Published online Aug 27, 2022. doi: 10.4254/wjh.v14.i8.1562
Peer-review started: October 22, 2021
First decision: December 27, 2021
Revised: January 14, 2022
Accepted: July 27, 2022
Article in press: July 27, 2022
Published online: August 27, 2022
Processing time: 307 Days and 14.4 Hours
Hepatocellular carcinoma (HCC) is the fifth most common cancer. Differential expression of microRNAs (miRNAs)-326, miRNA-424, and miRNA-511 has been associated with the diagnosis and prognosis of HCC in different populations. However, limited information is available regarding their expression in Egyptian HCC patients.
To assess the role of circulating miRNAs-326, miRNA-424, and miRNA-511 in Egyptian HCC patients.
This prospective observational study included 70 HCC patients and 25 healthy controls. The circulating levels of these three miRNAs were evaluated by real-time PCR. Receiver operating characteristic curve analysis was used to test the diagnostic accuracy of microRNA expression levels.
All miRNAs were differentially expressed in HCC patients; miRNAs326 and miRNA-424 were upregulated, while miRNA-511 was downregulated. Both miRNA-326 and miRNA-424 showed sensitivity and specificity of 97%, 71.4%, and 52%, 60%, respectively, to differentiate HCC from controls. Moreover, miRNA-326 was associated with survival and could differentiate between Child grades (A vs B); miRNA-424 significantly differentiated early vs intermediate stages of HCC; while miRNA-511 was significantly correlated with response to modified Response Evaluation Criteria in Solid Tumors (mRECIST).
We conclude that miRNA-326, miRNA-424, and miRNA-511 have diagnostic and prognostic roles in Egyptian patients with hepatitis C virus-related HCC and should be considered for better disease management.
Core Tip: Hepatocellular carcinoma (HCC) is the sixth most common malignancy worldwide. Recent cancer incidence data confirmed that the global age normalization rate of primary liver cancer is 10.1/100000, with a male/female ratio of 3:1. The diagnosis of HCC usually occurs in the late stages, resulting in an elevated death rate; this makes HCC the third deadliest malignancy. We examined whether circulating miRNAs326, miRNA-424, and miRNA-511 could serve as promising candidate non-invasive biomarkers for HCC. Such biomarkers could be used for targeted gene therapy research and may assist in monitoring the severity of HCC.
- Citation: Youssef SS, Elfiky A, Nabeel MM, Shousha HI, Elbaz T, Omran D, Marie MS, Elzahry MA, Abul-Fotouh A, Hashem A, Guda MF, Abdelaziz AO. Assessment of circulating levels of microRNA-326, microRNA-424, and microRNA-511 as biomarkers for hepatocellular carcinoma in Egyptians. World J Hepatol 2022; 14(8): 1562-1575
- URL: https://www.wjgnet.com/1948-5182/full/v14/i8/1562.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i8.1562
Liver cancer is the fifth most common cancer in men and is the second cause of cancer-related deaths worldwide[1]. Hepatocellular carcinoma (HCC) is a primary liver cancer and constitutes 80%-90% of all primary tumors of the liver[2,3]. It has a high incidence and mortality rate, with approximately 662.000 deaths every year[4]. Most HCC patients are diagnosed at advanced stages, contributing to the high rates of recurrence and development of metastasis[5]. In addition, the commonly used biomarker for HCC screening and diagnosis is alpha-fetoprotein (AFP), which has modest sensitivity and specificity[6] and is actually influenced by tumor size and cancer stage[7]. Due to these factors, it is crucial to identify more efficient biomarkers that can be used for the early diagnosis and prognosis of HCC.
MicroRNAs (miRNAs) are short, non-coding RNAs that regulate the transcription or degradation of certain target mRNAs[8-10]. They mediate important physiological processes such as cell differentiation, proliferation, and survival[11,12]. Aberrant regulation of miRNAs is associated with the initiation and progression of numerous cancers, including HCC[13]. Moreover, survival and response to chemotherapeutic drugs have been found to be linked to miRNAs[14-16]. Experimentally, a number of miRNAs have been proven to be related to HCC, and have been proposed as diagnostic and prognostic markers of HCC[17,18].
Aberrant expression of miRNA-424 has been documented in HCC tissues and cell lines[19,20]. It plays a tumor suppressor role[19]. A similar correlation was found for miRNA-326[21] and miRNA-511[22], and both were significantly associated with survival[23]. Using the Cancer Genome Atlas, Lu and colleagues studied the prognostic and diagnostic roles of 33 miRNA signatures and found 11 down-regulated and 22 upregulated miRNAs by comparing cancerous and non-cancerous samples. They observed the good role of miRNA-326, -424, and -511[24].
Most miRNA studies in HCC are experimental and are based on HCC tissues and cell lines. They were mainly performed to identify the etiopathogenetic role of miRNAs in HCC. However, few studies have shown their fundamental role in the diagnosis and prognosis of HCC. Hence, this cross-sectional study with prospective follow-up of HCC patients was performed to search for three miRNAs (326, 424, and 511) in blood to highlight their potential role in the diagnosis and prognosis of HCC.
This prospective observational study included 70 adult Egyptian patients who developed HCC on top of hepatitis C virus (HCV)-related liver cirrhosis and 25 healthy age- and sex-matched participants who were seronegative for HCV and HBV, and served as controls. HCC was diagnosed according to the American Association for the Study of Liver Diseases updated practice guidelines[25], or was histo-pathologically confirmed. All the patients were recruited from the multidisciplinary HCC clinic, the Endemic Medicine Department, Kasr al Ainy School of Medicine, Cairo University, and were HCC-treatment naïve at the time of enrollment. Exclusion criteria included; HBV co-infection, any cause of chronic liver disease other than HCV, any associated malignancies other than HCC, and prior treatment for HCV or HCC.
All patients were subjected to clinical assessment, laboratory investigations (complete blood count, prothrombin time and concentration, liver and kidney function tests, AFP, hepatitis markers), ultrasound examination, and triphasic CT with documentation of HCC site, size, and number as well as the presence of portal vein thrombosis (PVT). At the time of enrollment into the study, blood samples were obtained from each participant, and Child-Pugh-score[26] and Barcelona Clinic Liver Cancer (BCLC) stage[27] were assessed based on the clinical, laboratory, and imaging data obtained.
Whole blood samples were collected from each participant in 5-mL sterile RNAase-free vacutainer tubes containing EDTA. Blood samples were collected on ice and processed within 30 min of collection. To separate the plasma, each blood sample was centrifuged for 10 min at 1900 g at 4 ℃ (Heraeus, Labofuge 400 R; Germany). Plasma samples were carefully transferred into sterile RNase-free tubes, and then centrifuged for 10 min at 12000 g at 4 ℃ to remove cellular nucleic acid contamination, and haemolysed plasma samples were excluded. Samples were separated into aliquots and stored at 80 ℃ until processed.
Total RNA was extracted from 200 µL plasma using the miRNeasy Serum/plasma cell lysates kit (Qiagen, Germany), according to the manufacturer's instructions. RNA concentration and purity were monitored using a Nanodrop spectrophotometer device (ThermoScientific, Wilmington, DE, United States).
The reverse transcription reaction was performed using the TaqMan™ MicroRNA Reverse Transcription Kit, Cat number # 4366596, (Applied Biosystems, Foster City, CA, United States) according to the manufacturer's instructions. miRNA-326, miRNA-511, and miRNA-424 quantification was carried out using quantitative real-time PCR (qRT-PCR) (Stratagene Mx3000p; Agilent Technologies, Germany). The qRT-PCR for each sample was carried out in duplicate using TaqMan 2x universal master mix II (Applied Biosystems, Foster City, CA, United States) and TaqMan microRNA Assay Mix containing PCR primers and TaqMan probes for each miRNA. The expression level of RNU6B was used as an endogenous control for normalization. To determine miRNA relative expression, it was reported as a fold change (ΔCt and ΔΔCt calculations).
After blood sampling, all patients were managed according to the BCLC guidelines[27] after a case-by-case discussion. All HCC patients were followed up for a period of 24 mo. HCC response to treatment was evaluated according to the Modified Response Evaluation Criteria in Solid Tumors (mRECIST)[28]. The overall survival time of the patients was defined as the period from the initial presentation to the last follow-up or death.
Microsoft Excel 2016 and the Statistical Package for Social Science (SPSS; Windows, version 26, IBM Corp., Armonk, N.Y., United States) were used to analyze the data. Continuous, normally distributed variables were presented as mean ± SD, with a 95% confidence interval (CI), while non-normally distributed variables were summarized as the median and interquartile range (IQR). Categorical data were presented as frequencies and percentages. A P value of less than 0.05 was considered statistically significant. The Student t-test was performed to compare the means of normally distributed variables between groups, and the Mann-Whitney U test was used for non-normal variables. The Chi-square test and Fisher's exact test were used to determine the distribution of categorical variables between groups. The diagnostic performance of the studied miRNAs was assessed by receiver operating characteristic (ROC) curves. The area under the ROC (AUROC) was used as an index to compare the accuracy of tests. The optimal cut-off point value was taken from the maximum combined sensitivity and specificity. The sensitivity and specificity of relevant cut-offs are also displayed. The survival analysis was conducted using the "Log Rank (Mantel-Cox) Kaplan-Meier test".
The mean age of the studied cohort was 62.0 ± 7.6 years. Most of the patients were male (68.8%), non-diabetic (74.3%), non-smokers (80%), and Child A (82.9%). According to the BCLC staging system, most patients were in the early stage (48.6%). Most of the HCC lesions were single (70%), present in the right hepatic lobe (84.3%), and not associated with PVT (92.9%). That is why most of the patients were subjected to hepatectomy and microwave ablation (MWA), and most of them showed complete response according to mRECIST. All HCC patients were followed up for a period of 24 mo or until death. The mean survival time was 367.1±173.9 days. The median values of miRNA-326, miRNA-511, and miRNA-424 were 35, 1.2, and 5.1, respectively (Table 1).
| HCC | |||
| N = 70 | |||
| Demographic data | Age (yr) | 62.0 ± 7.6 | |
| Gender (%) | Female | 22 (31.4) | |
| Male | 48 (68.6) | ||
| Smoker (%) | No | 56 (80.0) | |
| Yes | 14 (20.0) | ||
| DM | No | 52 (74.3) | |
| (%) | Yes | 18 (25.7) | |
| BMI (kg/m2) | 27.7 (23.6- 31.9) | ||
| Laboratory investigation | Hb (g/dL) | 13.2 ± 1.9 | |
| WBC (× 103/µL) | 6.0 (4.6-8.0) | ||
| Platelets (× 103/µL) | 169.5 ± 77.9 | ||
| ALT (U/mL) | 52.5 (32.0- 74.3) | ||
| AST (U/mL) | 60.5 (38.0- 83.3) | ||
| Alb (g/dL) | 3.7 ± 0.5 | ||
| Bil.T (mg/dL) | 0.9 (0.6- 1.4) | ||
| AFP (ng/dL) | 38.8 (10.0- 370.0) | ||
| Creatinine (mg/dL) | 0.9 ± 0.2 | ||
| BCLC (%) | Early | 34 (48.6) | |
| Intermediate | 23 (32.9) | ||
| Late | 13 (18.6) | ||
| Performance status (%) | 0 | 43 (61.4) | |
| 1 | 24 (34.3) | ||
| 2 | 2 (2.9) | ||
| 3 | 1 (1.4) | ||
| Child grade (%) | A | 58 (82.9) | |
| B | 11 (15.7) | ||
| C | 1 (1.4) | ||
| Child score | 5.8 ± 1.0 | ||
| Number of HCCs (%) | Single | 49 (70.0) | |
| Two | 12 (17.1) | ||
| ≥ 3 | 9 (12.9) | ||
| Site of HCC (%) | Right lobe | 59 (84.3) | |
| Left lobe | 7 (10.0) | ||
| Both lobes | 4 (5.7) | ||
| HCC size (cm) | 4.0 (2.7- 5.0) | ||
| Portal vein | Patent | 65 (92.9) | |
| (%) | |||
| Thrombosed | 5 (7.1) | ||
| Abdominal lymphadenopathy | No | 65 (92.9) | |
| (%) | Yes | 5 (7.1) | |
| Decision of treatment | Best supportive care | 7 (10.0) | |
| Combined therapy | 1 (1.4) | ||
| (%) | Hepatectomy | 3 (4.3) | |
| Microwave ablation | 20 (28.5) | ||
| Radioembolization | 3 (4.3) | ||
| Sorafinib | 3 (4.3) | ||
| TACE | 29 (41.4) | ||
| Response to treatment according to mRECIST (%) | Stationary | 4 (7.8) | |
| Partial response | 7 (13.7) | ||
| Complete response | 34 (66.7) | ||
| Progressive disease | 6 (11.8) | ||
| Clinical decompensation (%) | Yes | 9 (12.9) | |
| No | 61 (54.5) | ||
| Alive or dead (%) | Dead | 34 (48.6) | |
| Alive | 36 (51.4) | ||
| Survival (days) | 367.1 ± 173.9 | ||
| miRNA-326 | 35.0 (12.5- 162.3) | ||
| miRNA-511 | 1.2 (0.3- 3.2) | ||
| miRNA-424 | 5.1 (2.9- 10.3) | ||
There was no significant association between the studied miRNAs levels and gender, smoking, or the patients' performance status. The serum level of miRNA-424 was significantly elevated in diabetic patients. There was a significant difference in miRNA-326 between Child grades A and B. Moreover, there was a significant difference in miRNA-424 between early and intermediate HCC (Table 2).
| miRNA-326 | miRNA-511 | miRNA-424 | |
| Gender | |||
| Male 48 (68.6%) | 33.6 (11.3-157.4) | 1.1 (0.4-3.4) | 5.4 (3.2-10.4) |
| Females 22 (31.4%) | 42.3 (11.9-168.2) | 1.1 (0.4-3.4) | 4.4 (1.9-8.3) |
| P value | 0.7 | 0.8 | 0.3 |
| Smoker | |||
| No 56 (80.0%) | 27.4 (8.4-159.0) | 1.1 (0.3-2.7) | 5.4 (2.9-10.6) |
| Yes 14 (20.0%) | 67.0 (28.0-177.0) | 1.2 (0.7-5.9) | 4.1 (2.7-7.0) |
| P value | 0.3 | 0.3 | 0.3 |
| Diabetes mellitus | |||
| No 52 (74.3%) | 29.4 (9.1- 119.1) | 0.9 (0.3-2.5) | 4.6 (2.6-9.0) |
| Yes 18 (25.7%) | 70.4 (16.6- 218.4) | 1.5 (0.9-5.1) | 6.5 (4.1-13.7) |
| P value | 0.1 | 0.08 | 0.04a |
| Child grade | |||
| A 58 (82.9%) | 64.4 (15.3-177.0) | 1.2 (0.4-3.8) | 5.1 (3.0-10.3) |
| B 11 (15.7%) | 16.1 (4.1-110.7) | 0.9 (0.0-1.4) | 5.2 (2.6-11.9) |
| C 1 (1.4%) | 5.7 (5.7-5.7) | 0.03 (0.03-0.03) | 0.9 (0.9-0.9) |
| P value | |||
| B vs A | |||
| C vs A | 0.03a | 0.1 | 0.7 |
| C vs B | 0.2 | 0.1 | 0.1 |
| 0.7 | 0.3 | 0.3 | |
| HCC stages | |||
| Early | 32.0 (7.5-121.0) | 1.0 (0.3-2.6) | 4.5 (2.3-8.0) |
| Intermediate | 69.8 (16.1-185.0) | 1.4 (0.6-4.6) | 5.3 (3.9-13.2) |
| Late | 26.2 (14.5-179.8) | 0.9 (0.1-2.9) | 5.9 (2.5-10.1) |
| P value | |||
| Intermediate vs Early | 0.2 | 0.1 | 0.04a |
| Late vs Early | 0.6 | 0.8 | 0.5 |
| Late vs Intermediate | 0.7 | 0.2 | 0.6 |
| Performance status | |||
| 0; 43 (61.4%) | 64.4 (15.8-153.3) | 1.2 (0.5-3.5) | 5.7 (3.1-10.2) |
| 1; 24 (34.3%) | 18.9 (7.1-184.5) | 0.6 (0.0-2.5) | 4.0 (1.5-10.4) |
| 2; 2 (2.9%) | 7.2 (0.5-7.8) | 3.1 (0.4-3.5) | 5.3 (4.8-5.5) |
| 3; 1 (1.4%) | 160.9 (160.9- 160.9) | 9.6 (9.6-9.6) | 22.0 (22.0-22.0) |
| P value | 0.2 | 0.2 | 0.2 |
On comparing the miRNAs levels between healthy participants and HCC patients, it was found that HCC patients showed significantly higher levels of miRNA-326 (P = 0.001) and significantly lower levels of miRNA-511 (P = 0.02) (Table 3).
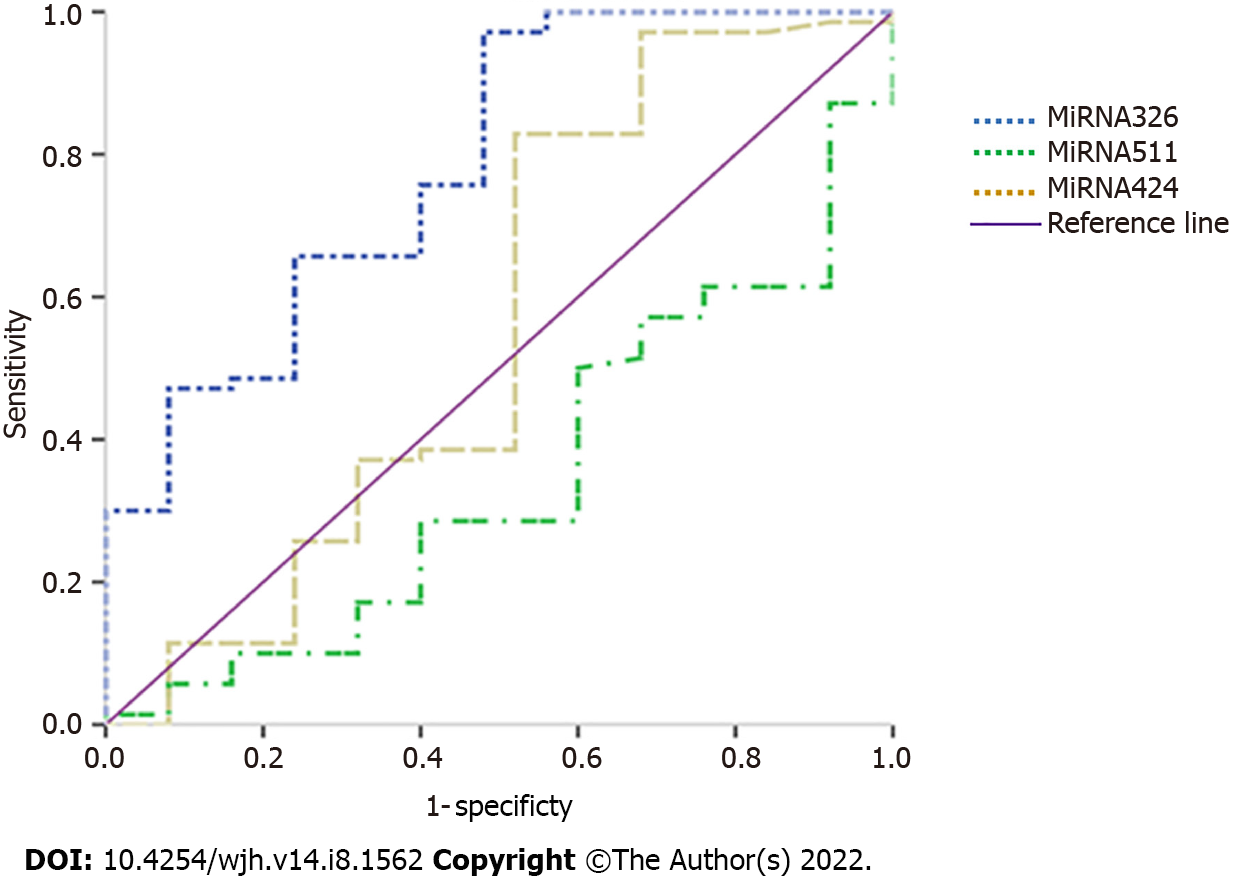
ROC analysis revealed the diagnostic performance of the studied miRNAs. miRNA-326 showed the best diagnostic performance, diagnosing HCC at a cut-off value of 1.165 with a sensitivity of 97.1%, specificity of 52%, positive predictive value (PPV) of 85%, and negative predictive value (NPV) of 86.7%. The area under curve (AUC) was 78.4% (67.7- 89.1), and the overall accuracy was 85.3% (P < 0.001) (Table 4, Figure 1).
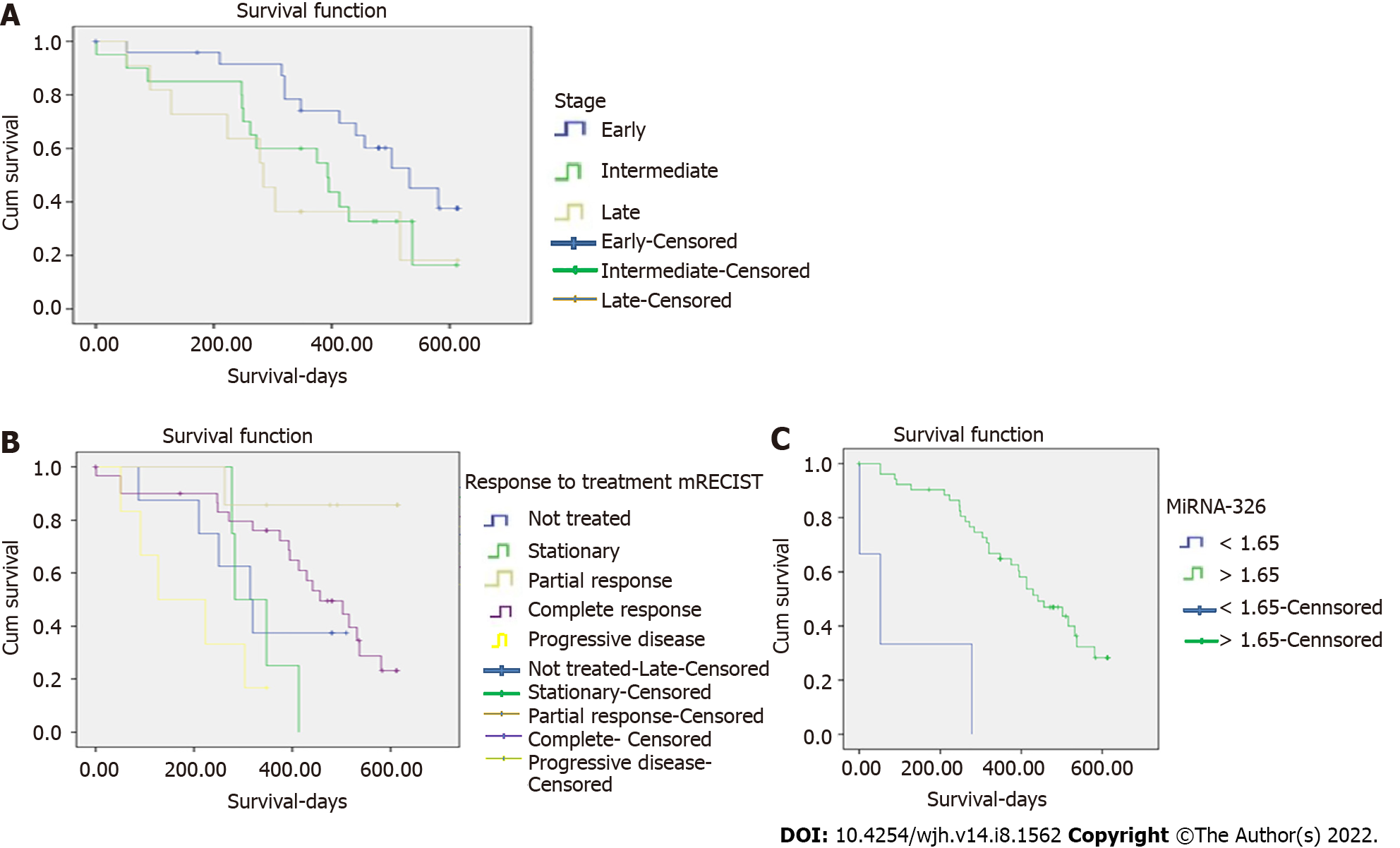
Despite finding no statistically significant differences in the studied miRNAs between the surviving and non-surviving HCC patients, we found that miRNA-326 >1.165 was significantly associated with overall survival (P = 0.001) (Table 5, Figure 2). Moreover, there was a significant association between the BCLC stage as well as the response to treatment according to mRECIST and overall survival (Figure 2). It was also found that miRNA-511 was significantly associated with the response to treatment according to mRECIST (Table 6).
| Dead, N = 34 | Median (95%CI)of the EstimatedSurvival Time | Log rank (Mantel-Cox) | P value | ||
| Gender | Female (%) | 9 (26.5) | 315.0 (134.4- 495.6) | 0.094 | 0.8 |
| Male (%) | 25 (73.5) | 441.0 (329.7- 552.3) | |||
| Smoker | No (%) | 29 (85.3) | 395.0 (299.9- 490.1) | 1.595 | 0.2 |
| Yes (%) | 5 (14.7) | 581.0 (336.4- 825.6) | |||
| DM | No (%) | 27 (79.4) | 413.0 (341.7- 484.3) | 0.259 | 0.6 |
| Yes (%) | 7 (20.6) | 429.0 (257.4- 600.6) | |||
| BCLC stages | Early (%) | 12 (35.3) | 532.0 (401.2- 662.8) | 5.594 | 0.05a |
| Intermediate (%) | 14 (41.2) | 393.0 (225.7- 560.3) | |||
| Late (%) | 8 (23.5) | 284.0 (196.6- 371.4) | |||
| Performance status | 0 (%) | 21 (61.8) | 395.0 (314.6 - 475.4) | 1.188 | 0.6 |
| 1 (%) | 12 (35.3) | 516.0 (404.0- 628.0) | |||
| 2 (%) | 1 (2.9) | 278.0 (278.0- 278.0) | |||
| 3 | 0 (0.0) | - | |||
| Child score | A (%) | 26 (76.5) | 429.0 (316.6- 541.4) | 0.574 | 0.8 |
| B (%) | 7 (20.6) | 375.0 (241.9- 508.1) | |||
| C (%) | 1 (2.9) | 516.0 (516.0- 516.0) | |||
| Response to treatment mRECIST | Stationary | 4 (11.8%) | 284.0 (215.4- 352.6) | 2.9 | 0.4 |
| Partial response | 1 (2.9%) | 412.0 (412.0- 412.0) | |||
| Complete response | 19 (55.9%) | 456.0 (346.7- 565.3) | |||
| Progressive disease | 5 (14.7%) | 128.0 (0.0- 285.2) | |||
| miRNA-326 >1.165 | 441.0 (336.7- 545.3) | 17.1 | 0.001b | ||
| miRNA-511<2.063 | 441.0 (329.5- 552.5) | 0.626 | 0.4 | ||
| miRNA-424>2.462 | 395.0 (300.7- 489.3) | 2.707 | 0.1 |
| Response to treatment mRECIST | Stationary, n = 4 (5.7%) | Partial response, n = 7 (10.0%) | Complete response, n = 34 (48.6%) | Progressive disease, n = 6 (8.6%) | P value |
| miRNA-326 | 11.5 (2.1- 142.6) | 31.1 (6.9- 209.4) | 27.4 (10.0- 155.2) | 69.6 (17.7- 197.9) | 0.5 |
| miRNA-511 | 3.8 (0.8- 10.5) | 1.2 (0.3- 4.2) | 1.0 (0.2- 1.5) | 0.6 (0.1- 1.3) | 0.05a |
| miRNA-424 | 4.9 (3.0- 8.6) | 4.8 (2.1- 5.1) | 5.4 (2.6- 13.1) | 4.6 (3.1- 15.2) | 0.8 |
Several experimental studies have shown that miRNAs play a regulatory role in the progression of HCC by controlling cell cycle progression, cell growth, and apoptosis. Our study focused on three important miRNAs: miRNA-326, miRNA-424, and miRNA-511. miRNA-326 upregulation is known to inhibit cell proliferation and colony formation, and influence the invasiveness and migratory properties of HCC. It has a tumor suppressor role, and its low expression was found to be associated with tumor, node, metastasis (TNM) staging, tumor differentiation, and lymph node metastases in HCC patients. Regarding miRNA-424, experimental studies found that it is downregulated in HCC and has a role in inhibiting tumor migration, proliferation, and invasion. It was associated with AFP, TNM, multinodularity, vascular invasion, and intrahepatic metastases, as well as poor survival[29,30] and predicted tumor recurrence in HCC patients following liver transplantation[31]. Few studies have mentioned the role of miRNA-511 in HCC proliferation and invasiveness.
In our study, miRNA-326 was upregulated, in contrast to the aforementioned studies in which miRNA-326 was down-regulated. Importantly, these studies were on HCC tissues and cell lines, while our study adopted an easier methodology of testing miRNA-326 in the blood. It seems that blood tests could give paradoxical results. The study by Moya et al[32] assessed different miRNAs as biomarkers for prostate cancer and found a similar paradox. This difference is attributed to the possibility of preferential retainment of oncomirs (these are miRNAs that are overexpressed in cancers) and the release of tumor suppressor miRNAs into the circulation to promote oncogenesis. Added to this, the authors highlighted the physiological cancer conditions that can cause leakage of molecules.
In our study, a statistically significant difference was noted while revising the miRNA signature of miRNA-326 and miRNA-424 between our HCV-related HCC patients and healthy controls. A clear diagnostic role was identified for both markers. We would like to document another crucial issue. We found no single miRNA to correlate with the different HCC parameters. For example, miRNA-424 differentiated early vs intermediate HCC significantly, miRNA-326 was a major factor related to survival, and miRNA-511 significantly correlated with response to treatment. miRNA-326 cut-off level > 1.165 showed statistically significant sensitivity and specificity when upregulated, while miRNA-511 cut-off < 2.063 showed statistically significant sensitivity and specificity when downregulated. This provides the main clue that no single miRNA can be used for the diagnosis and prognosis of HCC. Wang and Lei identified an eleven long non-coding RNA (lncRNA) signature for predicting HCC prognosis and incorporated seven miRNAs (including miRNA-326 and -424)[33]. In another study, the authors mentioned seven miRNAs that were found to have different expressions between tumors and in-vicinity non-tumorous tissues and found that they were significantly associated with survival (including miRNA-326 and -511)[23]. Similarly, a multidimensional signature stratified the HCC patients into low-risk and high-risk groups (P < 0.001) in the training set, validation set, and independent set, and all were statistically significant. It showed better survival prediction power when compared to TNM staging and included three miRNAs (-149, -424, and -579)[34]. In the study by Lu et al[24], five biomarkers (among 33 miRNAs) significantly correlated with patient survival. These markers were miRNA-326, -421, -511, -3677, and -424. They divided patients into low-risk and high-risk groups according to the miRNA signatures. None of the high-risk patients survived for 5 years. Thus, it seems that using a combination of miRNAs gives better results than using one biomarker.
The mechanism of action of miRNA in HCC has been proposed in several experimental studies. Concerning miRNA-326, different mechanisms of action were proposed, including NF-kB expression[35], targeting MAPK1 and CSF1 as regulated by circASAP1[21], and targeting LIM and SH3 protein 1 (LASP1)[36]. Also, it acts by suppression of PDK1 in the PDK1/AKT/C-myc axis[37]. It acts as a sponge for circular RNA circ-0000517[38] and a regulator for the SMAD 6 axis[39]. HCC proliferation was found to be promoted via CircRNA-PTN that also sponges miRNA-326 and affects ErbB/PI3K in HCC cells[40]. Finally, HOXD-AS1 binds directly to miRNA-326, thereby targeting gene SLC27A4, which also plays a role in HCC progression and metastasis promotion[41]. Proposed mechanisms of action of miRNA-424 include the direct targeting of C-Myb[19], AKT3 and E2F3[30], E2F7 expression[42], and acts as a sponge to different lncRNAs such as lncRNA CASC9[43], LINL 00657[44], lncRNA CDKN2B-AS1[45], and lncRNA LINC00511[34]. All these targets are correlated with poor prognosis, promotion of cell viability, and migration. Also, it affected angiogenesis by activating the VEGFR-2 signaling pathway[46]. Serum clinical samples from HCC patients and healthy volunteers showed that miRNA-424 significantly decreased in HCC patients and correlated with poor overall survival and disease-free survival[47]. With regard to miRNA-511, studies proposed its action through sponging LINC01559[22], regulation of the AKT1 axis[48], and targeting PIK3R3 in the PIK3R3/AKT/mTOR signaling pathway[49].
Finally, we would like to highlight the aggressive behavior of HCC. Our patients were predominantly BCLC-A, Child-Pugh score A, performance status 0-1, and the majority had single lesions without evidence of vascular invasion or lymph node metastases. Moreover, we had good overall response rates (complete and partial response rates were nearly 60%). Nearly half of patients died during the follow-up period despite all these facts. This indicates the importance of primary prevention of HCV-related HCC through early management of its risk factors rather than managing HCC after its development.
Nonetheless, our study has limitations. We could not study more miRNAs that are correlated with HCC, and we could not correlate blood tests with tissue tests. However, our study is a clinical (not an experimental) one, and this helped to better correlate miRNA with different co-existing potentials and factors that could be present in actual life scenarios. In conclusion, we found a good diagnostic and prognostic role for our studied biomarkers (miRNA-326, -424, and -511) in HCC patients.
We conclude that miRNA-326, miRNA-424, and miRNA-511 have diagnostic and prognostic roles in Egyptian patients with HCV-related HCC and should be considered for better disease management.
Hepatocellular carcinoma (HCC) is lethal and is the fifth most common cancer. Differential expression of microRNAs (miRNAs)-326, miRNA-424, and miRNA-511 has been associated with the diagnosis and prognosis of HCC in different populations. However, limited information is available regarding their expression in Egyptian HCC patients.
This study aimed to assess the role of circulating miRNAs-326, miRNA-424, and miRNA-511 from plasma as a non-invasive route of detection and to explore the impact of these miRNAs in Egyptian HCC patients.
The study objectives were to assess the role of circulating miRNAs-326, miRNA-424, and miRNA-511 in Egyptian HCC patients by a non-invasive method using plasma samples.
This prospective observational study included 70 adult Egyptian patients who developed HCC on top of HCV-related liver cirrhosis and 25 healthy age- and sex-matched participants who were seronegative for HCV and HBV, and served as controls. HCC was diagnosed according to the American Association for the Study of Liver Diseases updated practice guidelines, or was histo-pathologically confirmed. All patients were recruited from the multidisciplinary HCC clinic, the Endemic Medicine Department, Kasr al Ainy School of Medicine, Cairo University, and were HCC-treatment naïve at the time of enrollment. Exclusion criteria included; HBV co-infection, any cause of chronic liver disease other than HCV, any associated malignancies other than HCC, and prior treatment for HCV or HCC. Data collection: All patients were subjected to clinical assessment, laboratory investigations [complete blood count, prothrombin time and concentration, liver and kidney function tests, alpha-fetoprotein (AFP), hepatitis markers], ultrasound examination, and triphasic CT with documentation of HCC site, size, and number as well as the presence of PVT. At the time of enrolment into the study, blood samples were obtained from each participant, and Child-Pugh-score and BCLC stage were assessed based on the clinical, laboratory, and imaging data obtained. Whole blood samples were collected from each participant in 5-mL sterile RNAase-free vacutainer tubes containing EDTA. Blood samples were collected on ice and processed within 30 min of collection. To separate the plasma, each blood sample was centrifuged for 10 min at 1900g at 4 ℃. Plasma samples were carefully transferred into sterile RNase-free tubes. Plasma samples were then centrifuged for 10 min at 12000g at 4 ℃ to remove cellular nucleic acid contamination, and haemolysed plasma samples were excluded. Samples were separated into aliquots and immediately stored at 80 ℃ until processed. RNA extraction: Total RNA was extracted from 200 µL plasma using the miRNeasy Serum/plasma cell lysates kit, according to the manufacturer's instructions. RNA concentration and purity were monitored using a Nanodrop spectrophotometer device. Quantitative RT-PCR: The reverse transcription reaction was performed using TaqMan™ MicroRNA Reverse Transcription Kit, according to the manufacturer's instructions. miRNA-326, miRNA-511, and miRNA-424 quantifications were carried out using quantitative real-time PCR. The qRT-PCR for each sample was carried out in duplicate using TaqMan 2x universal master mix II and TaqMan microRNA Assay Mix containing PCR primers and TaqMan probes for each miRNA. The expression level of RNU6B was used as an endogenous control for normalization. To determine miRNA relative expre
The mean age of the studied cohort was 62.0 ± 7.6 years. Most of the patients were male (68.8%), non-diabetic (74.3%), non-smokers (80%), and Child A (82.9%). According to the BCLC staging system, most patients were in the early stage (48.6%). Most of the HCC lesions were single (70%), present in the right hepatic lobe (84.3%), and not associated with PVT (92.9%). That is why most of the patients were subjected to hepatectomy and microwave ablation (MWA), and most of them showed complete response according to mRECIST. All HCC patients were followed up for a period of 24 mo or until death. The mean survival time was 367.1 ± 173.9 days. The median values of miRNA-326, miRNA-511, and miRNA-424 were 35, 1.2, and 5.1, respectively. Correlation of miRNAs with different parameters in HCC patients: There was no significant association between the studied miRNAs levels and gender, smoking, or the patients' performance status. The serum level of miRNA-424 was significantly elevated in diabetic patients. There was a significant difference in miRNA-326 between Child grades A and B. Moreover, there was a significant difference in miRNA-424 between early and intermediate HCC. Diagnostic efficiency of miRNAs in our patients: On comparing the miRNAs levels between healthy participants and HCC patients, it was found that HCC patients showed significantly higher levels of miRNA-326 (P = 0.001) and significantly lower levels of miRNA-511 (P = 0.02). ROC analysis revealed the diagnostic performance of the studied miRNAs. miRNA-326 showed the best diagnostic performance, diagnosing HCC at a cut-off value of 1.165 with a sensitivity of 97.1%, specificity of 52%, PPV of 85%, and NPV of 86.7%. The AUC was 78.4% (67.7-89.1), and the overall accuracy was 85.3% (P < 0.001). Association of miRNAs with response to treatment and survival: Despite finding no statistically significant differences in the studied miRNAs between surviving and non-surviving HCC patients, we found that miRNA-326 > 1.165 was significantly associated with overall survival (P = 0.001). Moreover, there was a significant association between the BCLC stage as well as the response to treatment according to mRECIST and overall survival. It was also found that miRNA-511 was significantly associated with response to treatment according to mRECIST.
We conclude that miRNA-326, miRNA-424, and miRNA-511 have diagnostic and prognostic roles in Egyptian patients with HCV-related HCC and should be considered for better disease management.
miRNA-326, miRNA-424, and miRNA-511 can be detected from plasma and have diagnostic and prognostic roles in Egyptian patients with HCV-related HCC and should be considered for better disease management.
The authors would like to acknowledge the National Research Centre, as well as Endemic Medicine and Hepato-gastroenterology Department, Faculty of Medicine, Cairo University, Cairo, Egypt, for providing laboratory space, equipment, and patient samples for achievement of the current work.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Tropical medicine
Country/Territory of origin: Egypt
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Tsoulfas G, Greece; Yao HR, China S-Editor: Ma YJ L-Editor: Webster JR P-Editor: Ma YJ
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20504] [Article Influence: 2050.4] [Reference Citation Analysis (20)] |
| 2. | Astarci HM, Özyalvaçli G, Sertçelik A. Karaciğerin primer ve metastatik karsinomlarının ayrımında pCEA, mCEA, AFP ve CK19'un tanısal değeri. Abant Tıp Dergisi. 2019;5:39-46. [DOI] [Full Text] |
| 3. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3852] [Article Influence: 963.0] [Reference Citation Analysis (3)] |
| 4. | McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7:418-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 903] [Article Influence: 100.3] [Reference Citation Analysis (0)] |
| 5. | Baek KK, Kim JH, Uhm JE, Park SH, Lee J, Park JO, Park YS, Kang WK, Lim HY. Prognostic factors in patients with advanced hepatocellular carcinoma treated with sorafenib: a retrospective comparison with previously known prognostic models. Oncology. 2011;80:167-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Attwa MH, El-Etreby SA. Guide for diagnosis and treatment of hepatocellular carcinoma. World J Hepatol. 2015;7:1632-1651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 169] [Article Influence: 16.9] [Reference Citation Analysis (4)] |
| 7. | Kudo M. Surveillance, diagnosis, treatment, and outcome of liver cancer in Japan. Liver Cancer. 2015;4:39-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 8. | He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4964] [Cited by in RCA: 5317] [Article Influence: 253.2] [Reference Citation Analysis (0)] |
| 9. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 10. | Guzel E, Okyay TM, Yalcinkaya B, Karacaoglu S, Gocmen M, Akcakuyu MH. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. North Clin Istanb. 2020;7:81-86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 11. | Bueno MJ, Pérez de Castro I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143-3148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 270] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 12. | Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 347] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 13. | Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1354] [Cited by in RCA: 1476] [Article Influence: 92.3] [Reference Citation Analysis (0)] |
| 14. | Zhang J, Wang Y, Zhen P, Luo X, Zhang C, Zhou L, Lu Y, Yang Y, Zhang W, Wan J. Genome-wide analysis of miRNA signature differentially expressed in doxorubicin-resistant and parental human hepatocellular carcinoma cell lines. PLoS One. 2013;8:e54111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao H, Sun Q, Yan F, Yan C, Li H, Ren X. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene. 2014;533:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 16. | Vaira V, Roncalli M, Carnaghi C, Faversani A, Maggioni M, Augello C, Rimassa L, Pressiani T, Spagnuolo G, Di Tommaso L, Fagiuoli S, Rota Caremoli E, Barberis M, Labianca R, Santoro A, Bosari S. MicroRNA-425-3p predicts response to sorafenib therapy in patients with hepatocellular carcinoma. Liver Int. 2015;35:1077-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 17. | Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P, Lee CG. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205-13215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 299] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 18. | Jin Y, Wong YS, Goh BKP, Chan CY, Cheow PC, Chow PKH, Lim TKH, Goh GBB, Krishnamoorthy TL, Kumar R, Ng TP, Chong SS, Tan HH, Chung AYF, Ooi LLPJ, Chang JPE, Tan CK, Lee CGL. Circulating microRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Sci Rep. 2019;9:10464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 19. | Yu L, Ding GF, He C, Sun L, Jiang Y, Zhu L. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One. 2014;9:e91661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 20. | Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia X, Zhao W, Huai W, Qiu Y, Sun L, Han L. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 21. | Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, Mao L, Luo CB, Yu SY, Huang XW, Cao Y, Fan J, Zhou J. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology. 2020;72:906-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 173] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 22. | Su Q, Wang H. Long non-coding RNA 01559 mediates the malignant phenotypes of hepatocellular carcinoma cells through targeting miR-511. Clin Res Hepatol Gastroenterol. 2021;45:101648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Zhang J, Chong CC, Chen GG, Lai PB. A Seven-microRNA Expression Signature Predicts Survival in Hepatocellular Carcinoma. PLoS One. 2015;10:e0128628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 24. | Lu M, Kong X, Wang H, Huang G, Ye C, He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:8775-8784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 25. | Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68:723-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2121] [Cited by in RCA: 3232] [Article Influence: 461.7] [Reference Citation Analysis (1)] |
| 26. | Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60:646-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5490] [Cited by in RCA: 5733] [Article Influence: 110.3] [Reference Citation Analysis (2)] |
| 27. | Forner A, Reig ME, de Lope CR, Bruix J. Current strategy for staging and treatment: the BCLC update and future prospects. Semin Liver Dis. 2010;30:61-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 764] [Cited by in RCA: 865] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 28. | Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis. 2010;30:52-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3353] [Cited by in RCA: 3293] [Article Influence: 219.5] [Reference Citation Analysis (36)] |
| 29. | Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C, Ren G, Wu H. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. 2019;224:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 30. | Yang H, Zheng W, Shuai X, Chang R-M, Yu L, Fang F, Yang L-Y. MicroRNA-424 inhibits Akt3/E2F3 axis and tumor growth in hepatocellular carcinoma. Oncotarget. 2015;6:27736. |
| 31. | Wu L, Yang F, Lin B, Chen X, Yin S, Zhang F, Xie H, Zhou L, Zheng S. MicroRNA-424 expression predicts tumor recurrence in patients with hepatocellular carcinoma following liver transplantation. Oncol Lett. 2018;15:9126-9132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Moya L, Meijer J, Schubert S, Matin F, Batra J. Assessment of miR-98-5p, miR-152-3p, miR-326 and miR-4289 Expression as Biomarker for Prostate Cancer Diagnosis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 33. | Wang A, Lei J. Identification of an 11-lncRNA signature with high performance for predicting the prognosis of hepatocellular carcinoma using bioinformatics analysis. Medicine (Baltimore). 2021;100:e23749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 34. | Wang RP, Jiang J, Jiang T, Wang Y, Chen LX. Increased long noncoding RNA LINC00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating miR-424 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:3291-3301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 35. | Bai ZZ, Li HY, Li CH, Sheng CL, Zhao XN. M1 Macrophage-Derived Exosomal MicroRNA-326 Suppresses Hepatocellular Carcinoma Cell Progression Via Mediating NF-κB Signaling Pathway. Nanoscale Res Lett. 2020;15:221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 36. | Hu S, Ran Y, Chen W, Zhang Y, Xu Y. MicroRNA-326 inhibits cell proliferation and invasion, activating apoptosis in hepatocellular carcinoma by directly targeting LIM and SH3 protein 1. Oncol Rep. 2017;38:1569-1578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 37. | Mo Y, He L, Lai Z, Wan Z, Chen Q, Pan S, Li L, Li D, Huang J, Xue F, Che S. Gold nano-particles (AuNPs) carrying miR-326 targets PDK1/AKT/c-myc axis in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol. 2019;47:2830-2837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 38. | He S, Yang J, Jiang S, Li Y, Han X. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020;20:404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 39. | He S, Guo Z, Kang Q, Wang X, Han X. Circular RNA hsa_circ_0000517 modulates hepatocellular carcinoma advancement via the miR-326/SMAD6 axis. Cancer Cell Int. 2020;20:360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 40. | Jia B, Yin X, Wang Y, Qian J, He Y, Yang C, Yu G, Guo B, Meng X. CircRNA-PTN Sponges miR-326 to Promote Proliferation in Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:4893-4903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 41. | Ji W, Wang Q, Yang J. LncRNA HOXD-AS1 promotes the metastasis of human hepatocellular carcinoma via modulating miR-326/SLC27A4. Cancer Cell Int. 2020;20:161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 42. | Zhao Y, Zhu C, Chang Q, Peng P, Yang J, Liu C, Liu Y, Chen X, Cheng R, Wu Y, Wu X, Hu L, Yin J. MiR-424-5p regulates cell cycle and inhibits proliferation of hepatocellular carcinoma cells by targeting E2F7. PLoS One. 2020;15:e0242179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 43. | Yao J, Fu J, Liu Y, Qu W, Wang G, Yan Z. LncRNA CASC9 promotes proliferation, migration and inhibits apoptosis of hepatocellular carcinoma cells by down-regulating miR-424-5p. Ann Hepatol. 2021;23:100297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 44. | Cao X, Zhang G, Li T, Zhou C, Bai L, Zhao J, Tursun T. LINC00657 knockdown suppresses hepatocellular carcinoma progression by sponging miR-424 to regulate PD-L1 expression. Genes Genomics. 2020;42:1361-1368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 45. | Shen X, Li Y, He F, Kong J. LncRNA CDKN2B-AS1 Promotes Cell Viability, Migration, and Invasion of Hepatocellular Carcinoma via Sponging miR-424-5p. Cancer Manag Res. 2020;12:6807-6819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 46. | Teng F, Zhang JX, Chang QM, Wu XB, Tang WG, Wang JF, Feng JF, Zhang ZP, Hu ZQ. LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 47. | Yao H, Liu X, Chen S, Xia W, Chen X. Decreased expression of serum miR-424 correlates with poor prognosis of patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:14830-14835. [PubMed] |
| 48. | Yang X, Liu L, Zou H, Zheng YW, Wang KP. circZFR promotes cell proliferation and migration by regulating miR-511/AKT1 axis in hepatocellular carcinoma. Dig Liver Dis. 2019;51:1446-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 49. | Cao G, Dong W, Meng X, Liu H, Liao H, Liu S. MiR-511 inhibits growth and metastasis of human hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol. 2015;36:4453-4459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |