Published online Feb 27, 2022. doi: 10.4254/wjh.v14.i2.304
Peer-review started: February 25, 2021
First decision: March 29, 2021
Revised: April 13, 2021
Accepted: January 29, 2022
Article in press: January 29, 2022
Published online: February 27, 2022
Processing time: 361 Days and 22 Hours
Non-alcoholic fatty liver disease (NAFLD) prevalence has increased drastically in recent decades, affecting up to 25% of the world’s population. NAFLD is a spectrum of different diseases that starts with asymptomatic steatosis and continues with development of an inflammatory response called steatohepatitis, which can progress to fibrosis. Several molecular and metabolic changes are required for the hepatocyte to finally vary its function; hence a “multiple hit” hypothesis seems a more accurate proposal. Previous studies and current knowledge suggest that in most cases, NAFLD initiates and progresses through most of nine hallmarks of the disease, although the triggers and mechanisms for these can vary widely. The use of animal models remains crucial for under
Core Tip: Non-alcoholic fatty liver disease (NAFLD) is a spectrum of different diseases that starts with asymptomatic steatosis, continues with steatohepatitis, and can progress to fibrosis. Current knowledge suggests that NAFLD initiates and progresses through most of nine hallmarks. Animal models remain crucial for understanding the disease and for developing tools based on biological knowledge. Metabolomics seems a valid tool for studying metabolic pathways and organ crosstalk in NAFLD. In this review, we provide a brief introduction to NAFLD hallmarks, the five groups of animal models available for studying NAFLD and the potential role of metabolomics in the study of experimental NAFLD.
- Citation: Martin-Grau M, Marrachelli VG, Monleon D. Rodent models and metabolomics in non-alcoholic fatty liver disease: What can we learn? World J Hepatol 2022; 14(2): 304-318
- URL: https://www.wjgnet.com/1948-5182/full/v14/i2/304.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i2.304
Non-alcoholic fatty liver disease (NAFLD) prevalence has increased drastically in the last decades, affecting up to 25% of the world’s population[1]. The rise of disorders such as obesity and type 2 diabetes mellitus, as well as changes in lifestyle and diet composition, have led to a worldwide increase in the incidence of NAFLD[2-5]. Given that NAFLD reduces life expectancy by four years and triggers the appearance of different comorbidities such as cardiovascular disease, kidney damage or osteoporosis[3-5], it seems vital for specialists to establish accurate and precise guidelines or strategies to address the disease[6]. Assuming that the first stages of NAFLD are reversible[2] and to control the disease worldwide, there is a need for new non-invasive methods based on diagnostic and predictive biomarkers to help diagnose NAFLD in these early stages and avoid of the biopsy, which remains the gold standard diagnostic method[7,8]. The use of animal models remains crucial for understanding the disease[9] and for developing tools based on biological knowledge. In this review, we will provide an updated summary on NAFLD development, the importance of experimental animals uses, the rodent models currently applied, and use of metabolomics as a new methodology for improving understanding and management of NAFLD.
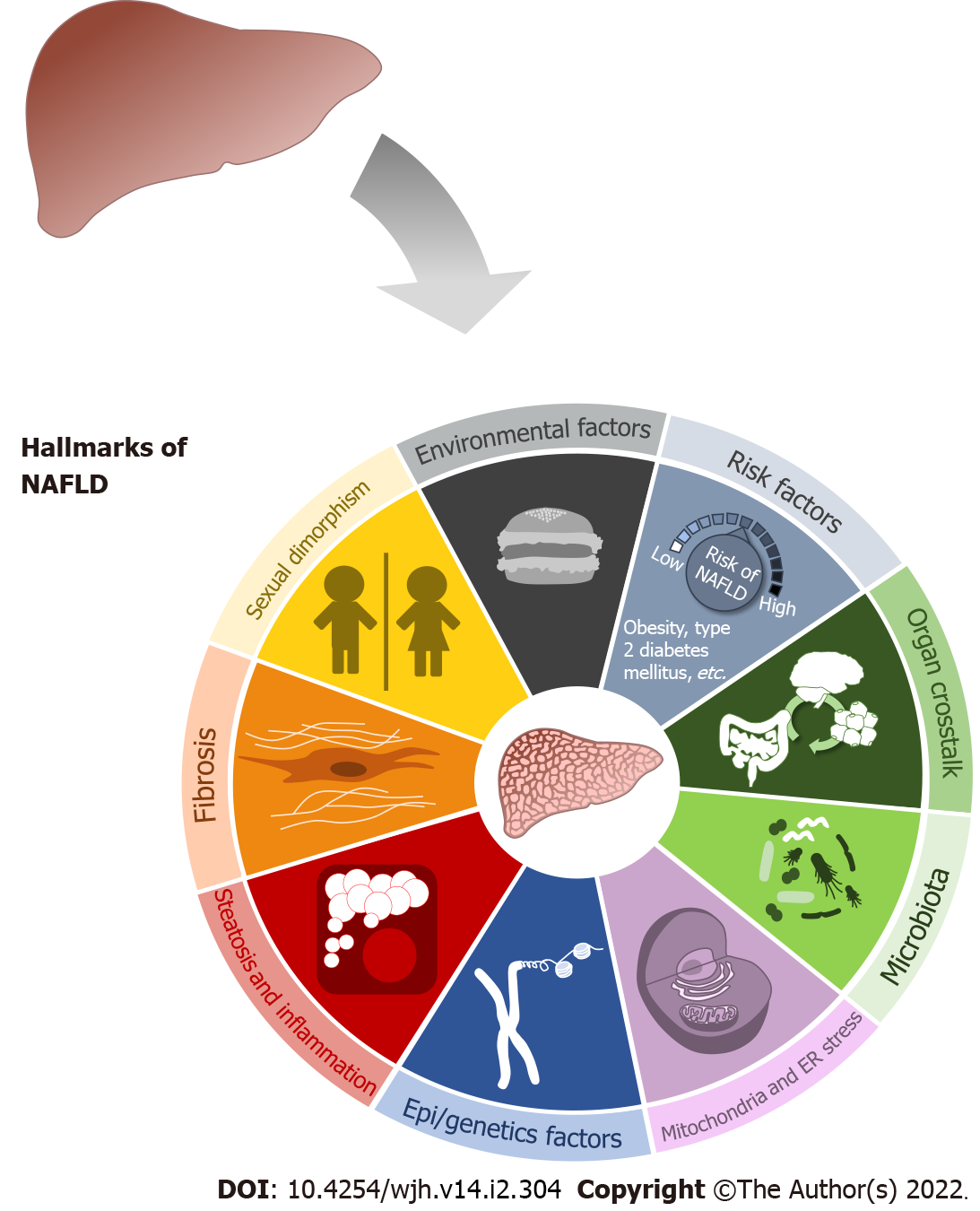
NAFLD is a spectrum of different diseases that starts with asymptomatic steatosis (NAFL) and continues with onset of an inflammatory response called steatohepatitis (NASH), which can progress to fibrosis. This hepatic fibrosis may produce cirrhosis and eventually, hepatocellular carcinoma (HCC)[2]. The first theory to explain NASH development, proposed in 1998, was known as the “two hits” hypothesis[10]. The first hit was fat storage in the hepatocytes, which would induce steatosis, the second hit being increased oxidative stress in the hepatocytes which would stimulate lipid peroxidation. It was believed this double hit was necessary to induce disease onset[10]. Currently, the “two hits” concept is considered old-fashioned by many experts. The hepatocyte needs several molecular and metabolic changes for its function to finally vary. Instead, it seems more precise to propose a “multiple hit” hypothesis[11]. This premise is intended to provide greater insight into NAFLD pathology and considers the different events that can take place in predisposed subjects during development of the disorder. Fat accumulation and synthesis of reactive oxygen species are essential events, yet other phenomena are also important and can be considered hallmarks of NAFLD initiation and progression (Figure 1).
Among environmental factors, the most prominent are dietary habits, physical activity, and socio-economic aspects. Increased calories intake, and consumption of high-sugar and high-fat diets increases the risk of developing not only NAFLD but also conditions such as obesity and type 2 diabetes mellitus[4,8,12]. Hallsworth et al[13] was the first to show an association between sedentary behavior and physical activity levels in patients with NAFLD, finding that these patients were on average more sedentary, walked less and spent less time on physical activity. Furthermore, it has been demonstrated that lifestyle interventions in diet and physical activity could improve the disease prognosis[12]. Finally, regarding socio-economic aspects, the role of educational level and family economic status in development of NAFLD is still under debate[4].
At the cellular level, important events such as mitochondrial dysfunction[14], endoplasmic reticulum (ER) stress[15,16], and activation of the inflammasome[17] contribute to fat accumulation in cells (steatosis) and inflammation. Genetic variants and epigenetic factors must also be taken into account in NAFLD progression[11,18]. A decade ago, PNPLA3 I148M was the first genetic variant reported to be associated with NAFLD. Currently, 13 genetic variants have been linked to increased risk of NAFLD or NASH, with the exception of the variant UCP2 866, which reduces the risk of NASH[18]. Some of these variants, such as TM6SF2, PNPLA3, NCAN, and PPP1R3B, have been linked to inherited NAFLD[8].
As a complete organ, the liver includes many non-parenchymal cells besides hepatocytes which contribute to the proper functioning of the organ. Among these are liver sinusoidal endothelial cells (LSECs), hepatic stellate cells (HSCs) and several immune cells, such as Kupffer cells[19,20]. Most of these cell types are essential to maintain homeostasis in the liver at the extracellular level, and alteration in their function has been associated with the NAFLD progression. LSECs maintain portal pressure and inhibit HSCs and Kupffer cells activation. During the first reversible stage of NAFLD, LSECs lose their functions, and, in turn, induce inflammation and fibrosis[21,22]. HSCs contribute to initiation and progression of liver fibrosis[19,23,24], one of the hallmarks of NAFLD evolution. Immune cells can be activated during liver disease creating a pro-inflammatory environment in the organ which contributes to NASH, fibrosis, cirrhosis, and HCC progression[19,25].
NAFLD illness is not limited to hepatic disease: the NAFLD liver interacts with other organs, creating an organ crosstalk[26], which provides further support for the “multiple hits” hypothesis. As a first example, adipose tissue (AT) dysfunction is related to NAFLD disease[11,26]. Excess fat consumption produces hypertrophy in adipocytes. AT can release several hormones or cytokines called adipokines which generate a pro-inflammatory environment[27]. This inflammatory state occurs first in the AT, then in the liver[28]. Furthermore, noncoding RNA[29] and extracellular vesicles[26,30] from the AT are linked to development of NAFLD and cell-to-cell communication. In the context of NAFLD, the gut-liver axis refers to the relationship between gut integrity, gut microbiota, and the liver[11,26,31]. Both organs are directly connected by the portal vein. In general, the gut presents different kinds of barriers and mechanisms to maintain its integrity. One function of these barriers is to control the passage of substances into the portal vein and the liver[31,32]. Further evidence suggests that the intestinal barriers are altered, and intestinal permeability is increased in NAFLD disease. Taking advantage of this altered permeability, bacteria can translocate more easily into the blood, enter the portal vein and finally reach the liver[32]. Increased gut permeability and bacterial translocation are associated with liver cirrhosis[31]. The gut microbiota is also altered in NAFLD due to intestinal microbial dysbiosis[33]. It has been shown that bacteria phyla are modified under high-fat diet-induced liver steatosis in rodent models[34] and human studies of NAFLD, NASH, and cirrhosis[35]. Variations in bacteria composition lead to altered concentration of some metabolites. This phenomenon, added to reduced permeability, triggers the arrival of molecules such as lipopolysaccharides in the systemic circulation and activation of Toll-like receptor in cells. Moreover, metabolism of trimethylamine which can be oxidized in the liver ultimately forming trimethylamine N-oxide, has been linked to NAFLD progression and cardiovascular disease[33]. Additionally, the liver has been associated with the brain[26]. The arcuate nucleus of the hypothalamus regulates satiety. In 2005, De Souza et al[36] proved that a high-fat diet caused several proinflammatory-related changes in mRNA expression in the hypothalamus of Wistar. Furthermore, cirrhotic patients can develop hepatic encephalopathy, a neurological comorbidity associated with NAFLD disease[37,38]. Finally, the kidney and the liver have also been linked. The study of Musso et al[39] in 2014 revealed that NAFLD severity was correlated with severity of chronic kidney disease (CKD). Many pathways are shared between NAFLD and CKD, so progression of NAFLD will contribute to CKD progression and vice versa[40].
NAFLD affects more men than women[41,42], due to the protective role of estrogens against disease development[43-46]. Nonetheless, in women of a certain age and under certain risk factors, incidence is higher than in men and they experience a more aggressive disease course. These risk factors are: (1) Earlier age of menarche; (2) Polycystic ovary syndrome; (3) Gestational diabetes; and (4) Menopause[46-49]. Interestingly, sex differences extend beyond incidence rates: NAFLD appears to develop in distinct ways in males and females[50-54]. However, further studies on about molecular processes are needed for enhanced insight into sexual dimorphism in NAFLD[55].
NAFLD is a complex disease which affects many hepatic parameters, as well as functions of other organs. With current methodologies, it is virtually impossible to study the “multiple hits” hypothesis of NAFLD as a whole in humans, because this requires access to multiple tissues, biofluids, and controlled environments. Animal models therefore remain essential for studying initiation and progression of NAFLD, and present various advantages over clinical research: (1) The possibility to obtain multiple samples and carry out longitudinal studies; (2) Shorter time to disease onset; (3) The possibility of controlling the variables of our model; and (4) Use of genetically modified animals to study a specific gene or metabolic pathway alteration. Compared to in vitro studies, animal models can be used to study the whole liver and organ crosstalk between the liver and other organs[56].
Nevertheless, a perfect animal model[9,57] providing information on all potential triggers and causes of NAFLD is elusive. Therefore, it is vital to know the stage of the disease to be studied and which model reproduces the physiopathological characteristics we want to study. Focusing on model selection, among key common characteristics, a good model must imitate certain aspects of the human NAFLD disorder, be reliable and reproducible, have low mortality, and be compatible with simple and viable methods[9]. Development of obesity or insulin resistance, AT inflammation, alterations of intestinal physiology, and a specific liver phenotype (Table 1) are traits that mimic human NAFLD[58,59]. Several animal models can be used to study metabolic diseases, including NAFLD, but rodents are the most commonly used. Rodent models are preferred because they easily develop obesity, type 2 diabetes mellitus, and NAFLD[60]. In mice, the ideal model genetic background is the strain C57BL/6, and specifically the substrain C57BL/6J, as C57BL/6J mice are more insulin resistant than C57BL/6N mice[61], which allows for better isolation of the NAFLD process from other metabolic alterations. For rat models, Wistar or Sprague Dawley rats are usually chosen, although other models besides rats and mice, such as New Zeland white rabbits, Guinea pigs, or Tree shrews, have also been used[60]. Rabbits, and many non-rodent models like pigs, have the important advantage of longer pre-pubertal stages, which allow them to mimic the subclinical NAFLD situation in children with greater precision than would be possible with mice or rats[59,62]. Also, pigs are anatomically and metabolically more similar to humans than rodent models. Nonetheless, these non-murine species have some drawbacks, such as they involve more complicated and less generally established genetic approaches, and housing larger animals can be more difficult from a logistic and economic point of view[59]. Models of smaller size and shorter lifetimes than mice and rats have also been explored. For example, use of zebrafish as a NAFLD model is recently increasing an inexpensive model in which NAFLD develops quickly[63].
| Rodent models | Obesity | Insulin resistance | Steatosis | NASH | Fibrosis | HCC |
| Dietary | ||||||
| Deficient diet | ||||||
| MCD | No | Hepatic IR | Yes | Yes | Yes | No |
| CDAA | No | No | Yes | Yes | Yes | Yes |
| High-amount diet | ||||||
| HFD | Yes | Yes | Yes | Yes | Yes | No |
| HFHS | Yes | Yes | Yes | Yes | Yes | No |
| High fructose diet | No | Yes | Yes | No | No | No |
| HFHC | Yes | Yes | Yes | Yes | Yes | No |
| Atherogenic diet (cholesterol + cholate) | No | Hepatic IR | Yes | Yes | Yes | No |
| Cafeteria diet or Western diet | Yes | Yes | Yes | Yes | No | - |
| ALIOS | Yes | Yes | Yes | Yes | Yes | Yes |
| AMLN | Yes | Yes | Yes | Yes | Yes | No |
| DIAMOND | Yes | Yes | Yes | Yes | Yes | Yes |
| Genetic | ||||||
| ob/ob | Yes | Yes | Yes | No | No | No |
| db/db | Yes | Yes | Yes | No | No | No |
| KK-Ay | Yes | Yes | Yes | No | No | No |
| foz/foz | Yes | Yes | Yes | No | No | No |
| fa/fa | Yes | Yes | Yes | No | No | No |
| PTEN knockout | No | No | Yes | Yes | Yes | Yes |
| PPAR-α knockout | No | No | Yes | No | No | No |
| SREBP-1c transgenic | No | Yes | Yes | No | No | No |
| Chemicals | ||||||
| Tetracycline | No | No | Yes | Yes | Yes | - |
| CCl4 | No | No | Yes | Yes | Yes | Yes |
| TAA | - | - | Yes | Yes | Yes | Yes |
| STZ | - | - | - | Yes | - | - |
| DMN | - | - | No | Yes | Yes | Yes |
| DEN | No | - | Yes | Yes | Yes | Yes |
| Porphyrinogenic agents (DDC or GF) | - | - | Yes | Yes | - | - |
| MSG | Yes | Yes | Yes | Yes | No | Yes |
| Tunicamycin | - | - | Yes | Yes | - | - |
| Surgical | ||||||
| CBDL | - | - | Yes | Yes | Yes | - |
| Combined models | ||||||
| ob/ob + MCD diet | Yes | - | Yes | Yes | No | No |
| db/db + MCD diet | Yes | Yes | Yes | Yes | Yes | No |
| HFD + thermoneutral housing at 30 ºC | - | - | - | Yes | Yes | - |
| HFD + CCl4 | No | - | Yes | Yes | Yes | Yes |
| HFD + DEN | Yes | - | Yes | Yes | Yes | Yes |
| CDAA + CCl4 | No | - | Yes | Yes | Yes | Yes |
| STAM model | No | - | Yes | Yes | Yes | Yes |
Despite the wide variety of models, rodents are still the preferred species for experimental NAFLD research because of their small size, ease of maintenance, short life span, and available genetic resources. The current rodent models used in NAFLD can be stratified into five main groups, depending on the disease inducer: dietary, genetic, chemical, surgical, and combined (mix of different models). Pathological characteristics of these rodent models are summarized in (Table 1).
Dietary models, which can be classified as deficient or high amount diets, are an excellent option for studying NAFLD disease. Deficient diets are not generally found in humans, as they are based on absence of essential elements. However, in animals, methionine and choline-deficient diet (MCD) or choline-deficient, L-amino defined diet are effective in generating liver damage[64,65]. The diets most closely resembling humans experience are the high amount calorie diets with an excessively high amount of specific nutrients, mainly fats and sugars[9,57,59,66]. Different diets can be defined by the high concentration of nutrients or how they are combined. Among these are high-fat diets, high-cholesterol and cholate diets (atherogenic diet), high-fat high-cholesterol diets, high-sugar diets based on fructose or sucrose, and high-fat high-sugar diets. Their effects on NAFLD development are shown in (Table 1). Lastly, there are different animal models of NAFLD based on diets that promote NASH in a short period: (1) American lifestyle-induced obesity syndrome model (ALIOS model); (2) Amylin liver NASH model (AMLN model); and (3) Diet-induced animal model of NAFLD mice (DIAMOND model)[57]. The ALIOS model is based on a high-fat diet (45% fats, 2% trans fats), drinking water with fructose and glucose, and a sedentary behavior (cages without wire racks), promoting obesity[67]. The AMLN model is based on a high-fat (40% fats, 18% trans fats), high-fructose (22%) and cholesterol (2%) diet[68,69]. ALIOS and AMLN are very similar, but with different fat percentages, and in the AMLN model fructose is given in pellet form rather than in drinking water[68]. A variant of the AMLN model called the Gubra amylin NASH (GAN) diet is currently used, with the same composition, but trans-fat-free diet and with increased saturated fatty acids[70]. The DIAMOND model is based on a high-fat (42%), high-carbohydrate and cholesterol (0.1%) diet but with an added high-fructose and glucose solution[71]. All these models are modified Western or Cafeteria diets (combination of fat and sugars) presenting more or less the same composition but in different proportions[63,72].
Genetic models allow us to study genetic and pathophysiological consequences of alterations in certain genes potentially involved in NAFLD development. These models are based on mechanistic hypotheses and have the main limitation that every specific mutation in a single gene is not usually found in humans[9]. Nevertheless, they provide two major advantages over other models: first, the means to study disease mechanisms in NAFLD, and second, the opportunity to improve our knowledge of a specific mechanism in the disease models[73]. Nowadays, genetic engineering tools have facilitated generation of transgenic animals and knockouts, either by commercial houses or in academic laboratories[62,73-75]. There are many genetic models for NAFLD, each one based on different pathways affected in the disease[76]. The genetic models most commonly used in the study of NAFLD are reported in (Table 1).
The most widespread chemical models for studying NAFLD are those based on liver damage through tetracycline, carbon tetrachloride (CCl4), thioacetamide (TAA), and streptozotocin[9,65]. These models can produce significant liver damage depending on the experimental exposure time (days, weeks, or months) and the dose delivered, but in the focus is generally, on producing liver steatosis and fibrosis[63,65,74,77]. Treatment with the chemicals diethylnitrosamine (DEN) or dimethylnitrosamine (DMN) is typically used to induce HCC and the approach may be too aggressive for studying NAFLD alone[56,60,74]. The porphyrinogenic agents (3,5-diethoxycarbonyly-1,4-dihydrocollidine (DDC) and griseofulvin (GF) and the chemical monosodium glutamate (MSG) are less often used but can also induce steatosis and NASH[78]. The chemical Tunicamycin produces ER stress in the hepatocytes which can in turn induce steatosis[79,80]. Overall, chemical models represent a faster and more dramatic way to study liver damage, but the disease initiation and progression bears less resemblance to human NAFLD than diet or genetic models.
Hepatobiliary system surgery can induce NAFLD in experimental models. The most common surgical model is Bile Duct Ligation (BDL), which is used to produce fibrosis, cirrhosis and as a consequence, liver failure in rodents[65,74,81]. BDL can be performed in mice and rats[65], but this model is difficult to implement in mice, as several surgical complications can arise[56]. Surgical models are the least used models of NAFLD because of their complexity and lack of similarity to human NAFLD.
Genetic models do not usually develop NASH, fibrosis, or HCC spontaneously, so they are often supplemented with diet to achieve worse liver damage[9,57,62]. This is also the case with chemical models, in which the dose for inducing liver damage is often too aggressive but combining a low dose with some NAFLD-inducing diet modifications can help producing a model that progresses at a slower pace, which allows detection of the different stages of NAFLD progression[60,65,66,77]. These combined models genetic plus diet or chemical plus diet, are also a common option for studying NAFLD[76].
Currently, liver function is routinely controlled by blood analysis in which clinicians test for transaminases, albumin, platelets, bilirubin and clotting factors. Patients presenting abnormal levels of these parameters, especially transaminases, and whose medical history reveals risk factors for diabetes, obesity or metabolic syndrome, undergoes a non-invasive imaging method, mainly ultrasonography and elastography, to confirm the presence of steatosis and fibrosis in the liver. If the result is positive, the NAFLD fibrosis score and FIB-4 index scores can be applied. Depending on the score, patients are classified as at low, medium or high risk of fibrosis. The goal of these imaging methods is to detect whether fibrosis is present, due to the different follow-up required in patients with fibrosis. An invasive imaging method, biopsy, is performed on those with a high risk of fibrosis or with an unclear diagnosis under non-invasive imaging methods[8,82-84]. Nowadays, biopsy remains the gold-standard for diagnosis of hepatic steatosis, NASH and fibrosis, as histology confirms tissue damage[7,8]. Biopsy has a relatively high incidence of false negatives, since the fragment finally analyzed only represents about 1/50000 of the organ and analysis may vary between pathologists[7]. Moreover, non-invasive imaging methods also present disadvantages. Steatosis can only be detected at over 30% and these methods cannot determine whether NASH is present[85,86]. We are still far from achieving the main objective: NAFLD prevention and a rapid diagnosis. New non-invasive diagnostic methods are needed, and one alternative could be use of metabolomics in the search for new biomarkers.
Personalized medicine has become a fundamental strategy in the future of healthcare. The possibility of tailor-made treatments for patient groups will help streamline healthcare costs and enhance efficacy and safety of interventions. The transition to a personalized medicine model has been facilitated by recent advances in "omics" technologies that are allowing the degree of personalization in the diagnosis and treatment of different diseases to be increased to levels unimaginable just a few years ago[87]. Metabolomics is an emerging research area and can be considered, at a biochemical level, as the end of the “omic” cascade since changes in the metabolome constitute the organism's last response to genetic, chemical and environmental alterations[63].
Small biochemicals are the end products of all the regulatory processes present in a cell, tissue, or organism, including transcriptional and translational regulation and posttranslational modifications. Consequently, metabolic changes are among the best reporters of the organism's response to a disease process. The application of metabolomics to the study of metabolic diseases may increase our understanding of the pathophysiological processes involved, and thus help us to identify potential biomarkers. The identification and quantification of these low molecular weight molecules define the metabolic phenotype of these diseases and studying the metabolic changes that occur in response to different pathophysiological processes may help establish the mechanisms underlying the disease.
Metabolites can be measured in several body fluids or tissues, although plasma and urine are the most frequently used samples in metabolic research, they are readily available and have clinical relevance as a source of potential biomarkers. Almost all cells in the body communicate with plasma, either directly or through different tissues and biological fluids, releasing at least part of their intracellular content. By contrast, urine is produced by renal filtration of plasma and is widely considered to be among the most important samples for diagnosis as it contains not only many plasma components but also the catabolic products of different metabolic pathways.
Metabolic fingerprinting and metabolic profiling are two different approaches to the study of metabolites in biological samples. Metabolic fingerprinting does not aim to identify the entire set of metabolites but rather to compare patterns or fingerprints of metabolites that change in response to a disease state, pharmacological therapies, or environmental alterations. This approach can be used as a diagnostic tool to evaluate the disease state by comparing healthy controls and disease subjects. Nonetheless, qualitative and quantitative analyses are required to understand the mechanisms underlying a disease. Metabolite profiling focuses on the analysis of a group of metabolites related to a specific metabolic pathway. In this approach, target metabolites are selected beforehand and are assessed using specific analytical methods.
The analytic techniques used to study the metabolome are mass spectroscopy (MS), nuclear magnetic resonance (NMR), or a combination of both[88,89]. Each technique has its own strengths and weaknesses[88,90]. An advantage of NMR technique, is that it can be used to study tissues, including liver, without destroying the sample with the proton high-resolution magic-angle spinning probe (HR-MAS)[90,91].
Metabolomics is a very powerful tool for the study of metabolic diseases[90,92], yet applications of metabolomics to NAFLD is an understudied area. Nonetheless, some studies demonstrate the importance of measuring metabolites for better characterization of the disease. NAFLD is a metabolic illness, hence metabolomics as a technique offers the opportunity to better understand the metabolic alterations in NAFLD progression[87,92,93] and patient stratification[89]. MS and NMR have been used to study NAFLD progression in rodent models. Articles yielded from the keyword search using the term "metabolomics" and "rodent models" are shown in (Table 2). Metabolomics studies have been carried out in dietary, chemical, genetic and combined models of NAFLD. Including metabolic alterations could broaden the search for specific metabolomics biomarkers which would help in disease diagnosis.
| Rodent model | Produced by | Animals used | Biological sample | Platform used | Ref. |
| Dietary | HFD | C57BL/6 mice. 6-wk-old | Liver extract and serum | UPLC-QTOF-MS and GC-MS | Kim et al[94] |
| HFD and Paigen diet | BALB/c mice. 6-wk-old | Liver extract and urine | 1H-NMR | Klein et al[95] | |
| HFD and HCD | C57BL/6N mice. 6-wk-old | Urine | 1H-NMR | Jung et al[96] | |
| HFD and HCD | Wistar rats. 6-wk-old | Liver extract | 1H-NMR | Bertram et al[97] | |
| HFD | C57BL/6S1ac mice. 4-wk-old | Urine | 1H-NMR and UPLC-QTOF-MS | Li et al[98] | |
| HFD | C57BL/6J mice. 6-wk-old | Serum | UHPLC-QTOF-MS and GC-MS | Lai et al[99] | |
| High-fructose and saturated fatty acid diet | Sprague-Dawley rats | Liver extract | HR-MAS and 1H-NMR | Tranchida et al[100] | |
| HFHCC diet | C57BL/6J mice. 8-wk-old | Liver extract and plasma | GC-TOF MS and CSH-QTOF MS | Tu et al[101] | |
| HFD | Sprague-Dawley rats. 4-6-wk-old | Liver extract | LC-MS | Wan et al[102] | |
| HFD | Sprague-Dawley. 6-wk-old | Urine and feces | 1H-NMR | Chen et al[103] | |
| MCD | C57BL/6J mice. 8-wk-old | Feces | GC-MS | Ye et al[104] | |
| HFD | Swiss albino mice | Serum and feces | 1H-NMR | Carvalho et al[105] | |
| High fat-sucrose diet | Sprague-Dawley rats. 6-wk-old | Serum | HPLC-QTOF-MS | Xu et al[106] | |
| MCD and atherogenic diet | C57BL/6J mice. 10-wk-old | Liver extract | MS | Montandon et al[107] | |
| HFD | Sprague-Dawley rats. 6-8-wk-old | Serum | LC-MS | Cui et al[108] | |
| HFD | Sprague-Dawley, Fisher 344 and Brown-Norway rats. 5-wk-old | Liver extract | LC-MS | Boyce et al[109] | |
| Genetic | Db/db mice | C57BL/6J mice. 10-wk-old | Liver extract | 1H-NMR and UPLC-QTOF-MS | Kim et al[110] |
| Ob/ob mice | B6.Cg-Lepob/J mice. 8-wk-old | Liver extract | HR-MAS and1H-NMR | Gogiashvili et al[111] | |
| Chemical | DEN | Sprague-Dawley rats. 4-wk-old | Liver extract | 1H-NMR | Wang et al[112] |
| CCl4 | Wistar rats | Plasma | UPLC-QTOF-MS | Li et al[113] | |
| CCl4 | Sprague-Dawley rats. 4-wk-old | Urine | GC-TOF MS | Jiang et al[114] | |
| CCl4 | Wistar rats | Liver extract | GC-MS | Song et al[115] | |
| CCl4 | Sprague-Dawley rats | Urine | 1H-NMR | Wu et al[116] | |
| CCl4 | Wistar rats | Urine | GC-MS | Fang et al[117] | |
| CCl4 | Sprague-Dawley rats. 1-yr-old | Serum and urine | UPLC-QTOF-MS | Chang et al[118] | |
| CCl4 | Sprague-Dawley rats. 7-wk-old | Serum | 1H-NMR | Li et al[119] | |
| CCl4 | Sprague-Dawley rats | Serum | 1H-NMR | Liu et al[120] | |
| DEN | Sprague-Dawley rats. 6-wk-old | Serum | 1H-NMR | Yang et al[121] | |
| Combined model | Combined (genetic + dietary) with HCD | Acyl knockouts mice on a C57BL6/J background. 4-wk-old | Serum | LC-MS | Zhao et al[122] |
Despite the diversity of models used in previous metabolomics studies on NAFLD rodent models (Table 2), some common findings can be extracted. Fatty acids are stored as triacylglycerols in the liver when not catabolized by β-oxidation. Consequently, fatty liver seems to be a rearrangement of lipids in the liver and not just fat storage. Most studies in liver tissue of rodent models have revealed massive accumulation of triacylglycerols (see liver extract studies in Table 2). The well-known adipocyte origin of some of these triacylglycerols suggests AT as a potential source of triacylglycerols deposited in the liver in NAFLD. Furthermore, almost all studies in NAFLD rodent models report alterations in other metabolites like glucose, lactate, pyruvate, and alanine, suggesting that NAFLD is involved in cytosolic glycolysis and oxidative stress[97,112,119]. Metabolism of branched-chain amino acids also seems to be altered in NAFLD. A previous study including human subjects and animal models in the context of hepatic insulin resistance demonstrated a link between BCAA and the tri-carboxylic acid cycle[106,108]. The integration of findings in human and rodent model studies seems very complex. In a translational human-animal study, Han et al[123] studied the progression of fatty liver and liver steatosis, finding changes in metabolic networks related to amino acids and bile acids. However, these results were significantly different between animals and humans. Among others, taurine, a well-known amino acid with protective and antioxidant properties, was increased in humans but not in rat models. Finally, consistent finding in different rodent and human studies on NAFLD is an increased level in serum of bile acids, important molecules which signal many processes in the liver and are involved in lipid and glucose homeostasis.
NAFLD is the most prevalent liver disease worldwide. Approaches from different perspectives have led to increased insight into many aspects of the disease. Knowledge of the disease has increased with the use of animal models, especially those in rodents. Although, the perfect animal model does not exist, some models perfectly mimic several aspects of NAFLD development and have become very useful tools to address the disease in the search for biomarkers of the early reversible stages. Studying metabolism in these models provides a direct reflection of what happens inside the cell. Metabolomics seems an important tool for studying metabolic pathways and crosstalk between organs affected in animal models of NAFLD, and for identifying and validating relevant biomarkers with biological understanding.
We are grateful to Clinical Hospital Research Foundation (INCLIVA) and Central Unit for Medical Research (UCIM) at University of Valencia for technical support during our research.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yang L S-Editor: Gao CC L-Editor: A P-Editor: Gao CC
| 1. | Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5322] [Cited by in RCA: 7539] [Article Influence: 837.7] [Reference Citation Analysis (0)] |
| 2. | Yu Y, Cai J, She Z, Li H. Insights into the Epidemiology, Pathogenesis, and Therapeutics of Nonalcoholic Fatty Liver Diseases. Adv Sci (Weinh). 2019;6:1801585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 3. | Allen AM, Therneau TM, Larson JJ, Coward A, Somers VK, Kamath PS. Nonalcoholic fatty liver disease incidence and impact on metabolic burden and death: A 20 year-community study. Hepatology. 2018;67:1726-1736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 263] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 4. | Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4054] [Cited by in RCA: 3795] [Article Influence: 542.1] [Reference Citation Analysis (2)] |
| 5. | Araújo AR, Rosso N, Bedogni G, Tiribelli C, Bellentani S. Global epidemiology of non-alcoholic fatty liver disease/non-alcoholic steatohepatitis: What we need in the future. Liver Int. 2018;38 Suppl 1:47-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 281] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 6. | Tanaka N, Kimura T, Fujimori N, Nagaya T, Komatsu M, Tanaka E. Current status, problems, and perspectives of non-alcoholic fatty liver disease research. World J Gastroenterol. 2019;25:163-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 116] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (4)] |
| 7. | Jennison E, Patel J, Scorletti E, Byrne CD. Diagnosis and management of non-alcoholic fatty liver disease. Postgrad Med J. 2019;95:314-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 8. | Perumpail BJ, Khan MA, Yoo ER, Cholankeril G, Kim D, Ahmed A. Clinical epidemiology and disease burden of nonalcoholic fatty liver disease. World J Gastroenterol. 2017;23:8263-8276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 543] [Cited by in RCA: 516] [Article Influence: 64.5] [Reference Citation Analysis (6)] |
| 9. | Zhong F, Zhou X, Xu J, Gao L. Rodent Models of Nonalcoholic Fatty Liver Disease. Digestion. 2020;101:522-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 10. | Day CP, James OF. Steatohepatitis: a tale of two "hits"? Gastroenterology. 1998;114:842-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2953] [Cited by in RCA: 3129] [Article Influence: 115.9] [Reference Citation Analysis (36)] |
| 11. | Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism. 2016;65:1038-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1490] [Cited by in RCA: 2119] [Article Influence: 235.4] [Reference Citation Analysis (1)] |
| 12. | Hallsworth K, Adams LA. Lifestyle modification in NAFLD/NASH: Facts and figures. JHEP Rep. 2019;1:468-479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 159] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 13. | Hallsworth K, Thoma C, Moore S, Ploetz T, Anstee QM, Taylor R, Day CP, Trenell MI. Non-alcoholic fatty liver disease is associated with higher levels of objectively measured sedentary behaviour and lower levels of physical activity than matched healthy controls. Frontline Gastroenterol. 2015;6:44-51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 14. | Simões ICM, Fontes A, Pinton P, Zischka H, Wieckowski MR. Mitochondria in non-alcoholic fatty liver disease. Int J Biochem Cell Biol. 2018;95:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 184] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 15. | Xia SW, Wang ZM, Sun SM, Su Y, Li ZH, Shao JJ, Tan SZ, Chen AP, Wang SJ, Zhang ZL, Zhang F, Zheng SZ. Endoplasmic reticulum stress and protein degradation in chronic liver disease. Pharmacol Res. 2020;161:105218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 16. | Lebeaupin C, Vallée D, Hazari Y, Hetz C, Chevet E, Bailly-Maitre B. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. J Hepatol. 2018;69:927-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 657] [Article Influence: 93.9] [Reference Citation Analysis (0)] |
| 17. | Wan X, Xu C, Yu C, Li Y. Role of NLRP3 Inflammasome in the Progression of NAFLD to NASH. Can J Gastroenterol Hepatol. 2016;2016:6489012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 128] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 18. | Chandrasekharan K, Alazawi W. Genetics of Non-Alcoholic Fatty Liver and Cardiovascular Disease: Implications for Therapy? Front Pharmacol. 2019;10:1413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Hunt NJ, Kang SWS, Lockwood GP, Le Couteur DG, Cogger VC. Hallmarks of Aging in the Liver. Comput Struct Biotechnol J. 2019;17:1151-1161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 20. | Sato K, Kennedy L, Liangpunsakul S, Kusumanchi P, Yang Z, Meng F, Glaser S, Francis H, Alpini G. Intercellular Communication between Hepatic Cells in Liver Diseases. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 21. | Wilkinson AL, Qurashi M, Shetty S. The Role of Sinusoidal Endothelial Cells in the Axis of Inflammation and Cancer Within the Liver. Front Physiol. 2020;11:990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 22. | Furuta K, Guo Q, Hirsova P, Ibrahim SH. Emerging Roles of Liver Sinusoidal Endothelial Cells in Nonalcoholic Steatohepatitis. Biology (Basel). 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev. 2017;121:27-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 1059] [Article Influence: 132.4] [Reference Citation Analysis (0)] |
| 24. | Khomich O, Ivanov AV, Bartosch B. Metabolic Hallmarks of Hepatic Stellate Cells in Liver Fibrosis. Cells. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 25. | Arrese M, Cabrera D, Kalergis AM, Feldstein AE. Innate Immunity and Inflammation in NAFLD/NASH. Dig Dis Sci. 2016;61:1294-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 383] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 26. | Zhang X, Ji X, Wang Q, Li JZ. New insight into inter-organ crosstalk contributing to the pathogenesis of non-alcoholic fatty liver disease (NAFLD). Protein Cell. 2018;9:164-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 27. | Boutari C, Perakakis N, Mantzoros CS. Association of Adipokines with Development and Progression of Nonalcoholic Fatty Liver Disease. Endocrinol Metab (Seoul). 2018;33:33-43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 28. | van der Heijden RA, Sheedfar F, Morrison MC, Hommelberg PP, Kor D, Kloosterhuis NJ, Gruben N, Youssef SA, de Bruin A, Hofker MH, Kleemann R, Koonen DP, Heeringa P. High-fat diet induced obesity primes inflammation in adipose tissue prior to liver in C57BL/6j mice. Aging (Albany NY). 2015;7:256-268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 215] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 29. | Ma M, Duan R, Zhong H, Liang T, Guo L. The Crosstalk between Fat Homeostasis and Liver Regional Immunity in NAFLD. J Immunol Res. 2019;2019:3954890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | Gao X, Salomon C, Freeman DJ. Extracellular Vesicles from Adipose Tissue-A Potential Role in Obesity and Type 2 Diabetes? Front Endocrinol (Lausanne). 2017;8:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 31. | Chen D, Le TH, Shahidipour H, Read SA, Ahlenstiel G. The Role of Gut-Derived Microbial Antigens on Liver Fibrosis Initiation and Progression. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 32. | Di Ciaula A, Baj J, Garruti G, Celano G, De Angelis M, Wang HH, Di Palo DM, Bonfrate L, Wang DQ, Portincasa P. Liver Steatosis, Gut-Liver Axis, Microbiome and Environmental Factors. A Never-Ending Bidirectional Cross-Talk. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 33. | Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol. 2020;20:40-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 663] [Article Influence: 110.5] [Reference Citation Analysis (0)] |
| 34. | Gómez-Zorita S, Aguirre L, Milton-Laskibar I, Fernández-Quintela A, Trepiana J, Kajarabille N, Mosqueda-Solís A, González M, Portillo MP. Relationship between Changes in Microbiota and Liver Steatosis Induced by High-Fat Feeding-A Review of Rodent Models. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 35. | Safari Z, Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell Mol Life Sci. 2019;76:1541-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 339] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 36. | De Souza CT, Araujo EP, Bordin S, Ashimine R, Zollner RL, Boschero AC, Saad MJ, Velloso LA. Consumption of a fat-rich diet activates a proinflammatory response and induces insulin resistance in the hypothalamus. Endocrinology. 2005;146:4192-4199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 799] [Cited by in RCA: 857] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 37. | Suraweera D, Sundaram V, Saab S. Evaluation and Management of Hepatic Encephalopathy: Current Status and Future Directions. Gut Liver. 2016;10:509-519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 38. | Tapper EB, Henderson JB, Parikh ND, Ioannou GN, Lok AS. Incidence of and Risk Factors for Hepatic Encephalopathy in a Population-Based Cohort of Americans With Cirrhosis. Hepatol Commun. 2019;3:1510-1519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 39. | Musso G, Gambino R, Tabibian JH, Ekstedt M, Kechagias S, Hamaguchi M, Hultcrantz R, Hagström H, Yoon SK, Charatcharoenwitthaya P, George J, Barrera F, Hafliðadóttir S, Björnsson ES, Armstrong MJ, Hopkins LJ, Gao X, Francque S, Verrijken A, Yilmaz Y, Lindor KD, Charlton M, Haring R, Lerch MM, Rettig R, Völzke H, Ryu S, Li G, Wong LL, Machado M, Cortez-Pinto H, Yasui K, Cassader M. Association of non-alcoholic fatty liver disease with chronic kidney disease: a systematic review and meta-analysis. PLoS Med. 2014;11:e1001680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 545] [Cited by in RCA: 525] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 40. | Targher G, Byrne CD. Non-alcoholic fatty liver disease: an emerging driving force in chronic kidney disease. Nat Rev Nephrol. 2017;13:297-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 236] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 41. | Kirsch R, Clarkson V, Shephard EG, Marais DA, Jaffer MA, Woodburne VE, Kirsch RE, Hall Pde L. Rodent nutritional model of non-alcoholic steatohepatitis: species, strain and sex difference studies. J Gastroenterol Hepatol. 2003;18:1272-1282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 192] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 42. | Suzuki A, Abdelmalek MF. Nonalcoholic fatty liver disease in women. Womens Health (Lond). 2009;5:191-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 43. | Kamada Y, Kiso S, Yoshida Y, Chatani N, Kizu T, Hamano M, Tsubakio M, Takemura T, Ezaki H, Hayashi N, Takehara T. Estrogen deficiency worsens steatohepatitis in mice fed high-fat and high-cholesterol diet. Am J Physiol Gastrointest Liver Physiol. 2011;301:G1031-G1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 127] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 44. | Tobari M, Hashimoto E. Characteristic Features of Nonalcoholic Fatty Liver Disease in Japan with a Focus on the Roles of Age, Sex and Body Mass Index. Gut Liver. 2020;14:537-545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 45. | Hashimoto E, Tokushige K. Prevalence, gender, ethnic variations, and prognosis of NASH. J Gastroenterol. 2011;46 Suppl 1:63-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 199] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 46. | Yuan L, Kardashian A, Sarkar M. NAFLD in women: Unique pathways, biomarkers and therapeutic opportunities. Curr Hepatol Rep. 2019;18:425-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 47. | Ballestri S, Nascimbeni F, Baldelli E, Marrazzo A, Romagnoli D, Lonardo A. NAFLD as a Sexual Dimorphic Disease: Role of Gender and Reproductive Status in the Development and Progression of Nonalcoholic Fatty Liver Disease and Inherent Cardiovascular Risk. Adv Ther. 2017;34:1291-1326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 411] [Cited by in RCA: 397] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 48. | DiStefano JK. NAFLD and NASH in Postmenopausal Women: Implications for Diagnosis and Treatment. Endocrinology. 2020;161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 49. | Lonardo A, Mantovani A, Lugari S, Targher G. NAFLD in Some Common Endocrine Diseases: Prevalence, Pathophysiology, and Principles of Diagnosis and Management. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 50. | Bisaccia G, Ricci F, Mantini C, Tana C, Romani GL, Schiavone C, Gallina S. Nonalcoholic fatty liver disease and cardiovascular disease phenotypes. SAGE Open Med. 2020;8:2050312120933804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 51. | Stöppeler S, Palmes D, Fehr M, Hölzen JP, Zibert A, Siaj R, Schmidt HH, Spiegel HU, Bahde R. Gender and strain-specific differences in the development of steatosis in rats. Lab Anim. 2013;47:43-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 52. | Lee YH, Kim SH, Kim SN, Kwon HJ, Kim JD, Oh JY, Jung YS. Sex-specific metabolic interactions between liver and adipose tissue in MCD diet-induced non-alcoholic fatty liver disease. Oncotarget. 2016;7:46959-46971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 53. | Norheim F, Hui ST, Kulahcioglu E, Mehrabian M, Cantor RM, Pan C, Parks BW, Lusis AJ. Genetic and hormonal control of hepatic steatosis in female and male mice. J Lipid Res. 2017;58:178-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 54. | Palmisano BT, Zhu L, Stafford JM. Role of Estrogens in the Regulation of Liver Lipid Metabolism. Adv Exp Med Biol. 2017;1043:227-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 318] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 55. | Kurt Z, Barrere-Cain R, LaGuardia J, Mehrabian M, Pan C, Hui ST, Norheim F, Zhou Z, Hasin Y, Lusis AJ, Yang X. Tissue-specific pathways and networks underlying sexual dimorphism in non-alcoholic fatty liver disease. Biol Sex Differ. 2018;9:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 56. | Delire B, Stärkel P, Leclercq I. Animal Models for Fibrotic Liver Diseases: What We Have, What We Need, and What Is under Development. J Clin Transl Hepatol. 2015;3:53-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 57. | Bertola A. Rodent models of fatty liver diseases. Liver Res. 2018;2:3-13. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (1)] |
| 58. | Takahashi Y, Soejima Y, Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2012;18:2300-2308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 379] [Cited by in RCA: 414] [Article Influence: 31.8] [Reference Citation Analysis (2)] |
| 59. | Jahn D, Kircher S, Hermanns HM, Geier A. Animal models of NAFLD from a hepatologist's point of view. Biochim Biophys Acta Mol Basis Dis. 2019;1865:943-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 60. | Van Herck MA, Vonghia L, Francque SM. Animal Models of Nonalcoholic Fatty Liver Disease-A Starter's Guide. Nutrients. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 263] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 61. | Fontaine DA, Davis DB. Attention to Background Strain Is Essential for Metabolic Research: C57BL/6 and the International Knockout Mouse Consortium. Diabetes. 2016;65:25-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 171] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 62. | Sanches SC, Ramalho LN, Augusto MJ, da Silva DM, Ramalho FS. Nonalcoholic Steatohepatitis: A Search for Factual Animal Models. Biomed Res Int. 2015;2015:574832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 136] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 63. | Willebrords J, Pereira IV, Maes M, Crespo Yanguas S, Colle I, Van Den Bossche B, Da Silva TC, de Oliveira CP, Andraus W, Alves VA, Cogliati B, Vinken M. Strategies, models and biomarkers in experimental non-alcoholic fatty liver disease research. Prog Lipid Res. 2015;59:106-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 64. | Hebbard L, George J. Animal models of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2011;8:35-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 384] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 65. | Nevzorova YA, Boyer-Diaz Z, Cubero FJ, Gracia-Sancho J. Animal models for liver disease - A practical approach for translational research. J Hepatol. 2020;73:423-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 162] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 66. | Lau JK, Zhang X, Yu J. Animal models of non-alcoholic fatty liver disease: current perspectives and recent advances. J Pathol. 2017;241:36-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 261] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 67. | Tetri LH, Basaranoglu M, Brunt EM, Yerian LM, Neuschwander-Tetri BA. Severe NAFLD with hepatic necroinflammatory changes in mice fed trans fats and a high-fructose corn syrup equivalent. Am J Physiol Gastrointest Liver Physiol. 2008;295:G987-G995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 330] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 68. | Clapper JR, Hendricks MD, Gu G, Wittmer C, Dolman CS, Herich J, Athanacio J, Villescaz C, Ghosh SS, Heilig JS, Lowe C, Roth JD. Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am J Physiol Gastrointest Liver Physiol. 2013;305:G483-G495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 225] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 69. | Gwag T, Reddy Mooli RG, Li D, Lee S, Lee EY, Wang S. Macrophage-derived thrombospondin 1 promotes obesity-associated non-alcoholic fatty liver disease. JHEP Rep. 2021;3:100193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 70. | Boland ML, Oró D, Tølbøl KS, Thrane ST, Nielsen JC, Cohen TS, Tabor DE, Fernandes F, Tovchigrechko A, Veidal SS, Warrener P, Sellman BR, Jelsing J, Feigh M, Vrang N, Trevaskis JL, Hansen HH. Towards a standard diet-induced and biopsy-confirmed mouse model of non-alcoholic steatohepatitis: Impact of dietary fat source. World J Gastroenterol. 2019;25:4904-4920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (3)] |
| 71. | Asgharpour A, Cazanave SC, Pacana T, Seneshaw M, Vincent R, Banini BA, Kumar DP, Daita K, Min HK, Mirshahi F, Bedossa P, Sun X, Hoshida Y, Koduru SV, Contaifer D Jr, Warncke UO, Wijesinghe DS, Sanyal AJ. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J Hepatol. 2016;65:579-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 365] [Cited by in RCA: 392] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 72. | Sampey BP, Vanhoose AM, Winfield HM, Freemerman AJ, Muehlbauer MJ, Fueger PT, Newgard CB, Makowski L. Cafeteria diet is a robust model of human metabolic syndrome with liver and adipose inflammation: comparison to high-fat diet. Obesity (Silver Spring). 2011;19:1109-1117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 440] [Cited by in RCA: 438] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 73. | Vandamme TF. Use of rodents as models of human diseases. J Pharm Bioallied Sci. 2014;6:2-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 236] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 74. | Liu Y, Meyer C, Xu C, Weng H, Hellerbrand C, ten Dijke P, Dooley S. Animal models of chronic liver diseases. Am J Physiol Gastrointest Liver Physiol. 2013;304:G449-G468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 75. | Haemmerle G, Lass A. Genetically modified mouse models to study hepatic neutral lipid mobilization. Biochim Biophys Acta Mol Basis Dis. 2019;1865:879-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 76. | Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1376] [Cited by in RCA: 2918] [Article Influence: 416.9] [Reference Citation Analysis (1)] |
| 77. | Hundertmark J, Tacke F. How effective are nonalcoholic fatty liver disease models for drug discovery? Expert Opin Drug Discov. 2020;15:1237-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 78. | Denk H, Abuja PM, Zatloukal K. Animal models of NAFLD from the pathologist's point of view. Biochim Biophys Acta Mol Basis Dis. 2019;1865:929-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 79. | Chen Y, Zhang H, Chen Y, Zhang Y, Shen M, Jia P, Ji S, Wang T. Resveratrol Alleviates Endoplasmic Reticulum Stress-Associated Hepatic Steatosis and Injury in Mice Challenged with Tunicamycin. Mol Nutr Food Res. 2020;64:e2000105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 80. | Lee JS, Zheng Z, Mendez R, Ha SW, Xie Y, Zhang K. Pharmacologic ER stress induces non-alcoholic steatohepatitis in an animal model. Toxicol Lett. 2012;211:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 81. | Geerts AM, Vanheule E, Praet M, Van Vlierberghe H, De Vos M, Colle I. Comparison of three research models of portal hypertension in mice: macroscopic, histological and portal pressure evaluation. Int J Exp Pathol. 2008;89:251-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 82. | Stefan N, Häring HU, Cusi K. Non-alcoholic fatty liver disease: causes, diagnosis, cardiometabolic consequences, and treatment strategies. Lancet Diabetes Endocrinol. 2019;7:313-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 586] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 83. | Long MT, Gandhi S, Loomba R. Advances in non-invasive biomarkers for the diagnosis and monitoring of non-alcoholic fatty liver disease. Metabolism. 2020;111S:154259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 84. | Vilar-Gomez E, Chalasani N. Non-invasive assessment of non-alcoholic fatty liver disease: Clinical prediction rules and blood-based biomarkers. J Hepatol. 2018;68:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 447] [Article Influence: 63.9] [Reference Citation Analysis (0)] |
| 85. | Li Q, Dhyani M, Grajo JR, Sirlin C, Samir AE. Current status of imaging in nonalcoholic fatty liver disease. World J Hepatol. 2018;10:530-542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (3)] |
| 86. | Koplay M, Sivri M, Erdogan H, Nayman A. Importance of imaging and recent developments in diagnosis of nonalcoholic fatty liver disease. World J Hepatol. 2015;7:769-776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (2)] |
| 87. | Dumas ME, Kinross J, Nicholson JK. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology. 2014;146:46-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 143] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 88. | Marshall DD, Powers R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog Nucl Magn Reson Spectrosc. 2017;100:1-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 165] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 89. | Holmes E, Wijeyesekera A, Taylor-Robinson SD, Nicholson JK. The promise of metabolic phenotyping in gastroenterology and hepatology. Nat Rev Gastroenterol Hepatol. 2015;12:458-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 90. | Emwas A, Salek RM, Griffin JL, Merzaban J. NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics. 2013;9:1048-1072. [DOI] [Full Text] |
| 91. | Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, Holmes E, Lindon JC, Nicholson JK. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5:1019-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 265] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 92. | Newgard CB. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017;25:43-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 543] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 93. | Piazzolla VA, Mangia A. Noninvasive Diagnosis of NAFLD and NASH. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 156] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 94. | Kim HJ, Kim JH, Noh S, Hur HJ, Sung MJ, Hwang JT, Park JH, Yang HJ, Kim MS, Kwon DY, Yoon SH. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J Proteome Res. 2011;10:722-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 312] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 95. | Klein MS, Dorn C, Saugspier M, Hellerbrand C, Oefner PJ, Gronwald W. Discrimination of steatosis and NASH in mice using nuclear magnetic resonance spectroscopy. Metabolomics. 2011;7:237-246. [DOI] [Full Text] |
| 96. | Jung JY, Kim IY, Kim YN, Kim JS, Shin JH, Jang ZH, Lee HS, Hwang GS, Seong JK. 1H NMR-based metabolite profiling of diet-induced obesity in a mouse mode. BMB Rep. 2012;45:419-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 97. | Bertram HC, Larsen LB, Chen X, Jeppesen PB. Impact of high-fat and high-carbohydrate diets on liver metabolism studied in a rat model with a systems biology approach. J Agric Food Chem. 2012;60:676-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 98. | Li ZY, Ding LL, Li JM, Xu BL, Yang L, Bi KS, Wang ZT. ¹H-NMR and MS based metabolomics study of the intervention effect of curcumin on hyperlipidemia mice induced by high-fat diet. PLoS One. 2015;10:e0120950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 99. | Lai YS, Chen WC, Kuo TC, Ho CT, Kuo CH, Tseng YJ, Lu KH, Lin SH, Panyod S, Sheen LY. Mass-Spectrometry-Based Serum Metabolomics of a C57BL/6J Mouse Model of High-Fat-Diet-Induced Non-alcoholic Fatty Liver Disease Development. J Agric Food Chem. 2015;63:7873-7884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 100. | Tranchida F, Rakotoniaina Z, Shintu L, Tchiakpe L, Deyris V, Yemloul M, Stocker P, Vidal N, Rimet O, Hiol A, Caldarelli S. Hepatic metabolic effects of Curcuma longa extract supplement in high-fructose and saturated fat fed rats. Sci Rep. 2017;7:5880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 101. | Tu LN, Showalter MR, Cajka T, Fan S, Pillai VV, Fiehn O, Selvaraj V. Metabolomic characteristics of cholesterol-induced non-obese nonalcoholic fatty liver disease in mice. Sci Rep. 2017;7:6120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 102. | Wan W, Jiang B, Sun L, Xu L, Xiao P. Metabolomics reveals that vine tea (Ampelopsis grossedentata) prevents high-fat-diet-induced metabolism disorder by improving glucose homeostasis in rats. PLoS One. 2017;12:e0182830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 103. | Chen M, Lu B, Li Y, Wang Y, Zheng H, Zhong D, Liao Z, Wang M, Ma F, Liao Q, Xie Z. Metabolomics insights into the modulatory effects of long-term compound polysaccharide intake in high-fat diet-induced obese rats. Nutr Metab (Lond). 2018;15:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 104. | Ye JZ, Li YT, Wu WR, Shi D, Fang DQ, Yang LY, Bian XY, Wu JJ, Wang Q, Jiang XW, Peng CG, Ye WC, Xia PC, Li LJ. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J Gastroenterol. 2018;24:2468-2481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 105. | Carvalho DV, Silva LMA, Alves Filho EG, Santos FA, Lima RP, Viana AFSC, Nunes PIG, Fonseca SGDC, Melo TS, Viana DA, Gallão MI, Brito ES. Cashew apple fiber prevents high fat diet-induced obesity in mice: an NMR metabolomic evaluation. Food Funct. 2019;10:1671-1683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 106. | Xu Y, Han J, Dong J, Fan X, Cai Y, Li J, Wang T, Zhou J, Shang J. Metabolomics Characterizes the Effects and Mechanisms of Quercetin in Nonalcoholic Fatty Liver Disease Development. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 107. | Montandon SA, Somm E, Loizides-Mangold U, de Vito C, Dibner C, Jornayvaz FR. Multi-technique comparison of atherogenic and MCD NASH models highlights changes in sphingolipid metabolism. Sci Rep. 2019;9:16810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 108. | Cui H, Li Y, Cao M, Liao J, Liu X, Miao J, Fu H, Song R, Wen W, Zhang Z, Wang H. Untargeted Metabolomic Analysis of the Effects and Mechanism of Nuciferine Treatment on Rats With Nonalcoholic Fatty Liver Disease. Front Pharmacol. 2020;11:858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 109. | Boyce G, Shoeb M, Kodali V, Meighan T, Roberts JR, Erdely A, Kashon M, Antonini JM. Using liquid chromatography mass spectrometry (LC-MS) to assess the effect of age, high-fat diet, and rat strain on the liver metabolome. PLoS One. 2020;15:e0235338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 110. | Kim KE, Jung Y, Min S, Nam M, Heo RW, Jeon BT, Song DH, Yi CO, Jeong EA, Kim H, Kim J, Jeong SY, Kwak W, Ryu do H, Horvath TL, Roh GS, Hwang GS. Caloric restriction of db/db mice reverts hepatic steatosis and body weight with divergent hepatic metabolism. Sci Rep. 2016;6:30111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 111. | Gogiashvili M, Edlund K, Gianmoena K, Marchan R, Brik A, Andersson JT, Lambert J, Madjar K, Hellwig B, Rahnenführer J, Hengstler JG, Hergenröder R, Cadenas C. Metabolic profiling of ob/ob mouse fatty liver using HR-MAS 1H-NMR combined with gene expression analysis reveals alterations in betaine metabolism and the transsulfuration pathway. Anal Bioanal Chem. 2017;409:1591-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 112. | Wang J, Zhang S, Li Z, Yang J, Huang C, Liang R, Liu Z, Zhou R. (1)H-NMR-based metabolomics of tumor tissue for the metabolic characterization of rat hepatocellular carcinoma formation and metastasis. Tumour Biol. 2011;32:223-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 113. | Li Y, Wang L, Ju L, Deng H, Zhang Z, Hou Z, Xie J, Wang Y, Zhang Y. A Systematic Strategy for Screening and Application of Specific Biomarkers in Hepatotoxicity Using Metabolomics Combined With ROC Curves and SVMs. Toxicol Sci. 2016;150:390-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 114. | Jiang H, Song JM, Gao PF, Qin XJ, Xu SZ, Zhang JF. Metabolic characterization of the early stage of hepatic fibrosis in rat using GC-TOF/MS and multivariate data analyses. Biomed Chromatogr. 2017;31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 115. | Song YN, Dong S, Wei B, Liu P, Zhang YY, Su SB. Metabolomic mechanisms of gypenoside against liver fibrosis in rats: An integrative analysis of proteomics and metabolomics data. PLoS One. 2017;12:e0173598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 116. | Wu F, Zheng H, Yang ZT, Cheng B, Wu JX, Liu XW, Tang CL, Lu SY, Chen ZN, Song FM, Ruan JX, Zhang HY, Liang YH, Song H, Su ZH. Urinary metabonomics study of the hepatoprotective effects of total alkaloids from Corydalis saxicola Bunting on carbon tetrachloride-induced chronic hepatotoxicity in rats using 1H NMR analysis. J Pharm Biomed Anal. 2017;140:199-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 117. | Fang J, Wang L, Wang Y, Qiu M, Zhang Y. Metabolomics combined with pattern recognition and bioinformatics analysis methods for the development of pharmacodynamic biomarkers on liver fibrosis. Mol Biosyst. 2017;13:1575-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 118. | Chang H, Meng HY, Liu SM, Wang Y, Yang XX, Lu F, Wang HY. Identification of key metabolic changes during liver fibrosis progression in rats using a urine and serum metabolomics approach. Sci Rep. 2017;7:11433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 119. | Li Z, Li Y, Lu L, Yang Z, Xue W, Tian X, Zhang X. 1H-NMR Based Serum Metabolomics Study to Investigate Hepatoprotective Effect of Qin-Jiao on Carbon Tetrachloride-Induced Acute Hepatotoxicity in Rats. Evid Based Complement Alternat Med. 2017;2017:6091589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 120. | Liu XW, Tang CL, Zheng H, Wu JX, Wu F, Mo YY, Liu X, Zhu HJ, Yin CL, Cheng B, Ruan JX, Song FM, Chen ZN, Song H, Guo HW, Liang YH, Su ZH. Investigation of the hepatoprotective effect of Corydalis saxicola Bunting on carbon tetrachloride-induced liver fibrosis in rats by 1H-NMR-based metabonomics and network pharmacology approaches. J Pharm Biomed Anal. 2018;159:252-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 121. | Yang W, Zhou G, Zou S, Yang W, Liu A, Sun S, Xie B. Metabonomics of d-glucaro-1,4-lactone in preventing diethylnitrosamine-induced liver cancer in rats. Pharm Biol. 2018;56:643-648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 122. | Zhao S, Jang C, Liu J, Uehara K, Gilbert M, Izzo L, Zeng X, Trefely S, Fernandez S, Carrer A, Miller KD, Schug ZT, Snyder NW, Gade TP, Titchenell PM, Rabinowitz JD, Wellen KE. Dietary fructose feeds hepatic lipogenesis via microbiota-derived acetate. Nature. 2020;579:586-591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 412] [Cited by in RCA: 366] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 123. | Han J, Dzierlenga AL, Lu Z, Billheimer DD, Torabzadeh E, Lake AD, Li H, Novak P, Shipkova P, Aranibar N, Robertson D, Reily MD, Lehman-McKeeman LD, Cherrington NJ. Metabolomic profiling distinction of human nonalcoholic fatty liver disease progression from a common rat model. Obesity (Silver Spring). 2017;25:1069-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |