Published online Aug 18, 2017. doi: 10.4254/wjh.v9.i23.1001
Peer-review started: February 17, 2017
First decision: April 20, 2017
Revised: May 31, 2017
Accepted: June 19, 2017
Article in press: June 20, 2017
Published online: August 18, 2017
Processing time: 184 Days and 14.2 Hours
To assess the role of some circulating miRNAs (miR-23a, miR-203, miR338, miR-34, and miR-16) as tumor markers for diagnosis of hepatocellular carcinoma (HCC).
One hundred and seventy-one subjects were enrolled, 57 patients with HCC, 57 patients with liver cirrhosis (LC) and 57 healthy subjects as control group. Severity of liver disease was assessed by Child Pugh score. Tumor staging was done using Okuda staging system. Quantification of Micro RNA (miR-23a, miR-203, miR338, miR-34, and miR-16) was performed.
All studied miRNA showed significant difference between HCC and cirrhotic patients in comparison to healthy control. miR-23a showed statistically significant difference between HCC and cirrhotic patients being higher in HCC group than cirrhotic. miR-23a is significantly higher in HCC patients with focal lesion size equal or more than 5 cm, patients with multiple focal lesions and Okuda stage III. At cutoff value ≥ 210, miR-23a showed accuracy 79.3% to diagnose HCC patients with sensitivity 89.47% and specificity about 64.91%. At cut off level ≥ 200 ng/mL, serum alpha fetoprotein had 73.68% sensitivity, 52.63% specificity, 43.75% PPV, 80% NPV for diagnosis of HCC.
MicroRNA 23a can be used as a screening test for early detection of HCC. Also, it is related to larger size of tumour, late Okuda staging and multiple hepatic focal lesions, so it might be a prognostic biomarker.
Core tip: MicroRNA is promising as diagnostic and prognostic biomarkers. miR-23a can be used in screening of hepatocellular carcinoma (HCC) and it gives better results than alpha fetoprotein. It is also related to more progressive HCC so it can be used as predictor of prognosis.
- Citation: Mohamed AA, Ali-Eldin ZA, Elbedewy TA, El-Serafy M, Ali-Eldin FA, AbdelAziz H. MicroRNAs and clinical implications in hepatocellular carcinoma. World J Hepatol 2017; 9(23): 1001-1007
- URL: https://www.wjgnet.com/1948-5182/full/v9/i23/1001.htm
- DOI: https://dx.doi.org/10.4254/wjh.v9.i23.1001
MicroRNAs (miRNA) are small, non-coding RNAs that negatively regulate gene expression at the post-transcriptional level[1]. miRNA is known to regulate the cell cycle, apoptosis and metastasis[2]. Aberrant miRNA expression contributes to tumorigenesis and cancer progression[3]. miRNA is involved in various biological processes that underlie hepatic tumor formation[1]. Murakami et al[4] was the first to report that hepatic cmalignancy exhibited an abnormal expression pattern of miRNAs as the dysregulation of miRNA expression has been identified as a common characteristic of liver cancer. Later on, a number of studies have confirmed that miRNAs possess important regulatory roles in hepatocarcinogenesis and malignant transformation[5].
Circulating miRNAs that are released from cancerous tissues are stable and readily available for clinical analysis, and therefore may be useful for the first-line detection of cancer[6]. Studies concerning miRNAs appear to show a novel perspective for cancer diagnosis and treatment[2].
Hepatocellular carcinoma (HCC) is characterized by significant morbidity and high mortality rates worlwide[7]. Because of the difficulty of clinical diagnosis at the early stage, only 30%-40% of cases can undergo curative resection[8]. As there are currently no reliable tumor markers or imaging technologies that can accurately diagnose early HCC, the use of circulating miRNAs as a potential tool for HCC detection has become an emerging area of study[6,9]. Many circulating microRNAs were evaluated in liver diseases including miR-122, miR-21, miR-34a, miR-221, miR-23a, miR-216, miR-155, miR-186, miR-150, miR-130b, and miR-214[10]. Our aim is to detect the possibility of using some circulating miRNAs as tumor markers for diagnosis of HCC namely miR-23a, miR-203, miR-338-3p, miR-34, and miR-16.
This prospective cross sectional study included 171 subjects divided into 3 groups: Group I comprising 57 patients with HCC, Group II comprising 57 patients with liver cirrhosis, Group III 57 healthy subjects as a control group.
Informed written consent was obtained from all participants prior to enrollment in the study and approved by ethical committee of Faculty of Medicine, Tanta University. Patients with other cancers or metastatic liver cancer were excluded. All patients were submitted to detailed history and clinical assessment. Liver cirrhosis was diagnosed on the basis of history, clinical examination, laboratory findings, and abdominal ultrasonography. Severity of liver disease was assessed by Child Pugh score[11]. HCC was diagnosed by abdominal ultrasonography, abdominal triphasic computed tomography and serum Alpha fetoprotein (AFP). Tumor characteristics were detected including tumor size, focal lesion number, site, and portal vein invasion. Tumor staging was done using Okuda staging system[12].
Fasting venous blood samples (5 mL) were collected by trained laboratory technicians. Serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), albumin, total bilirubin levels and creatinine were measured by using SynchronCX4 clinical system. Serumα-fetoprotein levels and viral status (HCV-Ab and HBs Ag) were estimated by serological techniques (Axyam System, Abbott Laboratories). Prothrombin time measurements were performed for all patients; normal time was 12 s [100% concentration and International normalization ratio (INR) of 1]. Complete blood count was done using Automatic blood cell counter model PCE-210N (ERMA INC).
Total RNA was isolated according to the instructions of the supplier and was further purified using an RN easy mini kit (Qiagen, Valencia, CA, United States).
Quantification of Micro RNA was performed using Taq Man Gene Expression (Applied BiosystemsInc, Foster City, CA, United States). RNAU6 was used as house-keeping gene (endogenous reference cDNA) for all micro RNA in this study. Fractional threshold cycles (CT) were expressing the initial concentration of target sequence. Relative mRNA quantification was calculated using the arithmetic formula 2)-ΔCT, where ΔCT is the difference between the CT of a given target cDNA and an endogenous reference cDNA. Thus, this value yields the amount of the target normalized to an endogenous reference.
The collected data were tabulated and analyzed using SPSS version 23 software (SPSS Inc, Chicago, ILL Company). Categorical data were presented as number and percentages while quantitative data were expressed as mean, standard deviation and median. Comparison of continuous data between two groups was made by using unpaired t test for parametric data and Mann-Whitney test for non-parametric data. Comparison of continuous data between more than two groups was made by using one way ANOVA for parametric data and Kruskal-Wallis test for nonparametric data. χ2 test was used for comparison between categorical data. Receiving operating characteristic (ROC) analysis curves and the corresponding area under the curve were calculated for providing the accuracy of the microRNAs and AFP, in diagnosis of HCC. ROC curve was used for estimation of sensitivity (i.e., true positive rate), specificity (i.e., true negative rate), positive predictive value (PPV), negative predictive value (NPV) and cutoff values showing the best equilibrium between sensitivity and specificity were evaluated. The accepted level of significance in this work was stated at 0.05 (P < 0.05 was considered significant).
The demographic data and Child-Pugh scoring of the studied groups are shown in Table 1. Symptoms were elicited by 85.96% of the HCC. Out of the recruited patients, 73.68%, 14.04% and 12.28% were HCV patients, HBV patients and non-HCV non-HBV respectively. Regarding Okuda staging system, 12.28% of HCC patients presented in stage I, 40.35% of HCC patients presented in stage II and 47.37% of HCC patients presented in stage III. Imaging showed that all HCC occurred on top of cirrhosis, ascites was present in 82.46% of the HCC patients and portal vein thrombosis was found in 17.54%. Focal lesions were single in 56.14% of cases, affected the right lobe in 56.14% of cases and their size ranged from 3.2 to 14 cm with a mean of 7.61 ± 3.037 as shown in Table 2. Comparison between all studied groups as regard liver functions tests, and other investigations are shown in Table 3.
| Variable(s) | Group I | Group II | Group III | P value |
| HCC group (n = 57) | Cirrhotic group (n = 57) | Control group (n = 57) | ||
| Gender | ||||
| Male | 37 (64.91) | 39 (68.42) | 40 (70.18) | 0.8289 |
| Female | 20 (35.09) | 18 (31.58) | 17 (29.82) | |
| Age (mean ± SD) (yr) | 55.9 ± 5.194 | 54.88 ± 9.907 | 54.3 ± 6.34 | 0.5087 |
| Child Pugh classification | ||||
| A | 10 (17.54) | 15 (26.32) | - | 0.527 |
| B | 18 (31.58) | 16 (28.07) | - | |
| C | 29 (50.88) | 26 (45.61) | - |
| Variables | n (%) | |
| Size (mean ± SD) (range) cm | 7.61 ± 3.037 (3.2-14) | |
| Portal veins thrombosis | Yes | 10 (17.54) |
| No | 47 (82.46) | |
| No. of focal lesions | Single | 32 (56.14) |
| Multiple | 25 (43.86) | |
| Site of focal lesions | Right lobe | 32 (56.14) |
| Left lobe | 17 (29.82) | |
| Both lobes | 8 (14.04) | |
| Okuda stage | I | 7 (12.28) |
| II | 23 (40.35) | |
| III | 27 (47.37) |
| Variable(s) | Group I | Group II | Group III | P | P1 | P2 | P3 |
| HCC group (n = 57) | Cirrhotic group (n = 57) | Control group (n = 57) | |||||
| mean ± SD | mean ± SD | mean ± SD | |||||
| ALT (U/L) | 60.75 ± 32.63 | 59.04 ± 68.74 | 30.18 ± 5.48 | < 0.0001 | < 0.05 | < 0.001 | < 0.001 |
| AST (U/L) | 86.7 ± 35.1 | 66.77± 32.07 | 32.79 ± 7.2 | < 0.0001 | < 0.01 | < 0.001 | < 0.001 |
| Total bilirubin (mg/dL) | 4.48 ± 4.7 | 5.2 ± 5.59 | 0.77 ± 0.18 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| Serum albumin (g/dL) | 2.54 ± 0.38 | 2.72 ± 0.53 | 4.05 ± 0.47 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| INR | 1.48 ± 0.3 | 1.54 ± 0.72 | 0.99 ± 0.07 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| Serum α-feto protein (ng/mL) | 1418.55 ± 2953.2 | 41.61 ± 15.78 | 5.8 ± 1.65 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| Serum creatinine (mg/dL) | 2.2 ± 1.77 | 1.64 ± 1.23 | 0.95 ± 0.16 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| Hemoglobin (g/dL) | 9.72 ± 1.22 | 10.02 ± 0.89 | 12.62 ± 1.1 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| Platelet (× 109/L) | 98.33 ± 30.83 | 102.32 ± 33.24 | 220.93 ± 53.14 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| Total leucocytic count (× 109/L) | 3.17 ± 0.47 | 3.39 ± 0.50 | 6.83 ± 2 | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
Regarding miRNA values, the tested miR-23a, miR-203, miR-338, miR-34, and miR-16 showed a statistically significant difference between patients group I and II vs group III. It was found that 23a, 34 and 16 microRNAs were significantly higher in HCC group and cirrhotic group when compared with the control group; but 203 and 338 microRNAs were significantly lower in HCC group and cirrhotic group when compared with the control group. But only miRNA 23a showed statistically significant difference between group I and II, being higher in the HCC group than the cirrhotic group as shown in Table 4.
| Variable(s) | Group I | Group II | Group III | P | P1 | P2 | P3 |
| HCC group (n = 57) | Cirrhotic group (n = 57) | Control group (n = 57) | |||||
| Median (range) | Median (range) | Median (range) | |||||
| MicroRNA 23a | 214 (21-218) | 211 (21-218) | 26 (20-219) | < 0.0001 | < 0.05 | < 0.001 | < 0.001 |
| MicroRNA 34 | 214 (22-220) | 211 (24-218) | 25 (20-219) | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| MicroRNA 203 | 26 (21-219) | 26 (20-219) | 214 (21-225) | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| MicroRNA 338 | 26 (21-219) | 26 (21-219) | 212 (29-217) | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
| MicroRNA 16 | 214 (21-225) | 213 (24-218) | 25 (20-219) | < 0.0001 | > 0.05 | < 0.001 | < 0.001 |
When miRNAs were studied according to the focal lesion characteristics, it was found that 23a microRNA levels were higher in patients with focal lesion size equal to or more than 5 cm, in patients with multiple focal lesions; and in Okuda stage III as shown in Table 5.
| MicroRNA 16 | MicroRNA 338 | MicroRNA 203 | MicroRNA 34 | MicroRNA 23a | n | Variable(s) | |||||||||||
| P-value | Range | Median | P-value | Range | Median | P-value | Range | Median | P-value | Range | Median | P-value | Range | Median | |||
| 0.3910 | 24-217 | 212 | 0.3890 | 24-219 | 27 | 0.2571 | 22-217 | 24 | 0.0558 | 24-215 | 213 | 0.0008a | 21-213 | 211 | 11 | Less than 5 cm | Size of focal lesions |
| 21-225 | 214 | 21-215 | 25 | 21-219 | 26 | 22-220 | 215 | 23-2118 | 214 | 46 | Equal or more 5 cm | ||||||
| 0.3026 | 23-221 | 214 | 0.5352 | 21-219 | 26 | 0.5040 | 22-219 | 26 | 0.8975 | 24-220 | 214 | 0.0001a | 21-218 | 211 | 32 | Single | No. of focal lesions |
| 21-225 | 214 | 22-215 | 26 | 21-29 | 26 | 22-2119 | 215 | 214-218 | 215 | 25 | multiple | ||||||
| 0.9581 | 29-218 | 214 | 0.9247 | 22-29 | 26 | 0.7767 | 23-29 | 26 | 0.8255 | 22-219 | 214 | 0.0795 | 210-217 | 215 | 10 | Present | Portal vein thrombosis |
| 21-225 | 214 | 21-219 | 26 | 21-219 | 26 | 24-220 | 214 | 21-218 | 213 | 47 | Absent | ||||||
| 0.8881 | 22-225 | 214 | 0.8295 | 21-219 | 26 | 0.0807 | 21-217 | 27 | 0.3367 | 22-220 | 215 | 0.9736 | 21-218 | 214 | 36 | Less than 200 ng/mL | AFP level |
| 21-221 | 214 | 21-215 | 26 | 21-219 | 25 | 27-219 | 214 | 23-217 | 214 | 21 | Equal or more 200 ng/mL | ||||||
| 0.9347 | 25-217 | 214 | 0.1145 | 24-29 | 27 | 0.4488 | 23-217 | 26 | 0.4396 | 24-215 | 214 | 0.0001a | 24-213 | 211 | 7 | Stage I | Okuda stage |
| 23-221 | 214 | 21-219 | 24 | 22-219 | 27 | 29-220 | 214 | 21-218 | 211 | 23 | Stage II | ||||||
| 21-225 | 214 | 22-215 | 27 | 21-29 | 25 | 22-219 | 214 | 211-218 | 215 | 27 | Stage III | ||||||
| 0.1603 | 23-225 | 214 | 0.4405 | 21-219 | 26 | 0.4007 | 21-219 | 27 | 0.7827 | 24-220 | 214 | 0.5433 | 21-218 | 214 | 42 | HCV | Etiology of liver cirrhosis |
| 21-215 | 211 | 21-29 | 26 | 24-215 | 25 | 210-217 | 214 | 23-215 | 214 | 8 | HBV | ||||||
| 25-216 | 214 | 24-29 | 27 | 23-26 | 24 | 22-219 | 215 | 210-217 | 214 | 7 | None | ||||||
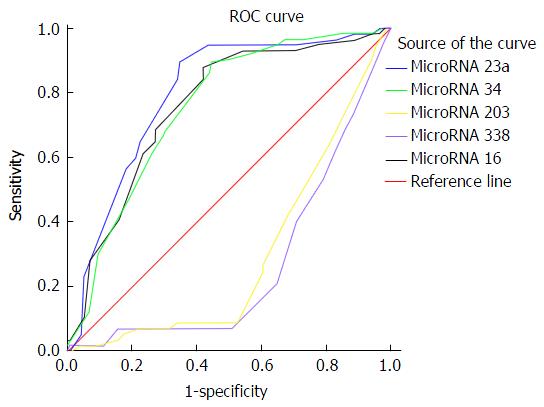
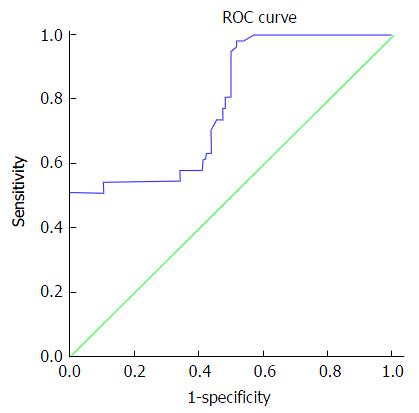
At a cut off level of 200 ng/mL, serum AFP in the studied patients had 73.68% sensitivity, 52.63% specificity, 43.75% PPV, and 80% NPV for the diagnosis of HCC. At a cut off level of 210ct, serum microRNA 23a had 89.47% sensitivity, 64.91% specificity, 56.04% PPV, and 92.5% NPV for diagnosis of HCC as shown in Table 6 and Figures 1 and 2.
| Variable(s) | Cutoff value | Sensitivity% | Specificity% | Positive predictive value (PPV%) | Negative predictive value (NPV%) | Accuracy |
| MicroRNA 23a | ≥ 210 | 89.47% | 64.91% | 56.04% | 92.5% | 79.3% |
| MicroRNA 34 | ≥ 210 | 89.47% | 55.26% | 50% | 91.3% | 79.3% |
| MicroRNA 203 | ≥ 24 | 80.7% | 10.53% | 31.08% | 52.17% | 29.6% |
| MicroRNA 338 | ≥ 25 | 63.16% | 16.67% | 27.48% | 47.5% | 26.4% |
| MicroRNA 16 | ≥ 210 | 87.72% | 57.89% | 51.02% | 90.4% | 75.7% |
| α-feto protein | ≥ 200 | 73.68% | 52.63% | 43.75% | 80% | 78.5% |
microRNA is differentially expressed in development of different types of malignancies, including hepatic malignancy[13], which suggests that microRNAs may have a role in carcinogenesis as new oncogenes or tumor-suppressor genes. The presence of microRNAs in serum was first reported in 2008 in cases with large B cell lymphoma[14]. They could be a potential biomarker for diagnosis of tumors[15]. The present study was designed to evaluate the role of some microRNA (23a, 34, 203, 338 and 16) in early diagnosis of HCC. To fulfill this aim 57 HCC patients, 57 patients with liver cirrhosis and 57 healthy controls were enrolled.
In the present work, serum microRNA 23a level was significantly higher in the HCC group in comparison to other groups. Also, it is significantly higher in liver cirrhosis group in comparison to healthy controls. A similar result was obtained by Li et al[16]. They reported up regulation of microRNA 23a in HCC patients in comparison to liver cirrhosis patients and healthy control. This up-regulation in liver cirrhosis than healthy controls and further up-regulation in HCC patients in comparison to viral hepatitis patients suggests a role of microRNA 23a in the pathogenesis of HCC. A study on resected human HCC tissues found that microRNA-23a down-regulates the expression of interferon regulatory factor-1 in HCC cells[17].
Serum microRNA 34 and microRNA 203 levels were similarly elevated in HCC and liver cirrhosis groups. Many authors found that microRNA 34 expression is increased in hepatic fibrosis[18], HCV infection[19], alcoholic liver disease[20], NAFLD[21,22] and HCC tissues[23-25].
On the contrary, studies on microRNA 203 in HCC tissues found that microRNA 203 is down-regulated in HCC tissue[26,27]. Moreover, studies correlate this down-regulation with recurrence of HCC in liver transplantation[26] and bad prognosis in HCC patients[27]. None of these studies was done on serum; they assessed tissue level of HCC in resected HCC tissues.
Serum microRNA 338 and microRNA 16levels are similarly reduced in HCC patients and liver cirrhosis patients. In line with these findings, many studies concluded reduced tissue expression of miR-338-3p in different types of cancers[28-30]. Studies on HCC showed that miR-338-3pmiR-338-3p was significantly down-regulated in HCC tissues and cell lines compared to the corresponding matched adjacent normal tissues[31,32].
On the contrary, other researchers found that circulating microRNA 338 level increased in HCC patients than liver cirrhosis and controls[33] but their study has a limitation of small sample size (37 HCC patients, 29 cirrhosis patients, and 31 healthy controls).
Serum microRNA 23a level at cutoff value ≥ 210 showed accuracy of 79.3% to differentiate HCC patients from cirrhotic patients and healthy control with high sensitivity about 90%, specificity about 65%, PPV 56% and NPV 92.9. These values are better than those elicited by alpha fetoprotein. The later at a cut off level of 200 ng/mL, had 73.68% sensitivity, 52.63% specificity, 43.75% PPV, and 80% NPV for the diagnosis of HCC. So, serum microRNA 23a can be used as a screening test to diagnose HCC as it showed high sensitivity. To the best of our knowledge no previous studies elicited such finding. A single study that used combination of 13 microRNA including 23a found that HCC on top of chronic HBV infection could be differentiated from chronic HCV infection and healthy control[16]. MicroRNA 23a levels were significantly higher in patients with focal lesion 5 cm or more in size, patients with multiple focal lesions; and Okuda stage III when compared with patients with less advanced HCC disease. Thus it could be used as a prognostic biomarker.
Other studied microRNA factors showed insignificant difference between HCC and liver cirrhosis patients, so they cannot be used as diagnostic markers of HCC. In conclusion, microRNA 23a can be used as a screening test for early detection of HCC. Also, it is related to larger size of tumour, late Okuda staging and multiple hepatic focal lesions, so it might be a prognostic biomarker. Validation study on large scale is needed to confirm these results.
MicroRNAs are small, non-coding RNAs that negatively regulate gene expression at the post transcriptional level including cell cycle, apoptosis and metastasis. Aberrant miRNA expression contributes to tumorigenesis and cancer progression. Number of studies have confirmed that miRNAs possess important regulatory roles in hepatocarcinogenesis and malignant transformation. Hepatocellular carcinoma (HCC) is characterized by significant morbidity and high mortality rates. Because of the difficulty of early clinical diagnosis, only 30%-40% of cases can undergo curative resection. Circulating miRNAs released from cancerous tissues are stable and readily available for clinical analysis and appear to show a novel perspective for cancer diagnosis.
Many microRNAs have been studied as biomarkers for diagnosis of malignancies. Yet, role of microRNA in early diagnosis of HCC is not confirmed.
This work demonstrates that miR-23a can be used in screening of liver cancer and it gives better results than alpha fetoprotein. This work showed first demonstration that microRNA 23a could be used as a promising biomarker for HCC patients, even though large scale examination is required. This manuscript would provide important clues for the development of microRNA as biomarkers for HCC.
MicroRNA 23a can be used as a screening test for early detection of HCC. Also, it is related to larger size of tumour, late Okuda staging and multiple hepatic focal lesions, so it might be a prognostic biomarker. Validation study on large scale is needed to confirm these results.
This manuscript would provide important clues for the development of microRNA as biomarkers for HCC.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Yu DY S- Editor: Ji FF L- Editor: A E- Editor: Li D
| 1. | Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX, Yuan Y, Zhuang SM. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene. 2013;32:3071-3079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 2. | Gong J, He XX, Tian A. Emerging role of microRNA in hepatocellular carcinoma (Review). Oncol Lett. 2015;9:1027-1033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 3. | Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 4. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 876] [Cited by in RCA: 896] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 5. | Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472-480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 435] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 6. | Qi J, Wang J, Katayama H, Sen S, Liu SM. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135-142. [PubMed] |
| 7. | World Health Organization. GLOBOCAN Estimated Cancer Incidence, Mortality and Prevalence Worldwide, 2012. |
| 8. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 498] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 9. | Chang-Hao Tsao S, Behren A, Cebon J, Christophi C. The role of circulating microRNA in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2015;20:78-104. [PubMed] |
| 10. | Callegari E, Elamin BK, Sabbioni S, Gramantieri L, Negrini M. Role of microRNAs in hepatocellular carcinoma: a clinical perspective. Onco Targets Ther. 2013;6:1167-1178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60:646-649. [PubMed] |
| 12. | Okuda K, Ohtsuki T, Obata H, Tomimatsu M, Okazaki N, Hasegawa H, Nakajima Y, Ohnishi K. Natural history of hepatocellular carcinoma and prognosis in relation to treatment. Study of 850 patients. Cancer. 1985;56:918-928. [PubMed] |
| 13. | Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586-591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 726] [Cited by in RCA: 750] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 14. | Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1258] [Cited by in RCA: 1333] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 15. | Huang FY, Wong DK, Seto WK, Lai CL, Yuen MF. Estradiol induces apoptosis via activation of miRNA-23a and p53: implication for gender difference in liver cancer development. Oncotarget. 2015;6:34941-34952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 16. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 17. | Yan Y, Liang Z, Du Q, Yang M, Geller DA. MicroRNA-23a downregulates the expression of interferon regulatory factor-1 in hepatocellular carcinoma cells. Oncol Rep. 2016;36:633-640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Li WQ, Chen C, Xu MD, Guo J, Li YM, Xia QM, Liu HM, He J, Yu HY, Zhu L. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J. 2011;278:1522-1532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 19. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 467] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 20. | Meng F, Glaser SS, Francis H, Yang F, Han Y, Stokes A, Staloch D, McCarra J, Liu J, Venter J. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol. 2012;181:804-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 21. | Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810-1820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 551] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 22. | Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 257] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 23. | Bernardi C, Soffientini U, Piacente F, Tonetti MG. Effects of microRNAs on fucosyltransferase 8 (FUT8) expression in hepatocarcinoma cells. PLoS One. 2013;8:e76540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Chiu LY, Kishnani PS, Chuang TP, Tang CY, Liu CY, Bali D, Koeberl D, Austin S, Boyette K, Weinstein DA. Identification of differentially expressed microRNAs in human hepatocellular adenoma associated with type I glycogen storage disease: a potential utility as biomarkers. J Gastroenterol. 2014;49:1274-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One. 2013;8:e61054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 26. | Chen HY, Han ZB, Fan JW, Xia J, Wu JY, Qiu GQ, Tang HM, Peng ZH. miR-203 expression predicts outcome after liver transplantation for hepatocellular carcinoma in cirrhotic liver. Med Oncol. 2012;29:1859-1865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | Liu Y, Ren F, Rong M, Luo Y, Dang Y, Chen G. Association between underexpression of microrna-203 and clinicopathological significance in hepatocellular carcinoma tissues. Cancer Cell Int. 2015;15:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 28. | Guo B, Liu L, Yao J, Ma R, Chang D, Li Z, Song T, Huang C. miR-338-3p suppresses gastric cancer progression through a PTEN-AKT axis by targeting P-REX2a. Mol Cancer Res. 2014;12:313-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 29. | Li P, Chen X, Su L, Li C, Zhi Q, Yu B, Sheng H, Wang J, Feng R, Cai Q. Epigenetic silencing of miR-338-3p contributes to tumorigenicity in gastric cancer by targeting SSX2IP. PLoS One. 2013;8:e66782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 30. | Chen X, Pan M, Han L, Lu H, Hao X, Dong Q. miR-338-3p suppresses neuroblastoma proliferation, invasion and migration through targeting PREX2a. FEBS Lett. 2013;587:3729-3737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 31. | Wang G, Sun Y, He Y, Ji C, Hu B, Sun Y. MicroRNA-338-3p inhibits cell proliferation in hepatocellular carcinoma by target forkhead box P4 (FOXP4). Int J Clin Exp Pathol. 2015;8:337-344. [PubMed] |
| 32. | Chen JS, Liang LL, Xu HX, Chen F, Shen SL, Chen W, Chen LZ, Su Q, Zhang LJ, Bi J. miR-338-3p inhibits epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma cells. Oncotarget. 2016; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Chen Y, Chen J, Liu Y, Li S, Huang P. Plasma miR-15b-5p, miR-338-5p, and miR-764 as Biomarkers for Hepatocellular Carcinoma. Med Sci Monit. 2015;21:1864-1871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |