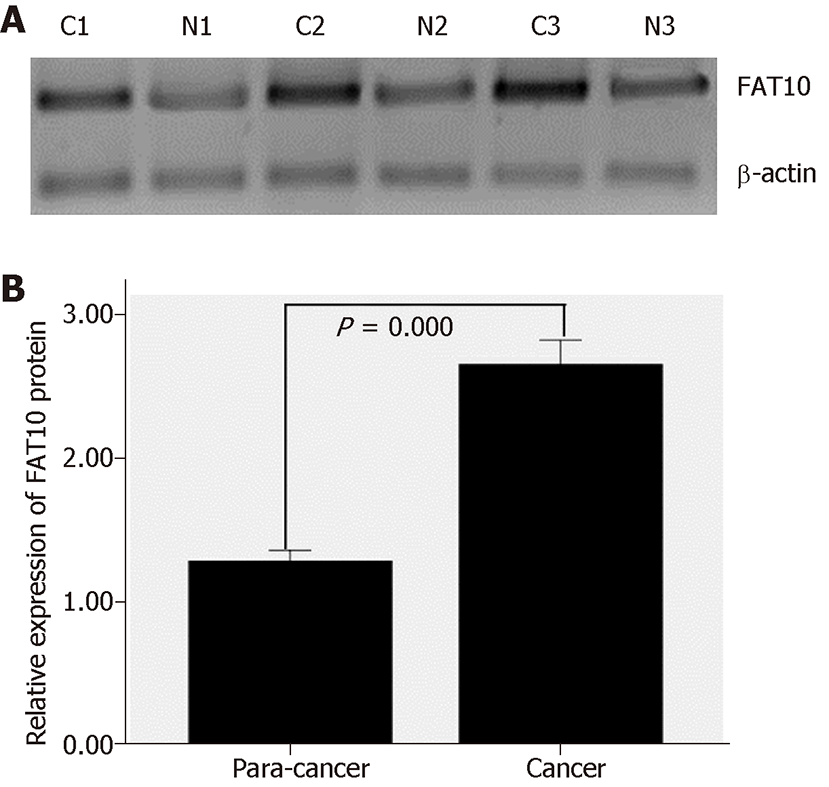
Western blotting showed that in 30 pairs of colorectal tissues, FAT10 expression was significantly higher in CRC than in tumor-adjacent tissues (t = 6.558, P = 0.000; Figure 2).
We assessed the relationship between FAT10 expression and some clinicopathological parameters of CRC, including age, sex, tumor size, clinical stage, tumor differentiation, lymph node metastasis, and p53 expression. FAT10 expression was associated with clinical stage and lymph node metastasis (Table 2). In addition, there was a positive correlation between p53 and FAT10 expression in CRC (Table 3).
| Clinicopathologic parameter | n | + | - | P value |
| Age, yr | 0.095 | |||
| < 50 | 8 | 3 | 5 | |
| ≥ 50 | 53 | 36 | 17 | |
| Gender | 0.476 | |||
| Male | 38 | 23 | 15 | |
| Female | 23 | 16 | 7 | |
| Tumor size, mm | 0.231 | |||
| < 50 | 41 | 28 | 13 | |
| ≥ 50 | 20 | 11 | 9 | |
| Tumor differentiation | 0.325 | |||
| High | 46 | 31 | 15 | |
| Low | 15 | 8 | 7 | |
| Clinical stage | 0.000 | |||
| I/II | 22 | 7 | 15 | |
| III/IV | 39 | 32 | 7 | |
| Lymph node metastasis | 0.000 | |||
| No | 23 | 8 | 15 | |
| Yes | 38 | 31 | 7 |
| FAT10 positive | FAT10 negative | |
| p53 positive | 30 | 4 |
| p53 negative | 9 | 18 |
FAT10 is a regulatory protein of the UBL family that regulates various cell processes including mitosis, chromosome stability, apoptosis, immune control, and 26S-proteasome-mediated protein degradation[21-25]. FAT10 can bind with a mitotic spindle assembly checkpoint protein, mitotic arrest deficiency 2 (MAD2), in a noncovalent manner. MAD2 is responsible for maintaining the integrity of the spindle during mitosis, and dysfunction of MAD2 can lead to chromosome instability, which is an important characteristic of many tumors[22,26,27].
Overexpression of the FAT10 gene has been found in some malignant tumors, including gastrointestinal and gynecological malignancies[28-30]. It is reported that interferon-γ and tumor necrosis factor (TNF)-α can increase the expression of the FAT10 gene[31-35], while FAT10 expression can be negatively regulated by p53, which plays an important role in regulating the cell cycle[36-38]. FAT10 is abnormally highly expressed in some malignant tumors and highly expressed in premetaphase of the cell cycle; MAD2 dysfunction causes abnormal mitotic division and chromosome instability; and expression of FAT10 is positively regulated by TNF-α (a putative tumor promoter)[32] and negatively regulated by p53 (a guardian of the genome)[37]. These results suggest that FAT10 plays an important role in cell cycle regulation and tumorigenesis.
Our results showed that the positive expression rate of FAT10 protein gradually increased from normal mucosal tissue to tumor-adjacent tissue and CRC. Consistent with this finding, Western blotting indicated that FAT10 protein expression was significantly higher in CRC tissue than in tumor-adjacent tissue. Collectively, these findings suggest that increased FAT10 expression plays an important role in colorectal carcinogenesis.
In addition, we analyzed the relationship between FAT10 expression and some clinicopathological parameters of CRC (including age, sex, tumor size, clinical stage, tumor differentiation, lymph node metastasis, and p53 expression). FAT10 expression was associated with clinical stage and lymph node metastasis. FAT10 expression was significantly higher in stage III/IV than in stage I/II CRC, and CRC with lymph node metastasis expressed more FAT10. Moreover, FAT10 expression was positively correlated with p53 expression. These results indicate that FAT10 expression is closely related to the degree of malignancy of CRC and the invasion and proliferation of cancer cells.
In summary, FAT10 is a UBL family member that is closely associated with the development of a wide variety of tumors. This study demonstrated that FAT10 is highly expressed in CRC, and FAT10 expression is closely related to clinical stage and lymph node metastasis. However, our current study is a proof-of-principle, and additional research needs to be performed to confirm our results. Therefore, further exploration of the role of FAT10 in the development of CRC and the underlying mechanisms, especially its relationship with the cell cycle, will be important for understanding the value of FAT10 in CRC diagnosis, prognosis and therapy.
The worldwide mortality rate of colorectal cancer (CRC) is about one half of its morbidity. Ubiquitin is a key regulatory factor in the cell cycle and widely exists in eukaryotes. Human leukocyte antigen F-associated transcript 10 (FAT10), also known as diubiquitin, is an 18-kDa protein with 29% and 36% homology with the N and C termini of ubiquitin, respectively.
The function of FAT10 has not been fully elucidated, and some studies have shown that it plays an important role in various cell processes.
The objective of this study is to examine FAT10 expression and to analyze the relationship between FAT10 expression and the clinicopathological parameters of CRC.
Immunohistochemistry and Western blotting were used to measure FAT10 expression in 61 cases of CRC and para-cancer colorectal tissues. In addition, the relationship between FAT10 expression and the clinicopathological parameters of CRC was statistically analyzed.
Immunohistochemical analysis showed that the positive rate of FAT10 expression in CRC was significantly higher than in tumor-adjacent tissue and normal colorectal mucosal tissue. Western blotting indicated that FAT10 expression was significantly higher in CRC than in tumor-adjacent tissue. FAT10 expression was closely associated with clinical stage and lymphatic spread of CRC.
FAT10 expression is highly upregulated in CRC and is closely associated with clinical stage and lymphatic spread of CRC.
Further exploration of the role of FAT10 in the development of CRC and the underlying mechanisms, especially its relationship with the cell cycle, will be important for understanding the value of FAT10 in CRC diagnosis, prognosis and therapy.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chadokufa S, Shin T, Toyonaga T S- Editor: Wang JL L- Editor: Filipodia E- Editor: Wu YXJ
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13214] [Article Influence: 1468.2] [Reference Citation Analysis (3)] |
| 2. | Bhutani MS, Uthamanthil R, Suzuki R, Shetty A, Klumpp SA, Nau W, Stafford RJ. Endoscopic ultrasound-guided inoculation of transmissible venereal tumor in the colon: A large animal model for colon neoplasia. Endosc Ultrasound. 2016;5:85-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 3. | Cartana ET, Gheonea DI, Cherciu IF, Streaţa I, Uscatu CD, Nicoli ER, Ioana M, Pirici D, Georgescu CV, Alexandru DO, Şurlin V, Gruionu G, Săftoiu A. Assessing tumor angiogenesis in colorectal cancer by quantitative contrast-enhanced endoscopic ultrasound and molecular and immunohistochemical analysis. Endosc Ultrasound. 2018;7:175-183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Eshtiaghpour D, Iskander JM, Singh IM, Chung DS, Eysselein VE, Reicher S. Time-of-day effect and the yield of endoscopic ultrasound fine needle aspiration. Endosc Ultrasound. 2016;5:196-200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 5. | Montagnani F, Di Leonardo G, Pino M, Perboni S, Ribecco A, Fioretto L. Protracted Inhibition of Vascular Endothelial Growth Factor Signaling Improves Survival in Metastatic Colorectal Cancer: A Systematic Review. J Transl Int Med. 2017;5:18-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Malmstrøm ML, Săftoiu A, Vilmann P, Klausen TW, Gögenur I. Endoscopic ultrasound for staging of colonic cancer proximal to the rectum: A systematic review and meta-analysis. Endosc Ultrasound. 2016;5:307-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Wu D, Li JN, Qian JM. Endoscopic Diagnosis and Treatment of Precancerous Colorectal Lesions in Patients with Inflammatory Bowel Disease: How Does the Latest SCENIC International Consensus Intersect with Our Clinical Practice? J Transl Int Med. 2017;5:4-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Wang Y, Zhou Y, Hu Z. The Functions of Circulating Tumor Cells in Early Diagnosis and Surveillance During Cancer Advancement. J Transl Int Med. 2017;5:135-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Ersan V, Kutlu R, Erdem C, Karagul S, Kayaalp C. Colorectal Stenting for Obstruction due to Retrorectal Tumor in a Patient Unsuitable for Surgery. J Transl Int Med. 2017;5:186-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Altun M, Walter TS, Kramer HB, Herr P, Iphöfer A, Boström J, David Y, Komsany A, Ternette N, Navon A, Stuart DI, Ren J, Kessler BM. The human otubain2-ubiquitin structure provides insights into the cleavage specificity of poly-ubiquitin-linkages. PLoS One. 2015;10:e0115344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Hospenthal MK, Mevissen TET, Komander D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat Protoc. 2015;10:349-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 178] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 12. | Xie X, Li F, Wang Y, Wang Y, Lin Z, Cheng X, Liu J, Chen C, Pan L. Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2. Autophagy. 2015;11:1775-1789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 13. | Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM, Komander D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34:307-325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 14. | Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, Penengo L. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015;10:226-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 152] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 15. | Murdoch JD, Rostosky CM, Gowrisankaran S, Arora AS, Soukup SF, Vidal R, Capece V, Freytag S, Fischer A, Verstreken P, Bonn S, Raimundo N, Milosevic I. Endophilin-A Deficiency Induces the Foxo3a-Fbxo32 Network in the Brain and Causes Dysregulation of Autophagy and the Ubiquitin-Proteasome System. Cell Rep. 2016;17:1071-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 16. | Jain CK, Arora S, Khanna A, Gupta M, Wadhwa G, Sharma SK. The ubiquitin-proteasome pathway an emerging anticancer strategy for therapeutics: a patent analysis. Recent Pat Anticancer Drug Discov. 2015;10:201-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Ponder EL, Bogyo M. Ubiquitin-like modifiers and their deconjugating enzymes in medically important parasitic protozoa. Eukaryot Cell. 2007;6:1943-1952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Madsen L, Schulze A, Seeger M, Hartmann-Petersen R. Ubiquitin domain proteins in disease. BMC Biochem. 2007;8 Suppl 1:S1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Jentsch S, Pyrowolakis G. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 2000;10:335-342. [PubMed] |
| 20. | Gao Y, Theng SS, Zhuo J, Teo WB, Ren J, Lee CG. FAT10, an ubiquitin-like protein, confers malignant properties in non-tumorigenic and tumorigenic cells. Carcinogenesis. 2014;35:923-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Merbl Y, Refour P, Patel H, Springer M, Kirschner MW. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell. 2013;152:1160-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 22. | Ren J, Wang Y, Gao Y, Mehta SB, Lee CG. FAT10 mediates the effect of TNF-α in inducing chromosomal instability. J Cell Sci. 2011;124:3665-3675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | Cajee UF, Hull R, Ntwasa M. Modification by ubiquitin-like proteins: significance in apoptosis and autophagy pathways. Int J Mol Sci. 2012;13:11804-11831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Yang Z, Wu D, Zhou D, Jiao F, Yang W, Huan Y. Induction of anti-tumor immunity by dendritic cells transduced with FAT10 recombinant adenovirus in mice. Cell Immunol. 2015;293:17-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Schmidtke G, Aichem A, Groettrup M. FAT10ylation as a signal for proteasomal degradation. Biochim Biophys Acta. 2014;1843:97-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Theng SS, Wang W, Mah WC, Chan C, Zhuo J, Gao Y, Qin H, Lim L, Chong SS, Song J, Lee CG. Disruption of FAT10-MAD2 binding inhibits tumor progression. Proc Natl Acad Sci U S A. 2014;111:E5282-E5291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 27. | Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc Natl Acad Sci U S A. 1999;96:4313-4318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 144] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 28. | Ji F, Jin X, Jiao CH, Xu QW, Wang ZW, Chen YL. FAT10 level in human gastric cancer and its relation with mutant p53 level, lymph node metastasis and TNM staging. World J Gastroenterol. 2009;15:2228-2233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Yuan R, Wang K, Hu J, Yan C, Li M, Yu X, Liu X, Lei J, Guo W, Wu L, Hong K, Shao J. Ubiquitin-like protein FAT10 promotes the invasion and metastasis of hepatocellular carcinoma by modifying β-catenin degradation. Cancer Res. 2014;74:5287-5300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 30. | Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL, Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, Choti M, Lee LA. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22:2592-2603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 150] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 31. | Aichem A, Groettrup M. The ubiquitin-like modifier FAT10 in cancer development. Int J Biochem Cell Biol. 2016;79:451-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 32. | Gao Y, Theng SS, Mah WC, Lee CG. Silibinin down-regulates FAT10 and modulate TNF-α/IFN-γ-induced chromosomal instability and apoptosis sensitivity. Biol Open. 2015;4:961-969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Lukasiak S, Schiller C, Oehlschlaeger P, Schmidtke G, Krause P, Legler DF, Autschbach F, Schirmacher P, Breuhahn K, Groettrup M. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27:6068-6074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 129] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 34. | Basler M, Buerger S, Groettrup M. The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense. Mol Immunol. 2015;68:129-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Aichem A, Kalveram B, Spinnenhirn V, Kluge K, Catone N, Johansen T, Groettrup M. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci. 2012;125:4576-4585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 36. | Zhang DW, Jeang KT, Lee CG. p53 negatively regulates the expression of FAT10, a gene upregulated in various cancers. Oncogene. 2006;25:2318-2327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 37. | Lim CB, Zhang D, Lee CG. FAT10, a gene up-regulated in various cancers, is cell-cycle regulated. Cell Div. 2006;1:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Maclaine NJ, Hupp TR. The regulation of p53 by phosphorylation: a model for how distinct signals integrate into the p53 pathway. Aging (Albany NY). 2009;1:490-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |