Published online Mar 27, 2024. doi: 10.4254/wjh.v16.i3.439
Peer-review started: November 8, 2023
First decision: November 30, 2023
Revised: December 22, 2023
Accepted: February 18, 2024
Article in press: February 18, 2024
Published online: March 27, 2024
Processing time: 140 Days and 4.8 Hours
Sterol O-acyltransferase 1 (SOAT1) is an important target in the diagnosis and treatment of liver cancer. However, the prognostic value of SOAT1 in patients with hepatocellular carcinoma (HCC) is still not clear.
To investigate the correlation of SOAT1 expression with HCC, using RNA-seq and gene expression data of The Cancer Genome Atlas (TCGA)-liver hepatocellular carcinoma (LIHC) and pan-cancer.
The correlation between SOAT1 expression and HCC was analyzed. Cox hazard regression models were conducted to investigate the prognostic value of SOAT1 in HCC. Overall survival and disease-specific survival were explored based on TCGA-LIHC data. Biological processes and functional pathways mediated by SOAT1 were characterized by gene ontology (GO) analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed genes. In addition, the protein-protein interaction network and co-expression analyses of SOAT1 in HCC were performed to better understand the regulatory mechanisms of SOAT1 in this malignancy.
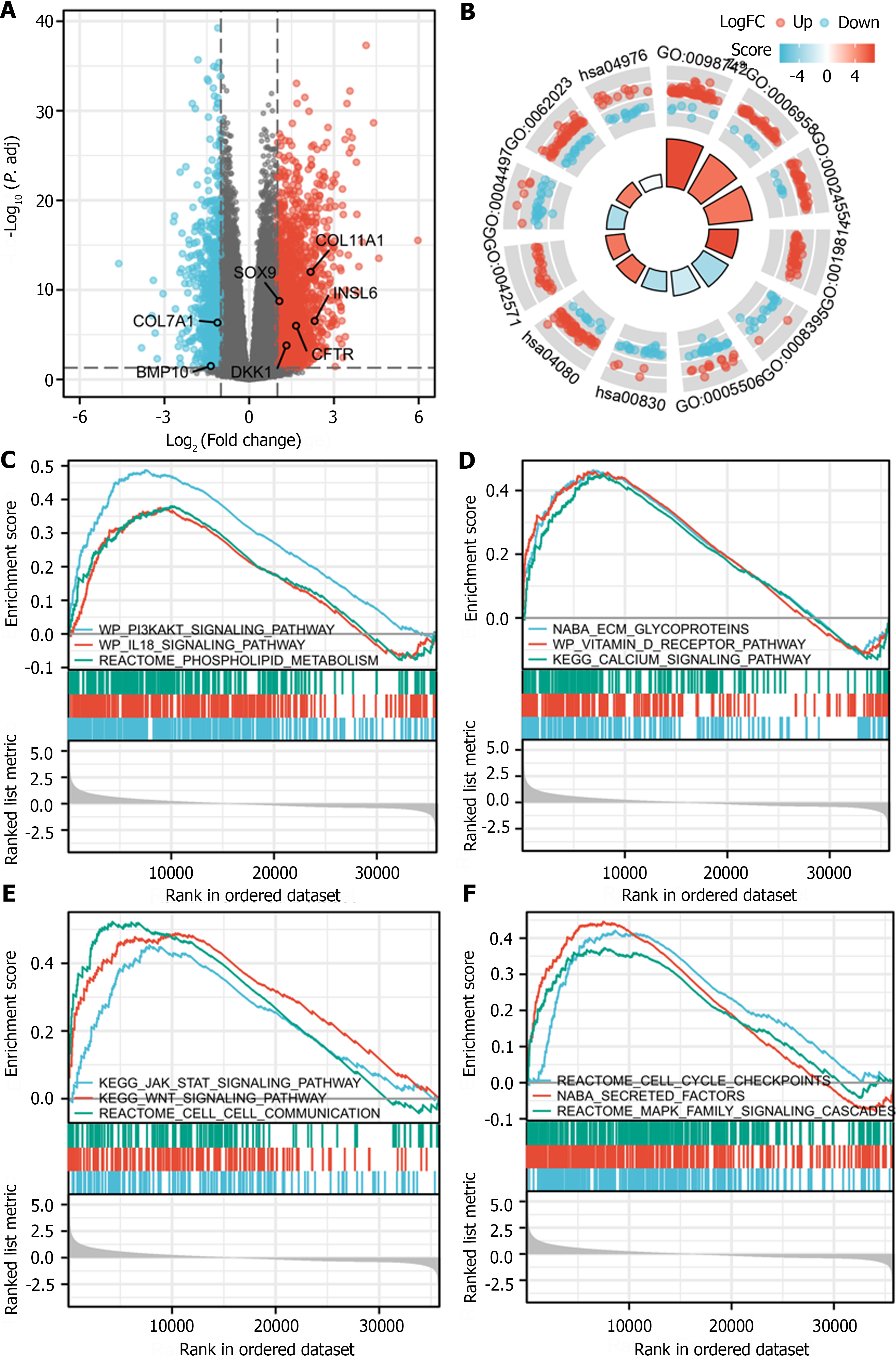
SOAT1 and SOAT2 were highly expressed in unpaired samples, while only SOAT1 was highly expressed in paired samples. The area under the receiver operating characteristic curve of SOAT1 expression in tumor samples from LIHC patients compared with para-carcinoma tissues was 0.748, while the area under the curve of SOAT1 expression in tumor samples from LIHC patients compared with GTEx was 0.676. Patients with higher SOAT1 expression had lower survival rates. Results from GO/KEGG and gene set enrichment analyses suggested that the PI3K/AKT signaling pathway, the IL-18 signaling pathway, the calcium signaling pathway, secreted factors, the Wnt signaling pathway, the Jak/STAT signaling pathway, the MAPK family signaling pathway, and cell–cell communication were involved in such association. SOAT1 expression was positively associated with the abundance of macrophages, Th2 cells, T helper cells, CD56bright natural killer cells, and Th1 cells, and negatively linked to the abundance of Th17 cells, dendritic cells, and cytotoxic cells.
Our findings demonstrate that SOAT1 may serve as a novel target for HCC treatment, which is helpful for the development of new strategies for immunotherapy and metabolic therapy.
Core Tip: As patients would greatly benefit from early detection of hepatocellular carcinoma, the complementary study of hepatocellular carcinoma-associated proteins in serum samples using state-of-the-art proteomics would be a very attractive direction for future exploration.
- Citation: Gan CJ, Zheng Y, Yang B, Cao LM. Comprehensive prognostic and immune analysis of sterol O-acyltransferase 1 in patients with hepatocellular carcinoma. World J Hepatol 2024; 16(3): 439-451
- URL: https://www.wjgnet.com/1948-5182/full/v16/i3/439.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i3.439
Liver cancer is one of the leading causes of death worldwide. Hepatocellular carcinoma (HCC) is the most devastating type of liver cancer[1], commonly diagnosed at an advanced stage, with a high rate of mortality and aggressive clinical course. The well-known risk factors for HCC include age, sex, alcohol consumption/abuse, environmental toxins, aflatoxin exposure, chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, and non-alcoholic fatty liver disease[2].
Liver transplantation, radical surgical resection, and radiofrequency ablation are commonly used in early-stage HCC. However, the majority of patients do not meet the criteria for radical treatment and are treated with systemic or local treatment instead[3]. Advanced HCC always presents a poor prognosis, although several new treatment modalities, such as immunotherapy and trans-arterial chemoembolization plus systemic treatments, have been proposed[4-6]. Therefore, exploring effective therapeutic targets for HCC is of great importance to both individuals and society.
Sterol O-acyltransferase (SOAT), known as acyl-CoA:cholesterol acyltransferase (ACAT), is located in the endoplasmic reticulum membrane. It plays an important role in cholesterol homeostasis and bile acid biosynthesis by catalyzing the conversion of cholesterol to cholesterol esters[7]. There are two SOAT isoforms in mammals, namely, SOAT1 and SOAT2. SOAT1 is a key enzyme with high expression levels. It is generally expressed in all tissues except the intestine and plays an important role by converting endoplasmic reticulum cholesterol into lipid droplet (LD) stored esters[8,9]. High SOAT1 expression has been shown in several tumor types (such as liver cancer, pancreatic cancer, and prostate cancer[10,11]) and associated with diagnosis and treatment[12-14]. Up-regulation of SOAT1 could further increase the expression levels of inflammatory factors and cause cardiovascular diseases such as atherosclerosis and coronary heart disease[15-17]. Cholesterol ester increases HCC growth by promoting the synthesis of phospholipids and hormones[18-21]. Proteomic evidence from early-stage HBV-HCC patients showed that HCC patients with more aggressive tumors and poor prognosis had disrupted cholesterol metabolism and increased SOAT1 expression[19]. The single nucleotide polymorphisms of SOAT1 have been closely related to cholesterol metabolism[22,23].
However, the relationship of SOAT1 expression with HCC remains unclear. In the current study, we explored whether SOAT1 is involved in the development of HCC, as well as the regulatory mechanisms of SOAT1[17]. Moreover, we further explored various biological processes and signaling pathways via which SOAT1 may potentially be involved in the pathogenesis of HCC.
The RNA-seq and gene expression data of The Cancer Genome Atlas (TCGA)-liver hepatocellular carcinoma (LIHC) and pan-cancer, including unpaired samples and paired samples, were extracted, filtered to remove missing and duplicated results, and transformed by log2 (TPM + 1) using the Xiantao tool (www.xiantao.love). SOAT1 gene expression was also analyzed using Clinical Proteomic Tumor Analysis Consortium samples. P < 0.05 was regarded as significant.
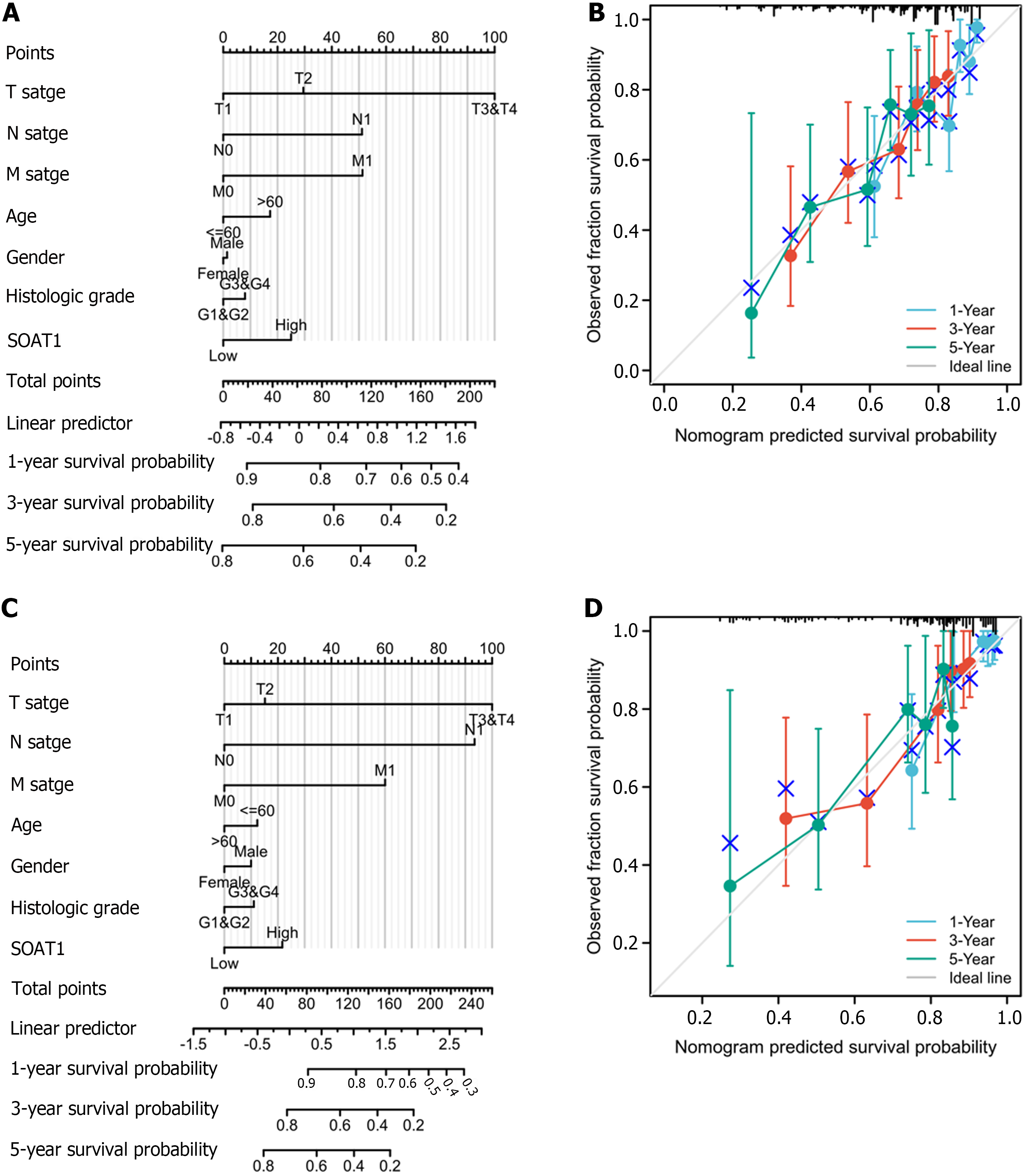
To investigate the prognostic value of SOAT1 expression, Cox proportional hazard regression models were generated to describe patients’ characteristics, including SOAT1 and SOAT2 expression levels and TNM stages. Overall survival (OS) and disease-specific survival (DSS) were also explored based on TCGA-LIHC data. P value < 0.05 was regarded as significant. To further investigate the prognostic value of SOAT1 expression, a nomogram and calibration curves were generated.
Receiver operation characteristic curve analysis was conducted to explore the diagnostic value of SOAT1 expression in TCGA-LIHC with and without GTEx and the area under the receiver operating characteristic curve (AUC) was calculated using the “pROC” package.
To validate the potential effects of SOAT1 expression on TCGA-LIHC progression, SOAT1 expression was determined in subgroups based on age, sex, and tumor stage. The RNA-seq data and related clinical data in level 3 HTSeq-fragments per kilobase per million mapped fragments formats were downloaded from the TCGA database, converted to transcripts per million formats, and then analyzed after log transformation. P value < 0.05 was considered as the cutoff criterion.
To analyze the relationship between SOAT1 expression and immune cells, single sample gene set enrichment analysis (GSEA) (the “GSVA” package in R) was performed, providing a critical assessment and integration of 24 immune cells for RNA-seq samples from TCGA-LIHC.
The differentially expressed genes (DEGs) between groups with different SOAT1 expression (cut-off value: 50%) in TCGA-LIHC were identified. Utilizing Limma, log2 (fold change) > 2 and P value < 0.05 were applied as the cut-off criteria.
Gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were conducted to investigate the DEGs between the high and low SOAT1 expression groups in TCGA-LIHC. GSEA was conducted utilizing the “clusterProfiler” package in R. P value < 0.05 was applied as the cut-off criterion.
To investigate the proteins that interact with SOAT1, the STRING database (https://string-db.org) was analyzed with a combined score of > 0.4. The nodes were analyzed with Cytoscape version 3.7.1. Protein–protein interaction (PPI) network analysis was conducted to obtain the hub genes using the Cytoscape plug-in MCODE.
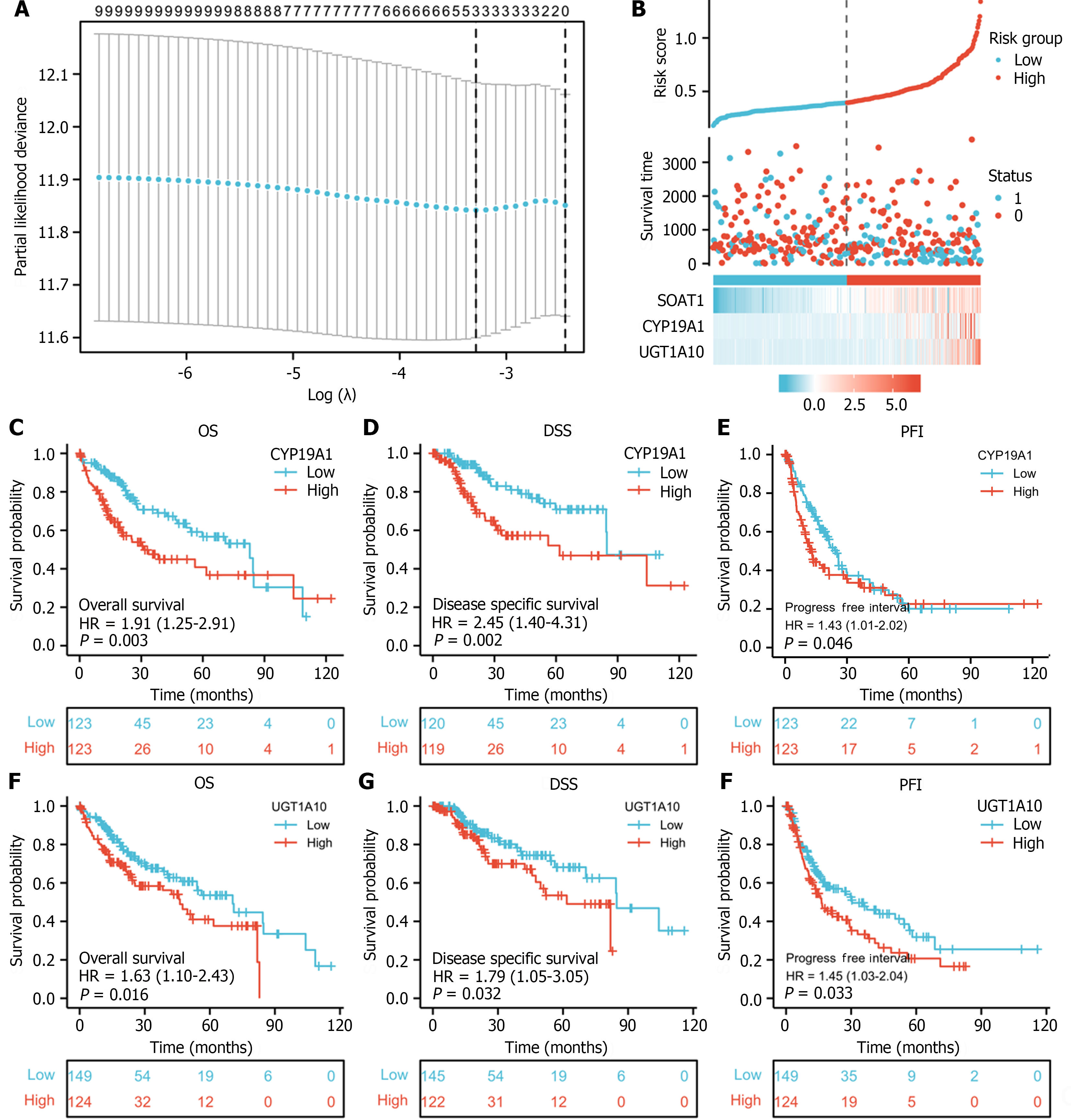
Lasso regression and risk score analysis were performed to investigate the association between SOAT1 expression, hub genes, and patient status. The association between survival and hub genes was analyzed to further show the prognostic value of SOAT1 expression in TCGA-LIHC.
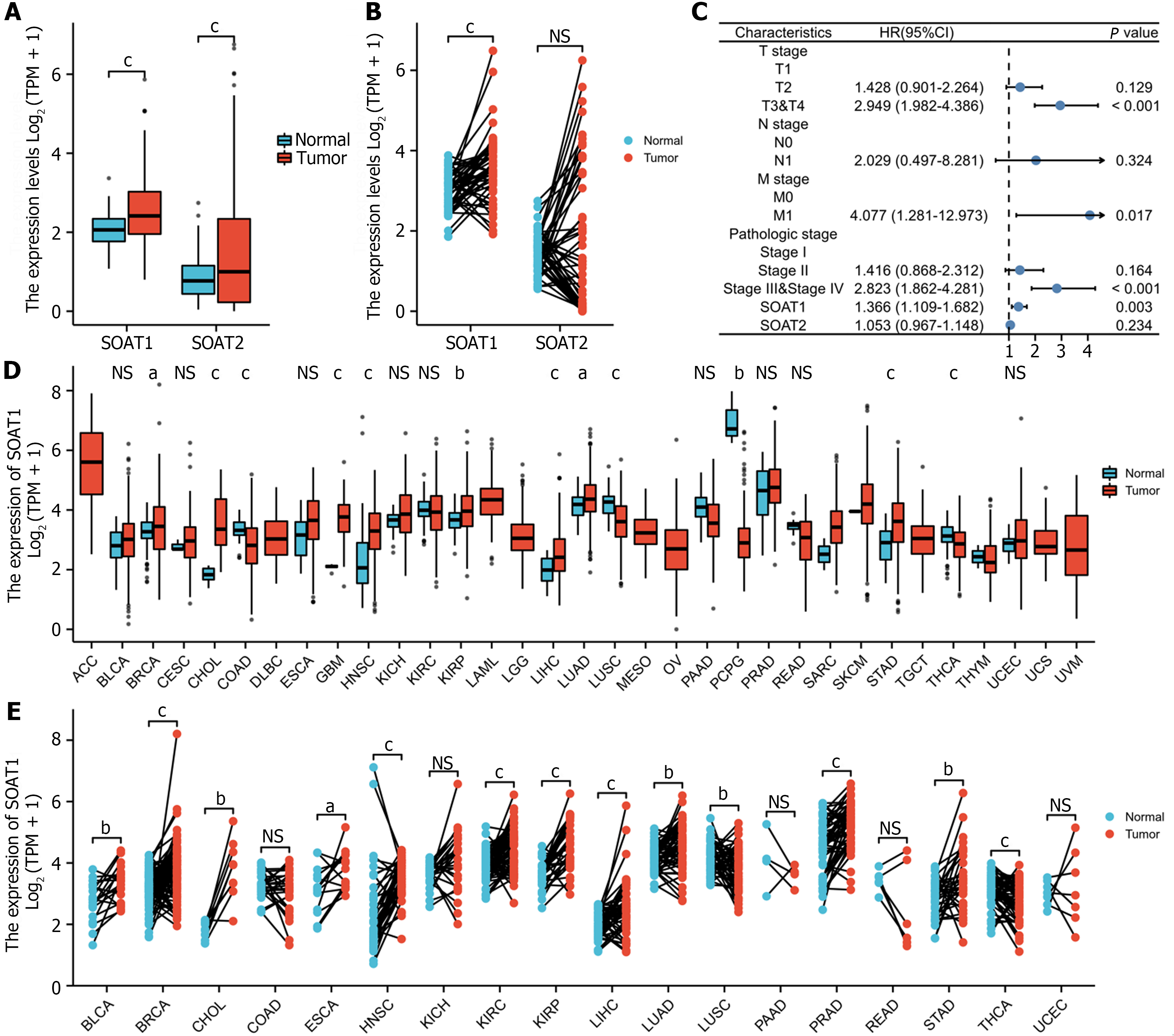
In the TCGA-LIHC cohort, SOAT1 and SOAT2 were highly expressed in unpaired samples, while only SOAT1 was highly expressed in paired samples (Figure 1A and B). The univariate analysis and multivariate analysis suggested that SOAT1 expression was an independent risk factor for HCC progression (Figure 1C; Supplementary Table 1). SOAT1 expression in pan-cancer, including unpaired and paired samples, was also investigated (Figure 1D and E).
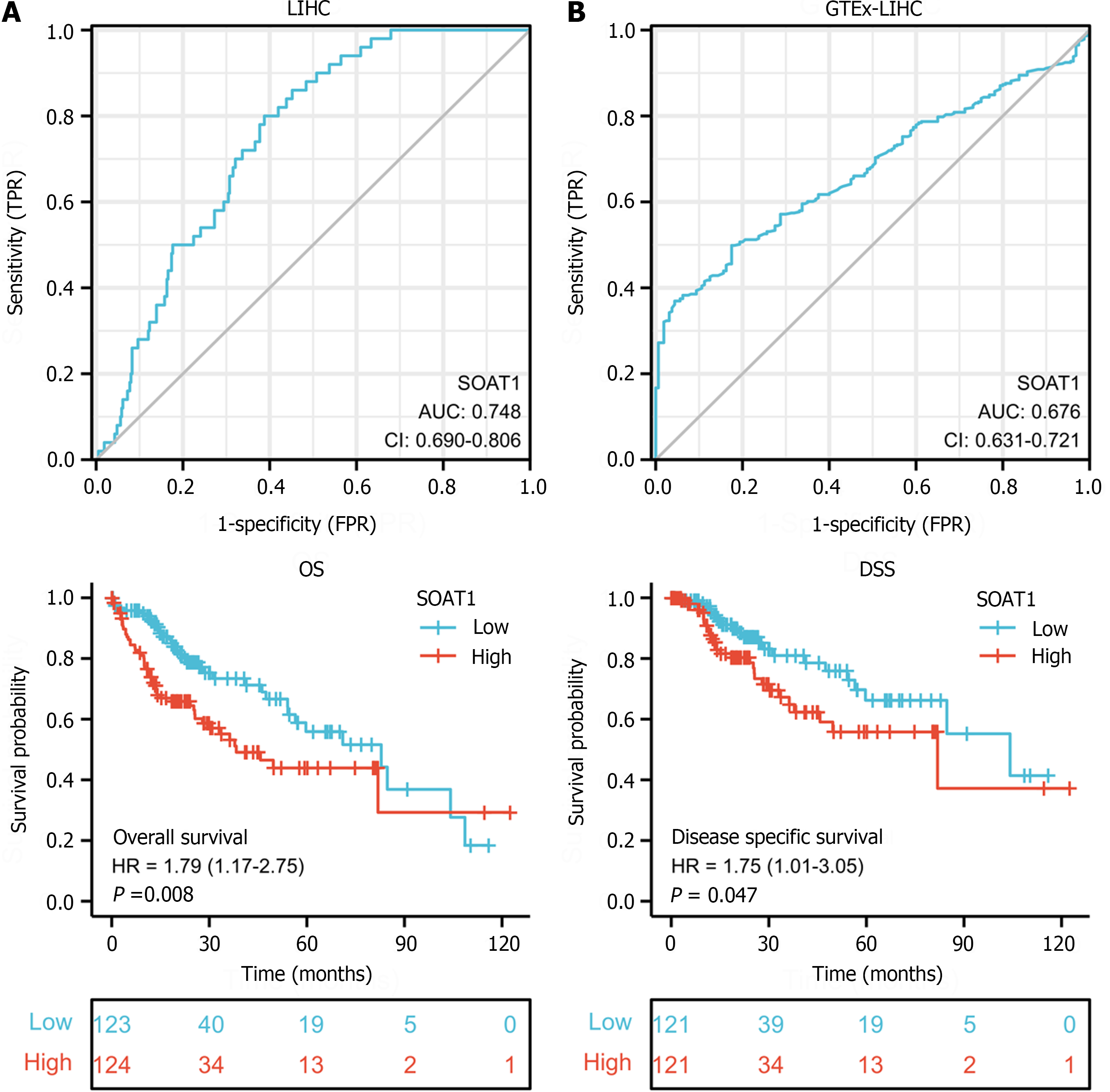
To explore the diagnostic value of SOAT1 expression in HCC, we performed receiver operating characteristic curve analysis. The AUC of SOAT1 expression in tumor samples from LIHC patients compared with para-carcinoma tissues was 0.748, while the AUC of SOAT1 expression in tumor samples from LIHC patients compared with GTEx was 0.676, suggesting that SOAT1 may be a potential diagnostic biomarker for HCC invasion (Figure 2A and B).
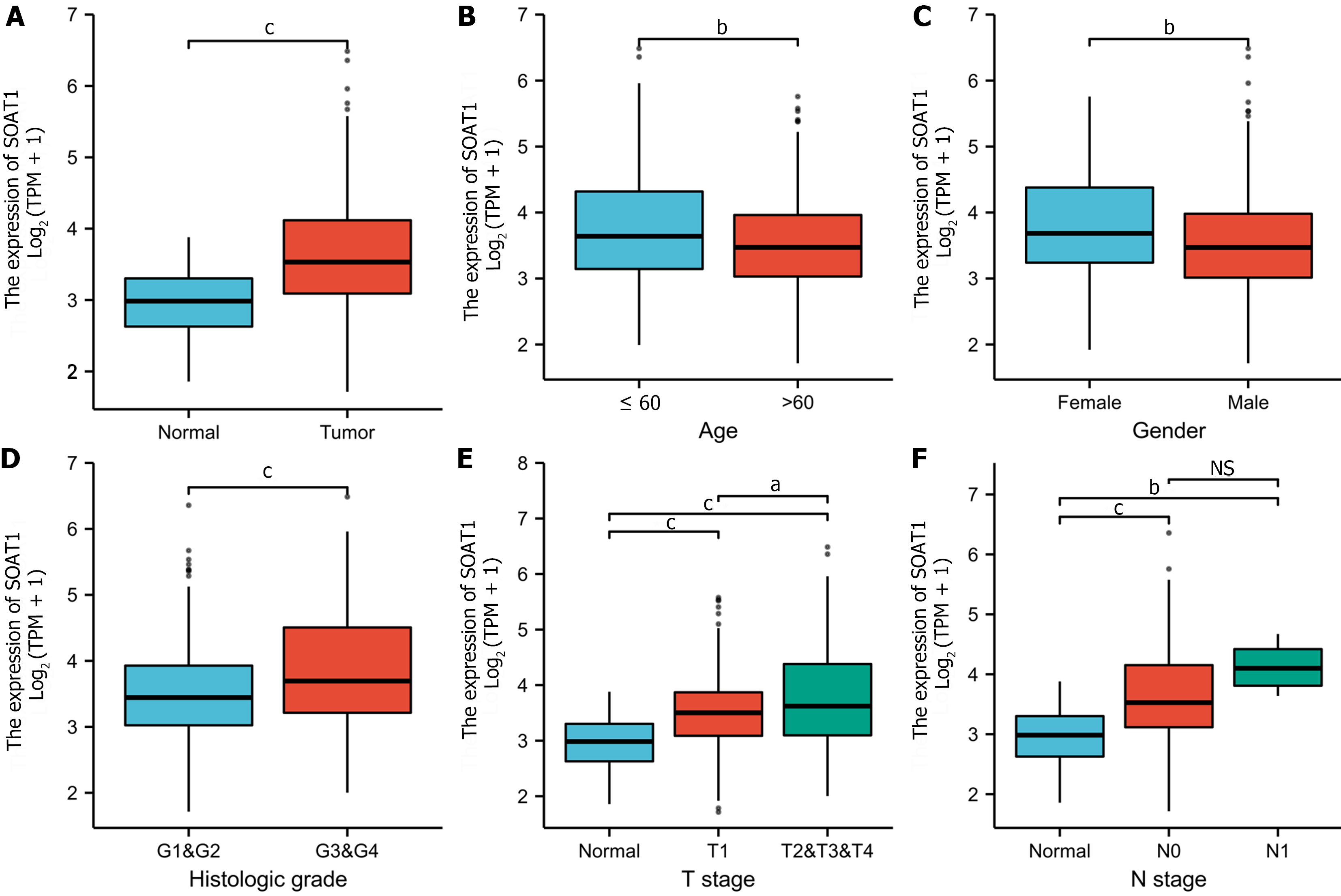
To clarify the prognostic value of SOAT1 expression in HCC, OS and DSS were analyzed. Patients with higher SOAT1 expression had lower survival rates (Figure 2C and D). SOAT1 expression was also associated with age, gender, histologic grade, T stage, and N stage (Figure 3). In addition, 1-, 3-, and 5-year OS and DSS analysis demonstrated that higher SOAT1 expression was associated with a worse prognosis (Figure 4).
After log transformation, DEGs between the group with high and low expression of SOAT1 in LIHC were identified. GO enrichment analysis, KEGG pathway enrichment analysis, and GSEA showed that these DEGs are mainly involved in the PI3K/AKT signaling pathway, the IL-18 signaling pathway, the calcium signaling pathway, secreted factors, the Wnt signaling pathway, the Jak/STAT signaling pathway, the MAPK family signaling pathway, and cell–cell communication (Figure 5).
Compared with healthy controls, patients with primary tumor showed significantly increased protein expression of SOAT1 (Supplementary Figure 1). To further analyze the association between SOAT1 expression and immune cells, single sample GSEA was conducted in LIHC, which showed that SOAT1 expression was positively associated with the abundance of macrophages, Th2 cells, T helper cells, CD56bright natural killer (NK) cells, and Th1 cells and negatively associated with the abundance of Th17 cells, dendritic cells, and cytotoxic cells (Supplementary Figure 2).
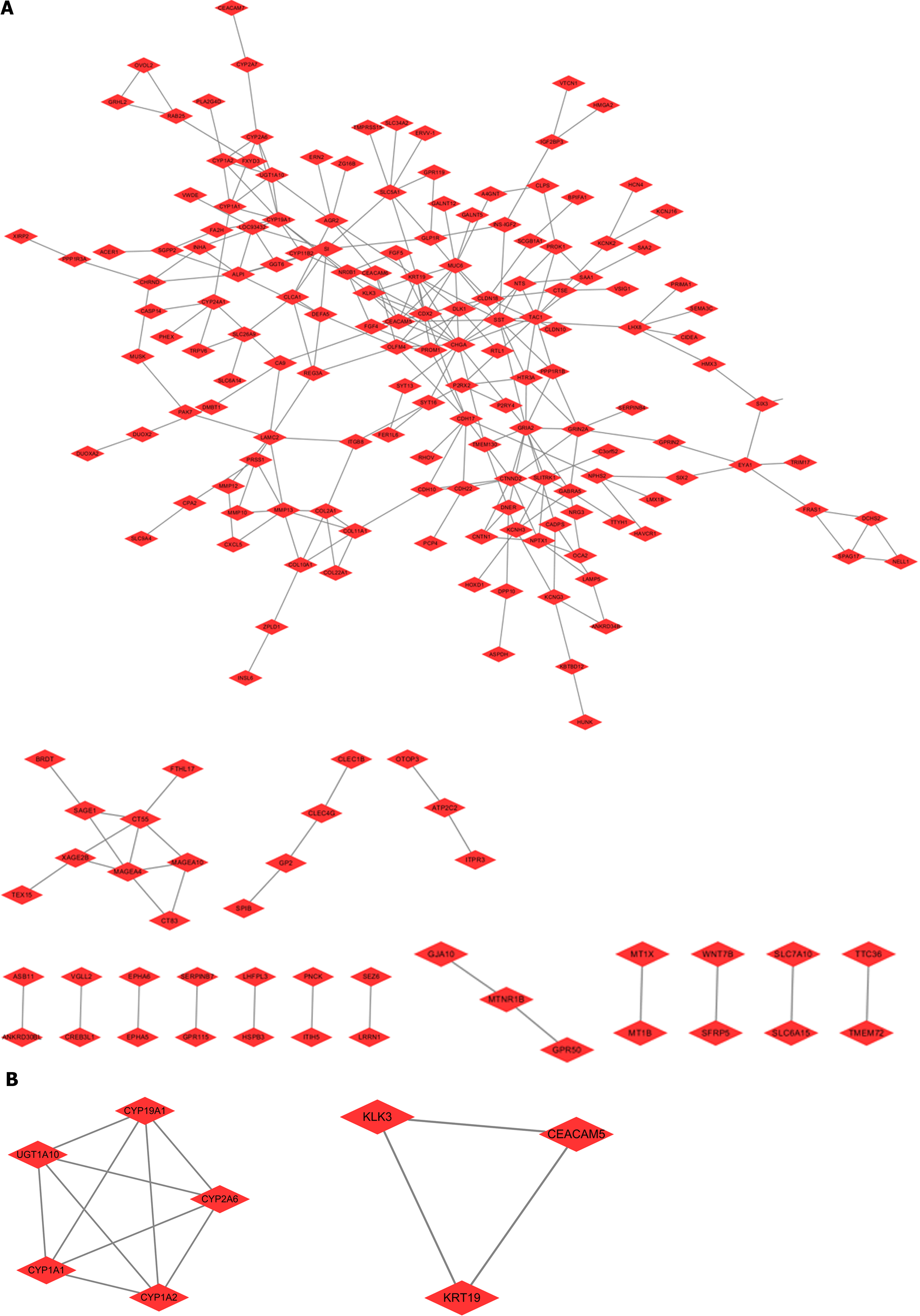
To clarify the proteins that interact with SOAT1 in TCGA-LIHC, the nodes with a comprehensive score more than 0.4 were studied using the STRING database. The hub genes were obtained from the Cytoscape plug-in MCODE, which included two modules in the network (including CYP19A1, CYP2A6, CYP1A2, CYP1A1, UGT1A10, KLK3, KRT19, and CEACAM5). These genes might be potential targets for HCC treatment (Figure 6).
To investigate the role of SOAT1 expression in LIHC progression, Lasso regression and risk score analysis were utilized. SOAT1 expression was highly correlated with survival time and with the expression of two hub genes, namely, CYP19A1 and UGT1A10 (Figure 7A and B). To further explore the prognostic value of these two hub genes, survival analysis was conducted, which showed that patients with higher expression of CYP19A1 and UGT1A10 had a worse prognosis, which was consistent with the prognostic value of SOAT1 expression.
Historically, chronic viral hepatitis was the main etiologies of HCC; however, nonalcoholic fatty liver disease and related metabolic factors have emerged as the fastest-growing risk factors for HCC in recent years. The relationship between lipids and HCC is complex, so more investigations are anticipated to continue over the next decade. Understanding the role of cholesterol in HCC development will contribute to developing new therapies. One way to further our unders
Previous studies have shown lower lipid levels in HCC patients compared to healthy controls[23], suggesting that cholesterol metabolism plays a pivotal role in the development of HCC[25,26]. Evidence from proteomic studies have found that HCC patients with abnormal cholesterol metabolism and high SOAT1 expression seemed to have a worse prognosis[19], suggesting that SOAT1 may have an effect on HCC by regulating lipid metabolism. A recent study has found that extracellular lipid loading promoted glioma-associated macrophage infiltration and new blood vessel formation in tumors, which was increased by an elevated continuous supply of lipids throughout the body[27]. It is direct evidence that LD+ glioblastoma cells are related to immunosuppressive glioma-associated macrophage infiltration. Since LDs are formed due to the aggregation of cholesterol esters, it is not surprising that SOAT1 expression is associated with M2 macrophage infiltration in HCC. There is a complex relationship between lipids and HCC. Altered lipid metabolism may be a result of HCC development. Cachexia commonly exists in cancer patients, characterized by reduced fat storage, increased carbohydrate utilization, and elevated protein degradation. The high growth rate of cancer cells may lead to hypoxia and increased energy requirements, ultimately promoting fatty-acid oxidation and depleting fat stores[28,29]. In addition, dysregulation of lipid metabolism may contribute to the development of HCC, due to impaired pro-tumorigenic insulin and insulin-like growth factor 1 signaling[30,31]. Additionally, research in mice and humans has showed that liver cells without fatty acid synthase might support c-MET oncogene-mediated liver tumor formation through up-regulation of SREBP2 via the cholesterol synthesis pathway[32].
Studies have demonstrated that SOAT1 plays a carcinogenic role through multiple pathways. Our OS and DSS analyses also showed that higher SOAT1 expression was associated with poor survival in patients with HCC. Therefore, further studies are warranted to explore the prognostic value of SOAT1 in HCC. Indeed, SOAT1 expression is associated with a poor prognosis in all HCC cases. Our 1-, 3- and 5-year OS and DSS analyses demonstrated that higher SOAT1 expression was associated with a worse prognosis (Figure 4), suggesting that SOAT1 may be a potential diagnostic biomarker for HCC invasion. Down-regulation of SOAT1 has been reported to inhibit proliferation and migration of HCC cells by reducing plasma membrane cholesterol content and inhibiting the integrin and TGF-β signaling pathways[19]. Consistently, integrin binding was also significantly enhanced, as determined by enrichment analysis of the GO and KEGG pathways of upregulated DEGs in HCC (Figure 5). Multiple genes, including CYP19A1, CYP2A6, CYP1A2, CYP1A1, UGT1A10, KLK3, KRT19, and CEACAM5 (Figure 6), whose encoded proteins may interact with SOAT1 in HCC, were identified via PPI network and co-expression analyses, which may be potential targets for HCC treatment. The higher the expression of CYP19A1 and UGT1A10, the worse the prognosis, which is consistent with the prognostic analysis of SOAT1 expression. SOAT1 expression was reported to be regulated by multiple mechanisms in tumors. Runt-related transcription factor 1 promotes SOAT1 expression in squamous cell carcinoma by binding to the promoter region of SOAT1[33]. Loss of p53 heterozygosity can promote the expression of SOAT1 by enhancing the transcription of SOAT1 in pancreatic ductal adenocarcinoma[10]. In addition, β-catenin has been reported to be directly bind to the SOAT1 promoter element and promote its transcription in colorectal cancer[21], as well.
The progression of HCC is complex and several factors are involved, including age, alcohol consumption, environmental toxins, HBV and HCV levels, and diet. In the present study, the prognostic value of SOAT1 in HCC was elucidated. Our findings suggest that SOAT1 may modestly alter the risk for HCC by regulating lipid metabolism, but the effect might be limited. Further studies are warranted to validate our results. The identification of other HCC proteins involved in this multigenic heterogeneous cancer type is an important objective for future research. Since early diagnosis of HCC is of great benefit to patients, complementary studies using the most advanced proteomic techniques on HCC-related proteins in serum samples can be a very attractive research direction in the future. That is, SOAT1 may be recognized as a new target to advance the development of immunotherapy and metabolic therapy.
Hepatocellular carcinoma (HCC) has a poor prognosis and heavy disease burden, but its treatment methods are not satisfactory.
High sterol O-acyltransferase 1 (SOAT1) expression has been shown to be associated with several tumor types (liver cancer, pancreatic cancer, and prostate cancer) and with diagnosis and treatment. However, the relationship between SOAT1 expression and HCC remains unclear. As patients would greatly benefit from early detection of HCC, the complementary study of HCC-associated proteins in serum samples using state-of-the-art proteomics would also be a very attractive direction for future research. Therefore, SOAT1 may serve as a novel target that drives the development of immunotherapy and metabolic therapy.
This study aimed to investigate the correlation between SOAT1 expression and HCC, using RNA-seq and gene expression data of The Cancer Genome Atlas (TCGA)-liver hepatocellular carcinoma (LIHC) and pan-cancer. Our findings demonstrate that SOAT1 may serve as a new target for HCC treatment and promote the development of new strategies for immunotherapy and metabolic therapy.
The correlation between SOAT1 expression and HCC was analyzed. Cox hazard regression models were used to investigate the prognostic value of SOAT1. Overall survival and disease-specific survival were also explored in TCGA-LIHC. Moreover, the biological processes and functional pathways regulated by SOAT1 were characterized using gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed genes. To better understand the regulatory mechanism of SOAT1 in HCC, protein–protein interaction network and co-expression analyses of SOAT1 in HCC were conducted.
SOAT1 and SOAT2 were highly expressed in unpaired samples, while only SOAT1 was highly expressed in paired samples. The area under the receiver operating characteristic curve of SOAT1 expression in tumor samples from LIHC patients compared with para-carcinoma tissues was 0.748, while the area under the curve of SOAT1 expression in tumor samples from LIHC patients compared with GTEx was 0.676. Patients with higher SOAT1 expression had lower survival rates. Results from GO/KEGG and gene set enrichment analyses suggested that the PI3K/AKT signaling pathway, the IL-18 signaling pathway, the calcium signaling pathway, secreted factors, the Wnt signaling pathway, the Jak/STAT signaling pathway, the MAPK family signaling pathway, and cell–cell communication were involved in such association. SOAT1 expression was positively associated with the abundance of macrophages, Th2 cells, T helper cells, CD56bright natural killer cells, and Th1 cells, and negatively linked to the abundance of Th17 cells, dendritic cells, and cytotoxic cells.
As patients would greatly benefit from early detection of hepatocellular carcinoma, the complementary study of hepatocellular carcinoma-associated proteins in serum samples using state-of-the-art proteomics would be a very attractive direction for future exploration.
The identification of other HCC proteins involved in this multigenic heterogeneous cancer type is an important objective for future research.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Rizzo A, Italy S-Editor: Liu JH L-Editor: Wang TQ P-Editor: Cai YX
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55842] [Article Influence: 7977.4] [Reference Citation Analysis (132)] |
| 2. | Beste LA, Leipertz SL, Green PK, Dominitz JA, Ross D, Ioannou GN. Trends in burden of cirrhosis and hepatocellular carcinoma by underlying liver disease in US veterans, 2001-2013. Gastroenterology. 2015;149:1471-1482.e5; quiz e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 359] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 3. | Rizzo A, Ricci AD, Brandi G. Systemic adjuvant treatment in hepatocellular carcinoma: tempted to do something rather than nothing. Future Oncol. 2020;16:2587-2589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 4. | Rizzo A, Ricci AD, Brandi G. Atezolizumab in advanced hepatocellular carcinoma: good things come to those who wait. Immunotherapy. 2021;13:637-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 5. | Rizzo A, Ricci AD, Brandi G. Trans-Arterial Chemoembolization Plus Systemic Treatments for Hepatocellular Carcinoma: An Update. J Pers Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 73] [Reference Citation Analysis (0)] |
| 6. | Santoni M, Rizzo A, Mollica V, Matrana MR, Rosellini M, Faloppi L, Marchetti A, Battelli N, Massari F. The impact of gender on The efficacy of immune checkpoint inhibitors in cancer patients: The MOUSEION-01 study. Crit Rev Oncol Hematol. 2022;170:103596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 118] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 7. | Ioannou GN. The Role of Cholesterol in the Pathogenesis of NASH. Trends Endocrinol Metab. 2016;27:84-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 376] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 8. | Chang CC, Sakashita N, Ornvold K, Lee O, Chang ET, Dong R, Lin S, Lee CY, Strom SC, Kashyap R, Fung JJ, Farese RV Jr, Patoiseau JF, Delhon A, Chang TY. Immunological quantitation and localization of ACAT-1 and ACAT-2 in human liver and small intestine. J Biol Chem. 2000;275:28083-28092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 186] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 9. | Kushwaha RS, Rosillo A, Rodriguez R, McGill HC Jr. Expression levels of ACAT1 and ACAT2 genes in the liver and intestine of baboons with high and low lipemic responses to dietary lipids. J Nutr Biochem. 2005;16:714-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 10. | Oni TE, Biffi G, Baker LA, Hao Y, Tonelli C, Somerville TDD, Deschênes A, Belleau P, Hwang CI, Sánchez-Rivera FJ, Cox H, Brosnan E, Doshi A, Lumia RP, Khaledi K, Park Y, Trotman LC, Lowe SW, Krasnitz A, Vakoc CR, Tuveson DA. SOAT1 promotes mevalonate pathway dependency in pancreatic cancer. J Exp Med. 2020;217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 11. | Stopsack KH, Gerke TA, Andrén O, Andersson SO, Giovannucci EL, Mucci LA, Rider JR. Cholesterol uptake and regulation in high-grade and lethal prostate cancers. Carcinogenesis. 2017;38:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 12. | Löhr M, Härtig W, Schulze A, Kroiß M, Sbiera S, Lapa C, Mages B, Strobel S, Hundt JE, Bohnert S, Kircher S, Janaki-Raman S, Monoranu CM. SOAT1: A Suitable Target for Therapy in High-Grade Astrocytic Glioma? Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Chan LK, Ng IO. Proteomic profiling in liver cancer: another new page. Transl Gastroenterol Hepatol. 2019;4:47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Khatib SA, Wang XW. Proteomic heterogeneity reveals SOAT1 as a potential biomarker for hepatocellular carcinoma. Transl Gastroenterol Hepatol. 2019;4:37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Abuzhalihan J, Wang YT, Ma YT, Fu ZY, Yang YN, Ma X, Li XM, Liu F, Chen BD. SOAT1 methylation is associated with coronary heart disease. Lipids Health Dis. 2019;18:192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Zhong N, Nong X, Diao J, Yang G. piRNA-6426 increases DNMT3B-mediated SOAT1 methylation and improves heart failure. Aging (Albany NY). 2022;14:2678-2694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Wu N, Li RQ, Li L. SOAT1 deficiency attenuates atherosclerosis by regulating inflammation and cholesterol transportation via HO-1 pathway. Biochem Biophys Res Commun. 2018;501:343-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Wang YJ, Bian Y, Luo J, Lu M, Xiong Y, Guo SY, Yin HY, Lin X, Li Q, Chang CCY, Chang TY, Li BL, Song BL. Cholesterol and fatty acids regulate cysteine ubiquitylation of ACAT2 through competitive oxidation. Nat Cell Biol. 2017;19:808-819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 105] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 19. | Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, Xing B, Sun W, Ren L, Hu B, Li C, Zhang L, Qin G, Zhang M, Chen N, Huang Y, Zhou J, Liu M, Zhu X, Qiu Y, Sun Y, Huang C, Yan M, Wang M, Liu W, Tian F, Xu H, Wu Z, Shi T, Zhu W, Qin J, Xie L, Fan J, Qian X, He F; Chinese Human Proteome Project (CNHPP) Consortium. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567:257-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 623] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 20. | Riscal R, Skuli N, Simon MC. Even Cancer Cells Watch Their Cholesterol! Mol Cell. 2019;76:220-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 21. | Zhu Y, Gu L, Lin X, Zhang J, Tang Y, Zhou X, Lu B, Liu C, Prochownik EV, Li Y. Ceramide-mediated gut dysbiosis enhances cholesterol esterification and promotes colorectal tumorigenesis in mice. JCI Insight. 2022;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 22. | Wu DF, Yin RX, Aung LH, Hu XJ, Cao XL, Miao L, Li Q, Yan TT, Wu JZ, Pan SL. Polymorphism of rs1044925 in the acyl-CoA:cholesterol acyltransferase-1 gene and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Chen Y, Yang X, Chen Y, Chen G, Winkler CA, An P, Lyu J. Impacts of the SOAT1 genetic variants and protein expression on HBV-related hepatocellular carcinoma. BMC Cancer. 2021;21:615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Mallick P, Kuster B. Proteomics: a pragmatic perspective. Nat Biotechnol. 2010;28:695-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 283] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 25. | Lu M, Hu XH, Li Q, Xiong Y, Hu GJ, Xu JJ, Zhao XN, Wei XX, Chang CC, Liu YK, Nan FJ, Li J, Chang TY, Song BL, Li BL. A specific cholesterol metabolic pathway is established in a subset of HCCs for tumor growth. J Mol Cell Biol. 2013;5:404-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 26. | Teng CF, Hsieh WC, Yang CW, Su HM, Tsai TF, Sung WC, Huang W, Su IJ. A biphasic response pattern of lipid metabolomics in the stage progression of hepatitis B virus X tumorigenesis. Mol Carcinog. 2016;55:105-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Offer S, Menard JA, Pérez JE, de Oliveira KG, Indira Chandran V, Johansson MC, Bång-Rudenstam A, Siesjö P, Ebbesson A, Hedenfalk I, Sundgren PC, Darabi A, Belting M. Extracellular lipid loading augments hypoxic paracrine signaling and promotes glioma angiogenesis and macrophage infiltration. J Exp Clin Cancer Res. 2019;38:241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24:472-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 375] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 29. | Shaw JH, Wolfe RR. Fatty acid and glycerol kinetics in septic patients and in patients with gastrointestinal cancer. The response to glucose infusion and parenteral feeding. Ann Surg. 1987;205:368-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 132] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1393] [Cited by in RCA: 1597] [Article Influence: 122.8] [Reference Citation Analysis (0)] |
| 31. | Renehan AG, Frystyk J, Flyvbjerg A. Obesity and cancer risk: the role of the insulin-IGF axis. Trends Endocrinol Metab. 2006;17:328-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 343] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 32. | Che L, Chi W, Qiao Y, Zhang J, Song X, Liu Y, Li L, Jia J, Pilo MG, Wang J, Cigliano A, Ma Z, Kuang W, Tang Z, Zhang Z, Shui G, Ribback S, Dombrowski F, Evert M, Pascale RM, Cossu C, Pes GM, Osborne TF, Calvisi DF, Chen X, Chen L. Cholesterol biosynthesis supports the growth of hepatocarcinoma lesions depleted of fatty acid synthase in mice and humans. Gut. 2020;69:177-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 140] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 33. | Jain P, Nattakom M, Holowka D, Wang DH, Thomas Brenna J, Ku AT, Nguyen H, Ibrahim SF, Tumbar T. Runx1 Role in Epithelial and Cancer Cell Proliferation Implicates Lipid Metabolism and Scd1 and Soat1 Activity. Stem Cells. 2018;36:1603-1616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |