Published online Dec 27, 2022. doi: 10.4254/wjh.v14.i12.1997
Peer-review started: September 5, 2022
First decision: September 30, 2022
Revised: October 12, 2022
Accepted: November 22, 2022
Article in press: November 22, 2022
Published online: December 27, 2022
Processing time: 109 Days and 7.7 Hours
Hepatitis B virus (HBV) infection is a major factor responsible for HBV+ hepatocellular carcinoma (HCC).
An immunological classification of HBV+ HCC may provide both biological in
Based on the enrichment of 23 immune signatures, we identified two immune-specific subtypes (Imm-H and Imm-L) of HBV+ HCC by unsupervised clustering. We showed that this subtyping method was reproducible and predictable by analyzing three different datasets.
Compared to Imm-L, Imm-H displayed stronger immunity, more stromal com
Our data suggest that “hot” tumors have a better prognosis than “cold” tumors in HBV+ HCC and that “hot” tumors respond better to immunotherapy.
Core Tip: First, for the first time, we identified immune-specific subtypes of hepatitis B virus (HBV) + hepatocellular carcinoma (HCC) based on immune signature scores and demonstrated that this new subtyping method was reproducible in three different datasets. Second, our subtyping method captures the comprehensive heterogeneity of HBV+ HCC in the tumor microenvironment, genomic integrity, protein expression profiles, DNA methylation profiles, tumor stemness, intratumor heterogeneity, and clinical outcomes. Third, our data suggest that it is copy number alterations but not tumor mutations responsible for the different immunity between the “hot” and “cold” tumor subtypes in HBV+ HCC. Finally, our identification of the immune-specific subtypes of HBV+ HCC may provide new insights into the tumor biology and identify the HBV+ HCC patients beneficial from immunotherapy.
- Citation: Li SW, Han LF, He Y, Wang XS. Immunological classification of hepatitis B virus-positive hepatocellular carcinoma by transcriptome analysis. World J Hepatol 2022; 14(12): 1997-2011
- URL: https://www.wjgnet.com/1948-5182/full/v14/i12/1997.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i12.1997
Hepatocellular carcinoma (HCC) is a major cancer of the liver that constitutes around 90% of liver cancer cases[1]. Although traditional therapeutic approaches, including surgery, chemotherapy, radiotherapy, and targeted therapy, are effective in improving the survival of HCC patients, the overall survival prognosis of HCC patients is generally unfavorable[2]. More recently, immunotherapy, such as immune checkpoint blockade (ICB), has shown success in the treatment of various cancers, including HCC[3]. However, only a small proportion of cancer patients respond well to immunotherapies to date[4]. To this end, certain predictive markers for cancer immunotherapy responses have been uncovered, e.g., PD-L1 expression[5], tumor mutation burden (TMB)[6], and mismatch repair deficiency[7]. In addition, the tumor immune microenvironment (TIME) plays an important role in immunotherapy responses[8]. Overall, the “hot” tumors infiltrated by a substantial number of tumor-infiltrating lymphocytes (TILs) are more responsive to immunotherapies, compared to the “cold” tumors lacking TILs[9]. Hence, an investigation of the TIME in HCC would aid in the prediction of immunotherapy responses.
With the recent emergence of large-scale cancer genomics data, such as the Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and International Cancer Genome Consortium (ICGC) (https://dcc.icgc.org/), many studies have investigated the TIME in HCC based on these data[10-12]. For example, Gao et al[10] identified four immune-relevant subtypes of HCC based on the enrichment of 13 signatures and revealed significantly different molecular and clinical characteristics among these subtypes. Sia et al[11] uncovered an immune subclass of HCC representing nearly 25% of HCC cases, based on gene expression profiles in tumor, stromal, and immune cells. Based on the enrichment of immune cell subpopulations, Farha et al[12] identified two immune clusters of HCC, and found that the cluster enriched with M0 macrophages had a worse prognosis.
Despite these previous studies[10-12], the discovery of immune-specific subtypes of hepatitis B virus-positive (HBV+) HCC is worth investigating, considering that HBV infection is a major cause of HCC[13]. In this study, to characterize the immunological landscape of HBV+ HCC, we identified its immune-specific subtypes by the unsupervised machine learning in transcriptomic data. Furthermore, we comprehensively compared the clinical and molecular features of these subtypes. Our analysis would provide new insights into the HBV+ HCC immunity and its associated clinical and molecular features, as well as potential clinical implications for the management of this disease.
We obtained the TCGA Hepatocellular Carcinoma (TCGA-LIHC) dataset, including transcriptomes (RSEM-normalized RNA-Seq gene expression profiles), somatic mutations (“maf” file), somatic copy number alterations (SCNAs) (“SNP6” files), normalized protein expression profiles by Reverse Phase Protein Array (RPPA), and clinical data, from the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/). We obtained other two HCC transcriptomic datasets (GSE14520 and GSE121248) from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). A description of these datasets is provided in Supplementary Table 1.
We evaluated the enrichment of an immune signature or pathway in a tumor by the single-sample gene-set enrichment analysis (ssGSEA)[14]. This method extends the GSEA method[15] to obtain the enrichment scores of input gene sets in specimens with input of gene expression matrices and marker or pathway gene sets. The marker or pathway gene sets of immune signatures or pathways are presented in Supplementary Table 2.
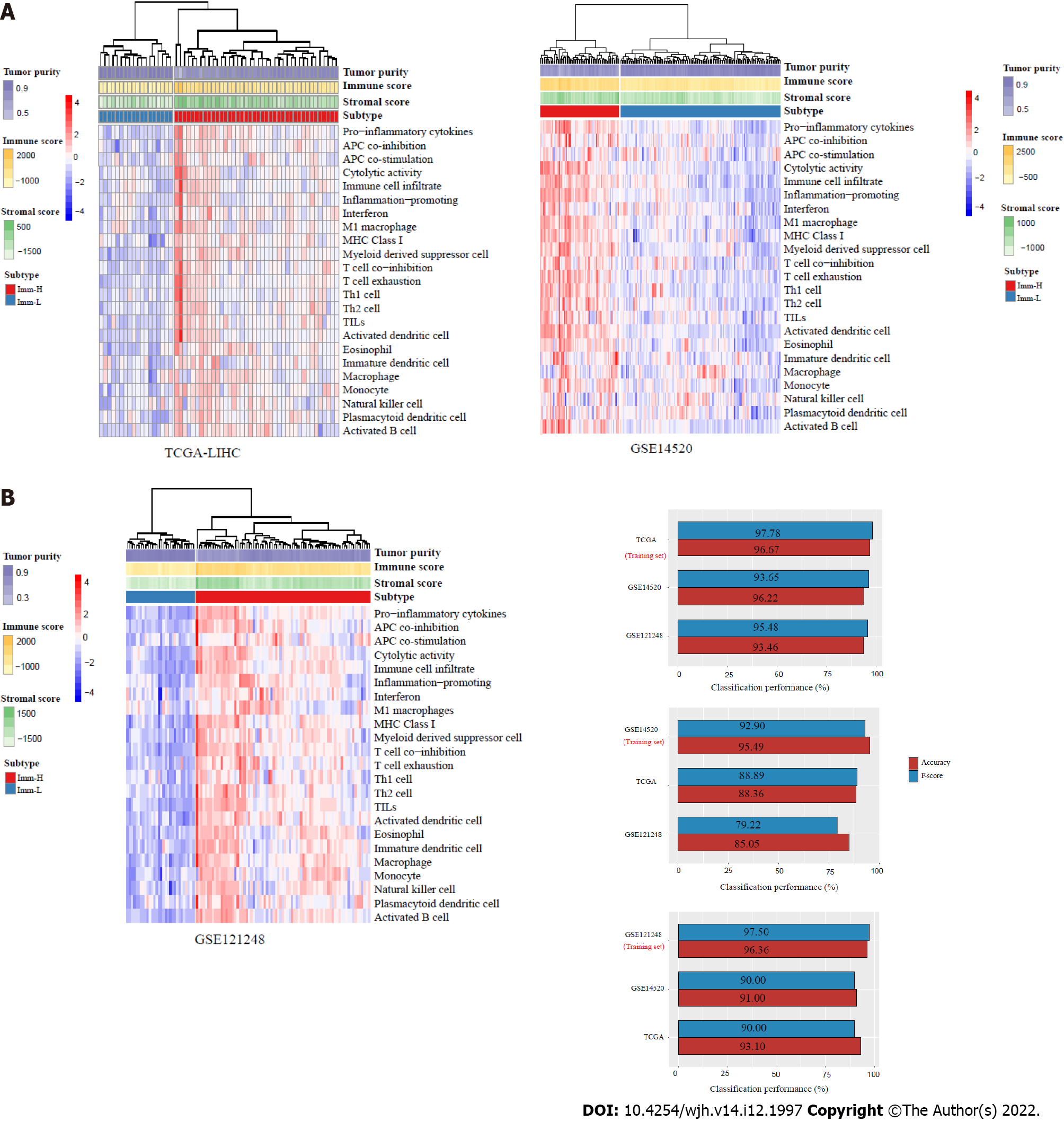
By hierarchical clustering, we identified immune-specific subtypes of HBV+ HCC based on the enrichment scores of 23 immune signatures. The 23 immune cell types included Pro-inflammatory cytokines, APC co-inhibition, APC co-stimulation, Cytolytic activity, Immune cell infiltrate, Inflammation-promoting, Interferon, M1 macrophage, MHC Class I, Myeloid-derived suppressor cell, T cell co-inhibition, T cell exhaustion, Th1 cell, Th2 cell, TILs, Activated dendritic cell, Eosinophil, Immature dendritic cell, Macrophage, Monocyte, Natural killer cell, Plasmacytoid dendritic cell, Activated B cell. Before clustering, we performed the Z-score normalization of the ssGSEA scores and converted them into distance matrices using the R function “dist” with the following parameter: Method = “euclidean.” We performed the hierarchical clustering using the function “hclust()” in the R package “Stats” with the following parameters: method = “ward.D2” and members = NULL.
We conducted classification with the random forest (RF) algorithm. In the RF, the size of trees was 500, and the features were the 23 immune signatures. The prediction performance, namely the accuracy and weighted F-score, were reported. We performed this procedure using the R package "randomForest".
We compared overall survival (OS) and disease-free survival (DFS) rates between two classes of samples with the Kaplan-Meier (K-M) method[16]. K-M curves were used to show the differences in survival rates, and log-rank tests were utilized to evaluate their significance.
A tumor’s TMB was defined as its total count of somatic mutations. We used GISTIC2[17] to calculate SCNA frequencies and amplitudes in the immune-specific subtypes of HBV+ HCC with the input of “SNP6” files. A tumor’s stemness score was its ssGSEA score of the stemness marker genes, as shown in Supplementary Table 1. We measured intratumor heterogeneity (ITH) levels with the DEPTH algorithm[18], which evaluates ITH levels based on gene expression profiles. We assessed immune scores and tumor purity of bulk tumors using ESTIMATE[19]. The immune scores indicate the levels of tumor immune infiltration, and tumor purity represents the fraction of tumor cells within a tumor bulk.
To identify pathways that are more enriched in one group compared to another group, we first uncovered the genes significantly upregulated in the group versus another group by Student's t tests using thresholds of false discovery rate (FDR) < 0.05 and mean gene expression levels’ fold change (FC) > 2. We then input the upregulated genes into the GSEA web tool[15] to obtain the Kyoto Encyclopedia of Genes and Genomes (KEGG)[20] pathways using a threshold of FDR < 0.05, which were more enriched in that group versus another class. Besides, we used the weighted gene co-expression network analysis (WGCNA)[21] to identify the gene modules of co-expressed genes. Based on the expression correlations between the hub genes in gene modules, we identified the gene ontology (GO) terms showing significant correlations with specific traits. The WGCNA analysis was performed with the R package “WGCNA” (version 1.68).
In comparisons of two classes of normally distributed data, including gene expression levels, protein expression levels, and the ratios of immune-stimulatory to immune-inhibitory signatures, we used two-tailed Student’s t tests. In comparisons of two classes of data that were not normally distributed, including immune scores, stemness scores, ITH scores, TMB, and global methylation levels, we performed one-tailed Mann–Whitney U tests. We utilized the Spearman method to calculate correlations between immune scores and protein expression levels or pathways’ enrichment scores, and reported correlation coefficients (ρ) and P values. We employed the Benjamini–Hochberg method[22] to calculate FDR for adjusting for multiple tests. We performed all statistical analyses in the R pro
This analysis identified two subtypes of HBV+ HCC according to the enrichment scores of 23 immune signatures by hierarchical clustering, consistently in three transcriptomic datasets (TCGA-LIHC, GSE14520, and GSE121248) (Figure 1A). We termed the subtypes Imm-H and Imm-L, respectively, which showed high and low immune signature enrichment, respectively (Figure 1A). To explore whether this classification is predictable, we took one of the three datasets as the training set and the rest as test sets, in turn, to predict the subtypes by RF based on the attribute values (ssGSEA scores of the 23 immune signatures). The 10-fold CV accuracies and weighted F-scores in the training sets were all above 90%. The prediction accuracies and weighted F-scores in test sets with TCGA-LIHC or GSE121248 as the training set were not less than 90% (Figure 1B). These results demonstrate that the subtyping is predictable.
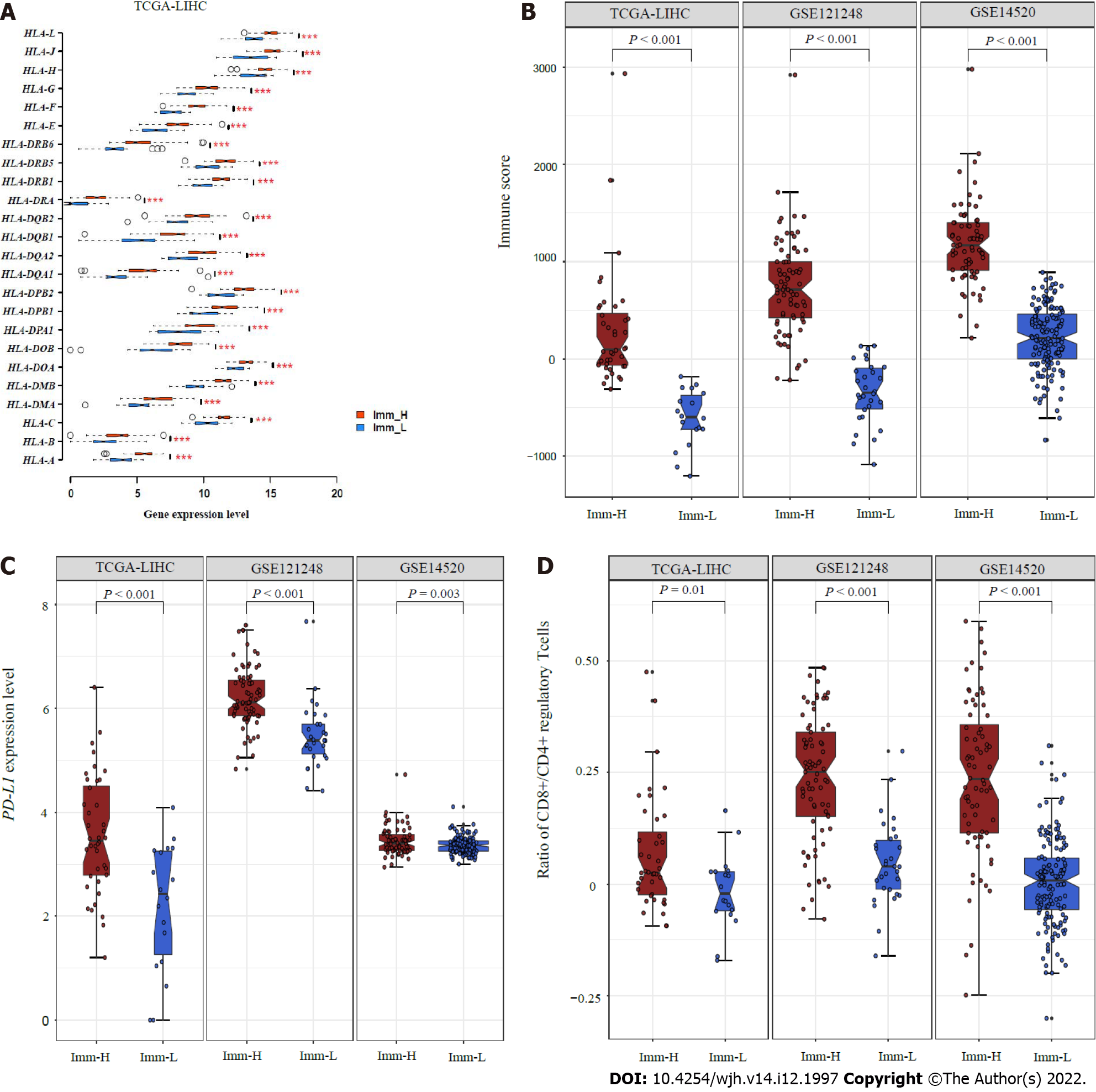
We further compared the expression levels of 25 genes encoding human leukocyte antigens between the subtypes. Of note, in TCGA-LIHC, all 25 genes were expressed at significantly higher levels in Imm-H than in Imm-L (FDR < 0.01; FC > 1.5) (Figure 2A). The immune scores were significantly higher in Imm-H than in Imm-L, consistently in the three datasets (P < 0.001) (Figure 2B). Furthermore, PD-L1, an antitumor immunosuppressive signature, was more highly expressed in Imm-H than in Imm-L (P < 0.01) (Figure 2C). Nevertheless, the ratios of immunostimulatory to immunosuppressive signatures (CD8+/CD4+ regulatory T cells), the base-2 Log-transformed values of the geometric mean expression levels of all marker genes of CD8+ T cells divided by those of CD4+ regulatory T cells, were significantly higher in Imm-H than in Imm-L (P < 0.05) (Figure 2D). Taken together, these results support that Imm-H has stronger anti-tumor immunity compared to Imm-L.
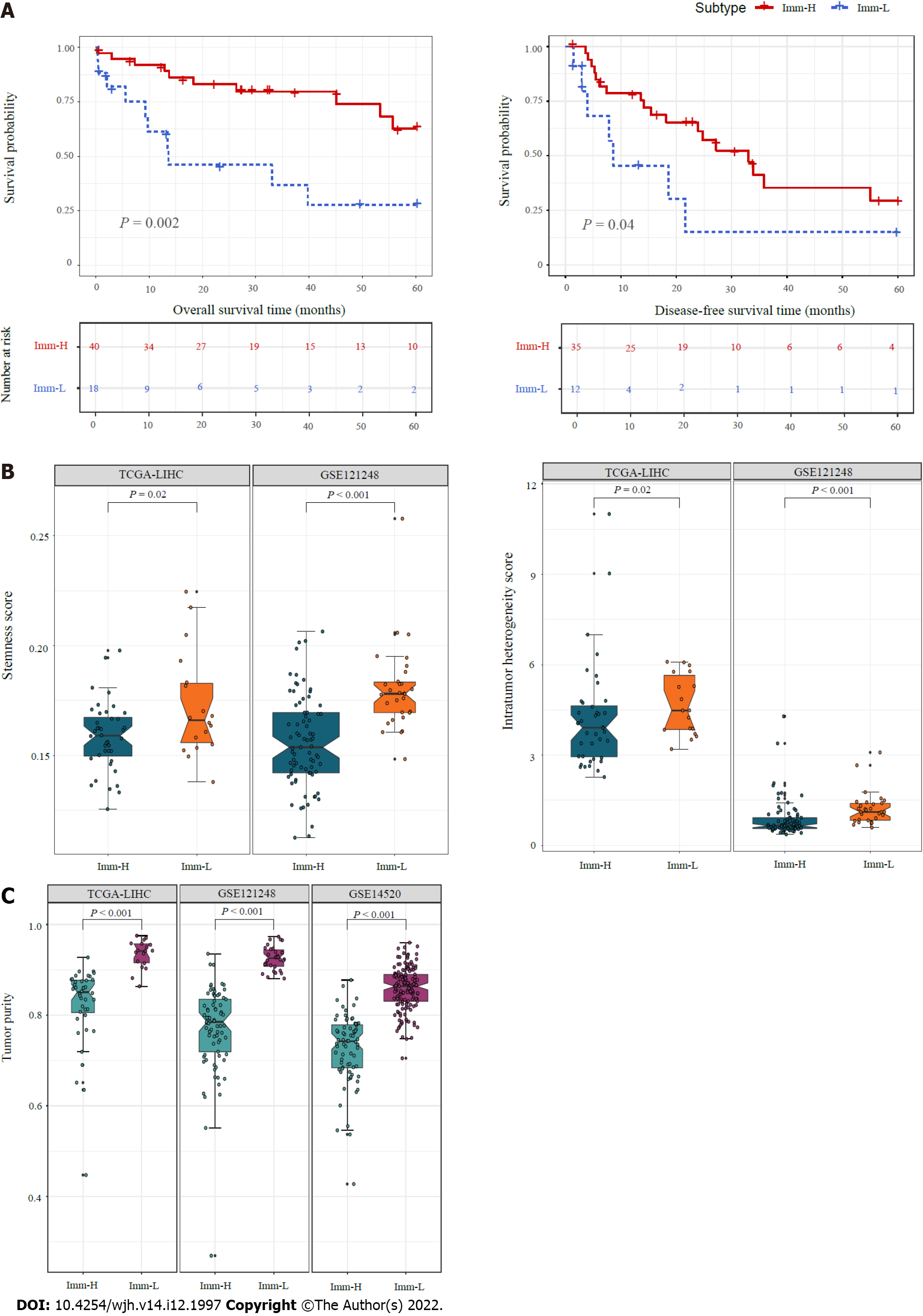
We compared 5-year OS and DFS prognosis between the immune-specific subtypes of HBV+ HCC in TCGA-LIHC, which had survival-related data available. Notably, Imm-H displayed significantly higher OS and DFS rates than Imm-L (P < 0.05) (Figure 3A). It supports the positive association between antitumor immune responses and survival prognosis in HBV+ HCC. We further compared several tumor progression-associated phenotypic features, including tumor stemness and ITH. We found that both stemness and ITH scores were markedly higher in Imm-L than in Imm-H in two of the three datasets (TCGA-LIHC and GSE121248) (P < 0.05) (Figure 3B). As expected, tumor purity was consistently higher in Imm-L than in Imm-H in the three datasets (Figure 3C). Altogether, these results indicate more favorable clinical outcomes in Imm-H than in Imm-L.
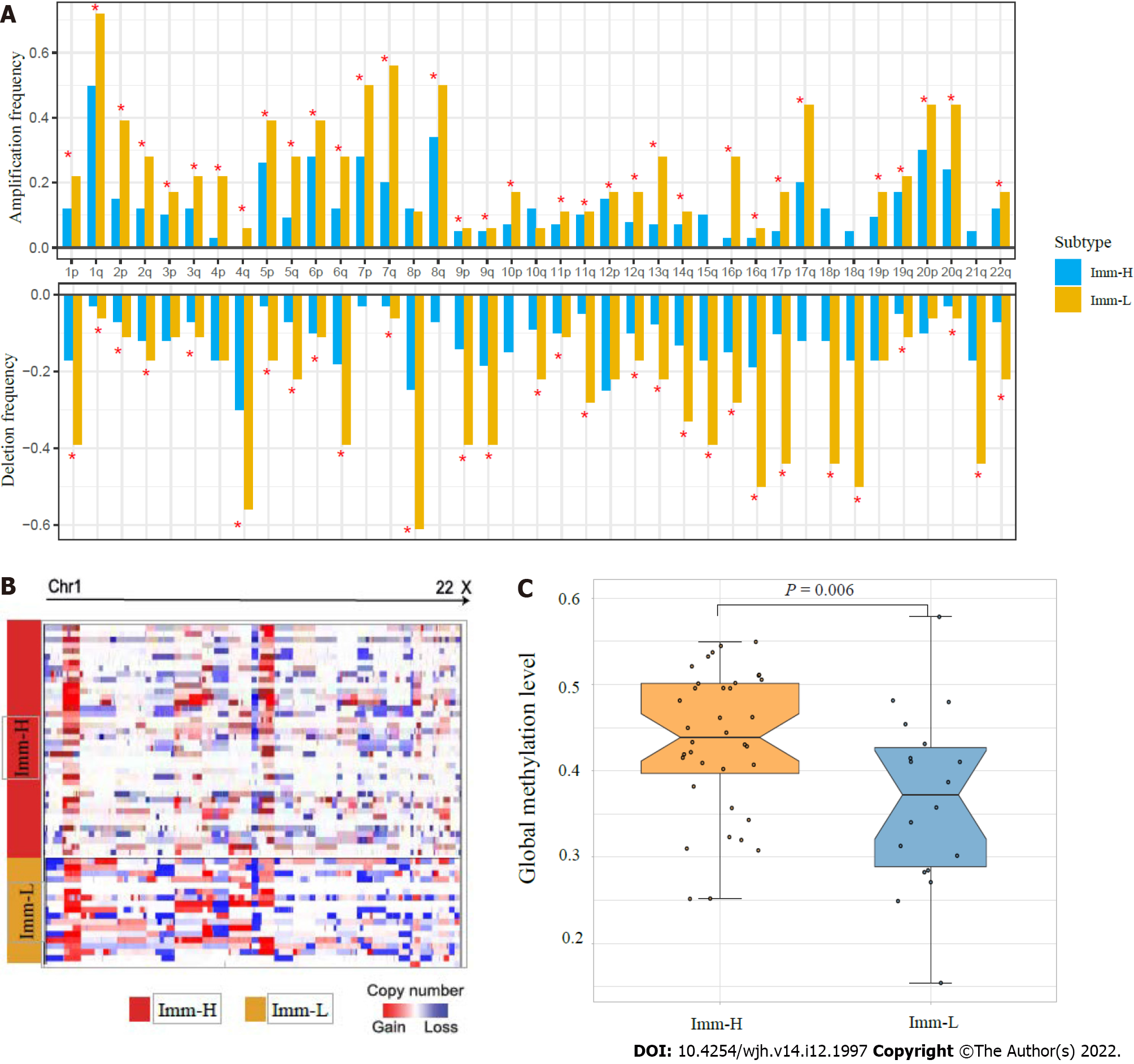
Tumor aneuploidy, also known as copy number alteration (CNA), is a typical genomic feature in tumors[23]. We found that Imm-L had higher frequencies of arm-level copy number amplification and deletion across chromosomes (Figure 4A). Moreover, Imm-L likely had higher amplitudes of copy number amplification and deletion across chromosomes as compared to Imm-H (Figure 4B). It suggested a higher level of genomic instability in Imm-L vs Imm-H, supporting a negative correlation between tumor aneuploidy and antitumor immunity[24]. Nevertheless, TMB showed no significant difference between Imm-H and Imm-L (Mann–Whitney U test, P = 0.86). Furthermore, we found that Imm-H had significantly higher global methylation levels[25] than Imm-L (P = 0.006) (Figure 4C). It conforms to a previous study showing that reduced DNA methylation levels promote antitumor immunosuppression[25].
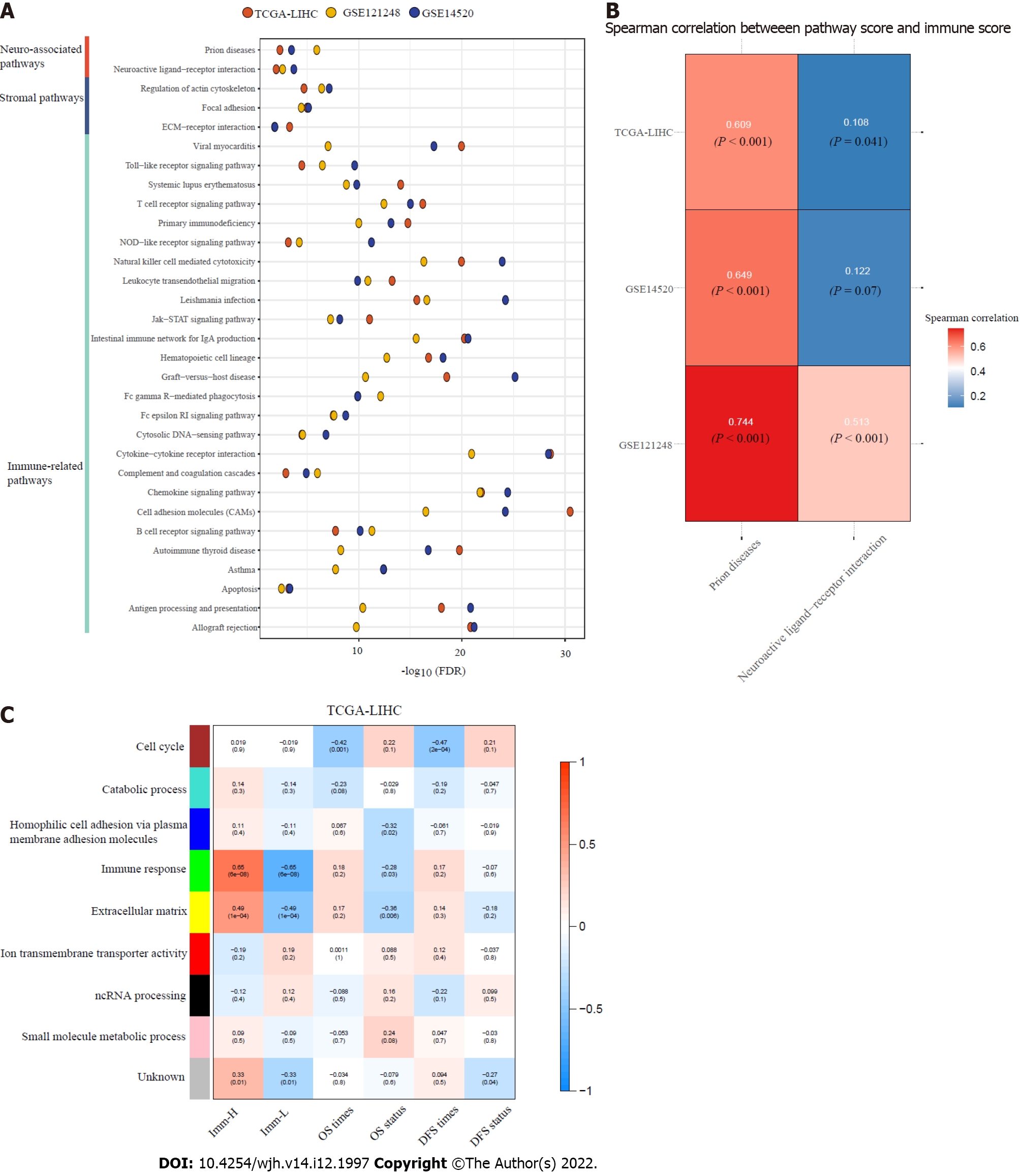
Based on the DEGs between Imm-H and Imm-L, we identified the KEGG pathways enriched in Imm-H and Imm-L common across the three datasets. Because there were no pathways enriched in Imm-L overlapping among the three datasets, we only attained the pathways enriched in Imm-H. We identified a total of 39 pathways highly enriched in Imm-H common in the three datasets. As expected, many immune-related pathways were on the list, including allograft rejection, antigen processing and presentation, apoptosis, asthma, autoimmune thyroid disease, B cell receptor signaling, cell adhesion molecules, chemokine signaling, complement and coagulation cascades, cytokine-cytokine receptor interaction, cytosolic DNA sensing, Fc epsilon RI signaling, Fc gamma R-mediated phagocytosis, graft vs host disease, hematopoietic cell lineage, intestinal immune network for IgA production, Jak-STAT signaling, leishmania infection, leukocyte transendothelial migration, natural killer cell-mediated cytotoxicity, NOD-like receptor signaling, primary immunodeficiency, T cell receptor signaling, systemic lupus erythematosus, Toll-like receptor signaling, and viral myocarditis (Figure 5A). Besides, several stromal pathways were included in the list, such as ECM receptor interaction, focal adhesion, and regulation of actin cytoskeleton. Interestingly, we found two neuro-associated pathways included in the pathway list: neuroactive ligand-receptor interaction, and prion diseases. It indicates that the activities of these neuro-associated pathways are positively correlated with antitumor immunity. Indeed, the enrichment scores of these pathways correlated positively with immune scores in these datasets (Spearman correlation, P < 0.05) (Figure 5B).
WGCNA[21] identified nine gene modules significantly differentiating HBV+ HCC by the immune-specific subtypes, OS, and/or DFS (Figure 5C). The gene modules upregulated in Imm-H while downregulated in Imm-L included the green module (with the representative GO term of immune response) and the yellow module (with the representative GO term of extracellular matrix) (P < 0.001). It is consistent with the results from the prior pathway analysis showing more highly enriched immune and stromal pathways in Imm-H versus Imm-L. Moreover, both modules were positively correlated with the OS prognosis (P < 0.05), consistent with the better OS in Imm-H relative to Imm-L. Besides, the blue module (with the GO term of homophilic cell adhesion via plasma membrane adhesion molecules) was significantly and positively correlated with the OS prognosis (P = 0.02), although it showed no significant enrichment difference between Imm-H versus Imm-L. It is reasonable that the positive association between the blue module and OS since reduced homophilic cell adhesion can promote tumor progression[26]. In contrast, the brown module had significant negative correlations with both OS and DFS time. The representative GO term for this module was cell cycle. It is justified since elevated cell cycle activity suggests tumor progression.
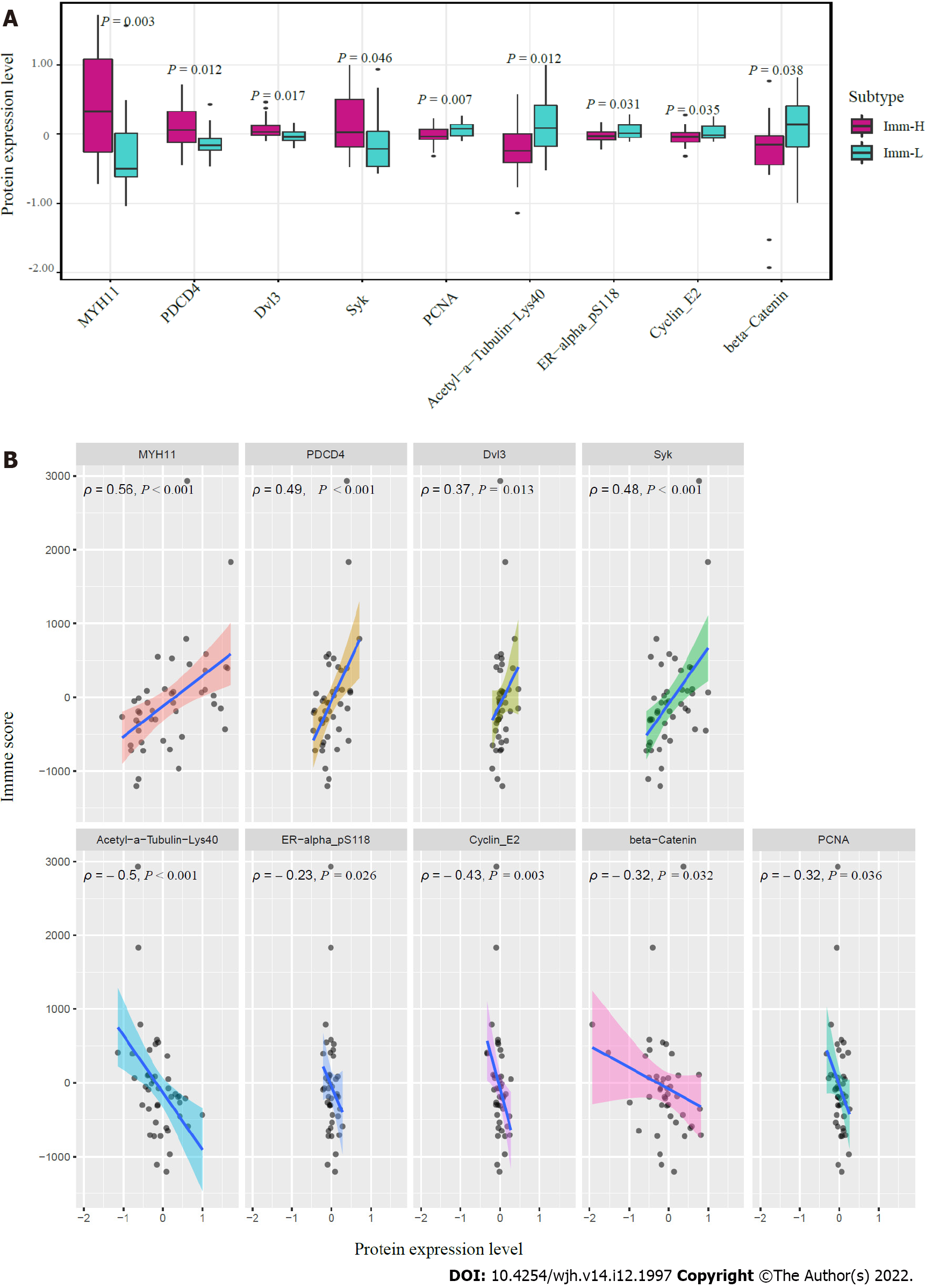
We compared the expression levels of 219 proteins between the immune-specific subtypes of HBV+ HCC in TCGA-LIHC. We identified nine proteins differentially expressed between Imm-H and Imm-L (P < 0.05) (Figure 6A). Among them, MYH11, PDCD4, Dvl3, and Syk were more highly expressed in Imm-H, while PCNA, Acetyl-a-Tubulin-Lys40, ER-α_pS118, Cyclin E2, and β-Catenin were more highly expressed in Imm-L. As expected, the proteins more enriched in Imm-H showed significantly positive expression correlations with immune scores in HBV+ HCC, and the proteins more enriched in Imm-L showed negative expression correlations with them (Spearman correlation, P < 0.05) (Figure 6B). Previous studies have shown that most of these proteins have associations with tumor immune regulation. For example, MYH11 is a smooth muscle myosin of the myosin heavy chain family, whose expression has been associated with antitumor immune infiltration in cancer[27]. PDCD4 is a tumor suppressor, whose expression in the tumor microenvironment is correlated with increased immune infiltration[28,29]. Syk is also a tumor suppressor and has a role in tumor immune regulation[30]. PCNA is involved in the DNA repair pathway in response to DNA damage, whose upregulation may promote tumor immune evasion[31,32]. This is consistent with its upregulation in Imm-L versus Imm-H. ER-α has been shown to induce antitumor immunosuppression[33], supporting our finding. Cyclin E2 is a positive regulator of cell cycle, which may inhibit antitumor immune responses and immunotherapy responses[34]. Again, this is consistent with our result. β-Catenin is an activator of the Wnt/β-catenin signaling pathway, whose overexpression suppresses antitumor immune responses[35], in line with our findings.
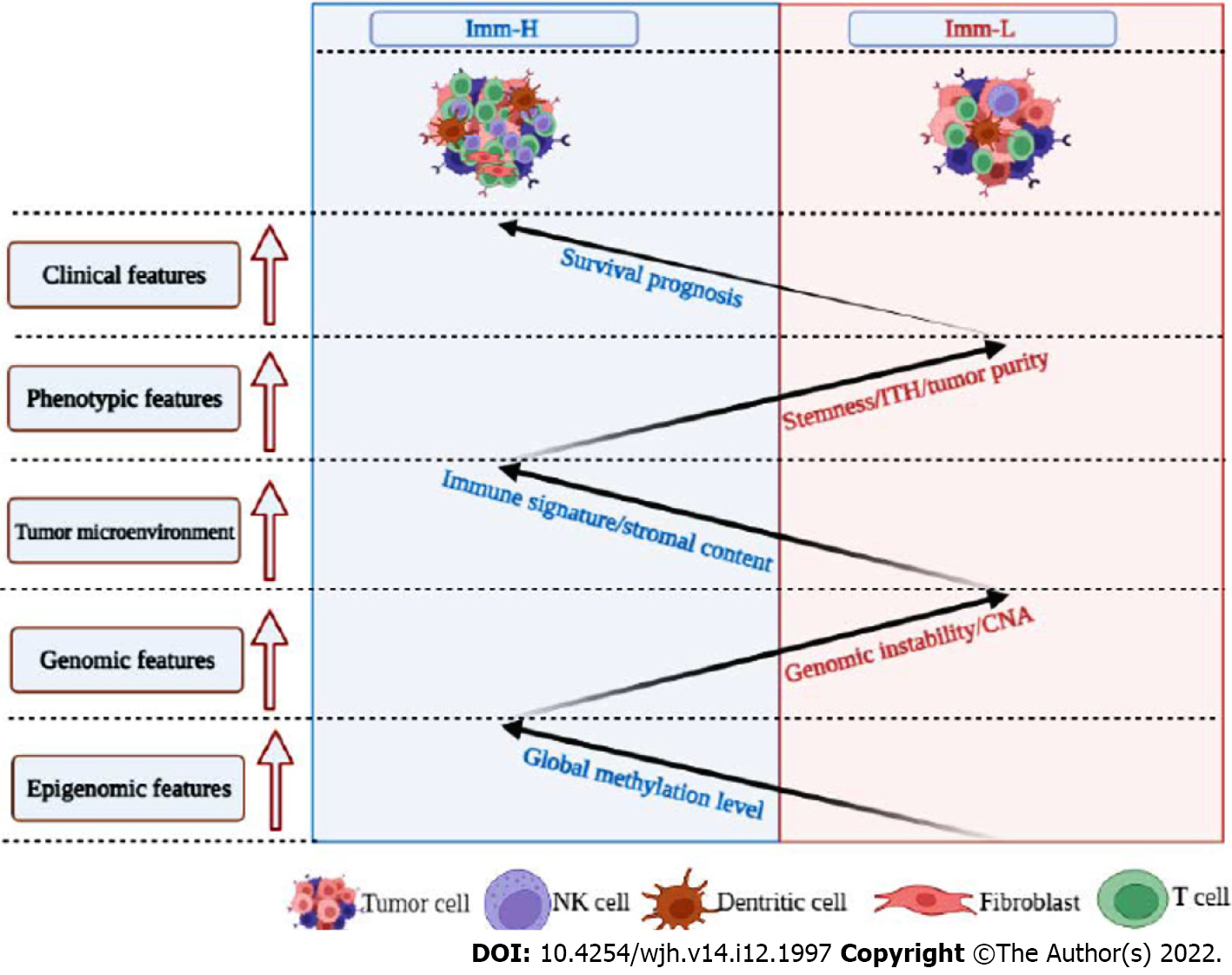
This study identified two immune-specific subtypes (Imm-H and Imm-L) of HBV+ HCC based on the enrichment of 23 immune signatures by unsupervised clustering. We showed that this subtyping method was reproducible as well as predictable by analyzing three different datasets. Furthermore, we demonstrated that both subtypes had significantly different clinical and molecular features. Compared to Imm-L, Imm-H displayed stronger immunity, more stromal components, lower tumor purity, lower stemness and ITH, lower-level CNAs, higher global methylation level, and better overall and disease-free survival prognosis (Figure 7). Our data support that “hot” tumors have a better prognosis than “cold” tumors in HBV+ HCC for their stronger antitumor immune responses. Similar findings were also observed in other cancers[36-38]. Intriguingly, although continual inflammatory responses in the liver caused by HBV infection is a major etiology for HBV+ HCC[39], higher immune/inflammatory responses are associated with a better prognosis in HBV+ HCC patients, as demonstrated by this analysis. It indicates that the relationship between immune/inflammatory responses and clinical outcomes in cancer is complex. Indeed, in some cancer types, such as glioma[40] and prostate cancer[41], the relationship between immune/inflammatory responses and clinical outcomes is negative. Thus, the relationship between immune responses and clinical outcomes in cancers is dependent on their tissue or cellular origins, the tumor microenvironment, the ratio of immunostimulatory over immunosuppressive signatures, as well as whether the immune response is the tumor progression-promoting inflammation or immune cell-mediated elimination of tumor cells.
Prior studies have shown that TMB and CNAs have a positive and negative association with antitumor immune responses, respectively[24]. However, our analysis suggests that TMB has no a significant association with antitumor immunity in HBV+ HCC, although CNAs have a negative association with antitumor immune responses. It suggests that it is CNAs but not TMB responsible for the significantly different immunity between the “hot” and “cold” tumor subtypes in HBV+ HCC. Furthermore, the significantly lower stemness and ITH of Imm-H compared to Imm-L suggest that stemness and ITH may lead to antitumor immunosuppression, consistent with previous findings[18,42,43].
Pathway analysis showed that two neuro-associated pathways (neuroactive ligand receptor interaction, and prion diseases) had higher enrichment in Imm-H than in Imm-L and that their upregulation was associated with increased tumor immune infiltration levels. The positive association between neuro-related pathways and antitumor immunity has been revealed in prior studies[44]. Interestingly, many studies have demonstrated the inverse relationship between cancer and Alzheimer’s disease (AD)[45]. AD is known as a progressive neurodegenerative disease as well as neuroinflammation disease[46]. A recent study has proposed that AD is an autoimmune disorder of innate immunity[47]. The present and prior data stimulate our imagination that the immune and inflammation could bridge the relationship between cancer and AD, such as hyperactivation of the immune system in AD patients reducing the risk of cancer.
Interestingly, besides the antitumor immune signatures, the immunosuppressive signature PD-L1, was also upregulated in Imm-H vs Imm-L. Because both PD-L1 expression[48] and ample TILs[9] are positive predictors of the response to ICB, Imm-H would respond better to immunotherapy than Imm-L. Thus, our subtyping method may stratify HBV+ HCC patients for immunotherapy. That is, immunotherapy may yield more propitious efficacy for Imm-H than for Imm-L HBV+ HCC patients.
HBV+ HCCs can be classified into two immune-specific subtypes in terms of their immune signature enrichment. Both subtypes have significantly different immunity, stromal contents, tumor purity, stemness, ITH, CNAs, methylation profiles, and survival prognosis. The immune-specific subtyping of HBV+ HCC may provide new biological insights as well as clinical implications for the management of this disease.
Hepatocellular carcinoma (HCC) is a major cancer of the liver that constitutes around 90% of liver cancer cases. Although traditional therapeutic approaches, including surgery, chemotherapy, radiotherapy, and targeted therapy, are effective in improving the survival of HCC patients, the overall survival prognosis of HCC patients is generally unfavorable. More recently, immunotherapy, such as immune checkpoint blockade, has achieved success in the treatment of various cancers, including HCC. However, only a small proportion of cancer patients respond well to immunotherapies to date.
Certain predictive markers for cancer immunotherapy responses have been uncovered, e.g., PD-L1 expression, tumor mutation burden (TMB), and mismatch repair deficiency. In addition, the tumor immune microenvironment (TIME) plays an important role in immunotherapy responses. Overall, the “hot” tumors infiltrated by a substantial number of tumor-infiltrating lymphocytes (TILs) are more responsive to immunotherapies, compared to the “cold” tumors lacking TILs. Hence, an investigation of the TIME in HCC would aid in the prediction of immunotherapy responses.
Despite these previous studies, the discovery of immune-specific subtypes of hepatitis B virus-positive (HBV+) HCC is worth investigating, considering that HBV infection is a major cause of HCC.
In this study, to characterize the immunological landscape of HBV+ HCC, we identified its immune-specific subtypes by the unsupervised machine learning in transcriptomic data. Furthermore, we comprehensively compared the clinical and molecular features of these subtypes.
Compared to Imm-L, Imm-H displayed stronger immunity, more stromal components, lower tumor purity, lower stemness and intratumor heterogeneity, lower-level copy number alterations, higher global methylation level, and better overall and disease-free survival prognosis.
Our immune-specific subtyping of HBV+ HCC may provide new biological insights as well as clinical implications for the management of this disease.
This study is interesting for several reasons. First, for the first time, we identified immune-specific subtypes of HBV+ HCC based on immune signature scores and demonstrated that this new subtyping method was reproducible in three different datasets. Second, our subtyping method captures the comprehensive heterogeneity of HBV+ HCC in the tumor microenvironment, genomic integrity, protein expression profiles, DNA methylation profiles, tumor stemness, intratumor heterogeneity, and clinical outcomes. Third, our data suggest that it is copy number alterations but not tumor mutations responsible for the different immunity between the “hot” and “cold” tumor subtypes in HBV+ HCC. Finally, our identification of the immune-specific subtypes of HBV+ HCC may provide new insights into the tumor biology and identify the HBV+ HCC patients beneficial from immunotherapy.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aynalem A, Ethiopia; Elpek GO, Turkey S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2066] [Cited by in RCA: 3157] [Article Influence: 526.2] [Reference Citation Analysis (37)] |
| 2. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3837] [Article Influence: 959.3] [Reference Citation Analysis (3)] |
| 3. | Yau T, Kang YK, Kim TY, El-Khoueiry AB, Santoro A, Sangro B, Melero I, Kudo M, Hou MM, Matilla A, Tovoli F, Knox JJ, Ruth He A, El-Rayes BF, Acosta-Rivera M, Lim HY, Neely J, Shen Y, Wisniewski T, Anderson J, Hsu C. Efficacy and Safety of Nivolumab Plus Ipilimumab in Patients With Advanced Hepatocellular Carcinoma Previously Treated With Sorafenib: The CheckMate 040 Randomized Clinical Trial. JAMA Oncol. 2020;6:e204564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 908] [Cited by in RCA: 960] [Article Influence: 192.0] [Reference Citation Analysis (0)] |
| 4. | Braun DA, Burke KP, Van Allen EM. Genomic Approaches to Understanding Response and Resistance to Immunotherapy. Clin Cancer Res. 2016;22:5642-5650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 5. | Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064-5074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1633] [Cited by in RCA: 1965] [Article Influence: 178.6] [Reference Citation Analysis (0)] |
| 6. | Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16:2598-2608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1259] [Cited by in RCA: 1703] [Article Influence: 212.9] [Reference Citation Analysis (0)] |
| 7. | Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-2520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6096] [Cited by in RCA: 7218] [Article Influence: 721.8] [Reference Citation Analysis (0)] |
| 8. | Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 2020;30:507-519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 453] [Cited by in RCA: 493] [Article Influence: 98.6] [Reference Citation Analysis (0)] |
| 9. | Haanen JBAG. Converting Cold into Hot Tumors by Combining Immunotherapies. Cell. 2017;170:1055-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 198] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 10. | Gao X, Huang H, Wang Y, Pan C, Yin S, Zhou L, Zheng S. Tumor Immune Microenvironment Characterization in Hepatocellular Carcinoma Identifies Four Prognostic and Immunotherapeutically Relevant Subclasses. Front Oncol. 2020;10:610513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 11. | Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, Putra J, Camprecios G, Bassaganyas L, Akers N, Losic B, Waxman S, Thung SN, Mazzaferro V, Esteller M, Friedman SL, Schwartz M, Villanueva A, Llovet JM. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology. 2017;153:812-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 690] [Cited by in RCA: 659] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 12. | Farha M, Jairath NK, Lawrence TS, El Naqa I. Characterization of the Tumor Immune Microenvironment Identifies M0 Macrophage-Enriched Cluster as a Poor Prognostic Factor in Hepatocellular Carcinoma. JCO Clin Cancer Inform. 2020;4:1002-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (1)] |
| 13. | Zamor PJ, deLemos AS, Russo MW. Viral hepatitis and hepatocellular carcinoma: etiology and management. J Gastrointest Oncol. 2017;8:229-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 14. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7222] [Cited by in RCA: 9187] [Article Influence: 765.6] [Reference Citation Analysis (0)] |
| 15. | Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-15550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27252] [Cited by in RCA: 37257] [Article Influence: 1862.9] [Reference Citation Analysis (0)] |
| 16. | Bland JM, Altman DG. Survival probabilities (the Kaplan-Meier method). BMJ. 1998;317:1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 544] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 17. | Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1825] [Cited by in RCA: 2498] [Article Influence: 178.4] [Reference Citation Analysis (0)] |
| 18. | Li M, Zhang Z, Li L, Wang X. An algorithm to quantify intratumor heterogeneity based on alterations of gene expression profiles. Commun Biol. 2020;3:505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 19. | Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3056] [Cited by in RCA: 6400] [Article Influence: 581.8] [Reference Citation Analysis (0)] |
| 20. | Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353-D361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4407] [Cited by in RCA: 5807] [Article Influence: 645.2] [Reference Citation Analysis (0)] |
| 21. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10254] [Cited by in RCA: 16305] [Article Influence: 959.1] [Reference Citation Analysis (0)] |
| 22. | Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 57:289-300. [RCA] [DOI] [Full Text] [Cited by in Crossref: 17030] [Cited by in RCA: 23021] [Article Influence: 3288.7] [Reference Citation Analysis (0)] |
| 23. | Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, Liu Y, Akbani R, Feng B, Donehower LA, Miller C, Shen Y, Karimi M, Chen H, Kim P, Jia P, Shinbrot E, Zhang S, Liu J, Hu H, Bailey MH, Yau C, Wolf D, Zhao Z, Weinstein JN, Li L, Ding L, Mills GB, Laird PW, Wheeler DA, Shmulevich I; Cancer Genome Atlas Research Network, Monnat RJ Jr, Xiao Y, Wang C. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23:239-254.e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 863] [Cited by in RCA: 777] [Article Influence: 111.0] [Reference Citation Analysis (0)] |
| 24. | Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 960] [Article Influence: 120.0] [Reference Citation Analysis (0)] |
| 25. | Jung H, Kim HS, Kim JY, Sun JM, Ahn JS, Ahn MJ, Park K, Esteller M, Lee SH, Choi JK. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat Commun. 2019;10:4278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 342] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 26. | Craig SE, Brady-Kalnay SM. Cancer cells cut homophilic cell adhesion molecules and run. Cancer Res. 2011;71:303-309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Wu SZ, Swarbrick A. Single-cell advances in stromal-leukocyte interactions in cancer. Immunol Rev. 2021;302:286-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Tran TT, Rane CK, Zito CR, Weiss SA, Jessel S, Lucca L, Lu BY, Oria VO, Adeniran A, Chiang VL, Omay SB, Hafler DA, Kluger HM, Jilaveanu LB. Clinical Significance of PDCD4 in Melanoma by Subcellular Expression and in Tumor-Associated Immune Cells. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Pin G, Huanting L, Chengzhan Z, Xinjuan K, Yugong F, Wei L, Shifang L, Zhaojian L, Kun H, Weicheng Y, Yingying L, Yongming Q, Yanan Y. Down-Regulation of PDCD4 Promotes Proliferation, Angiogenesis and Tumorigenesis in Glioma Cells. Front Cell Dev Biol. 2020;8:593685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Krisenko MO, Geahlen RL. Calling in SYK: SYK's dual role as a tumor promoter and tumor suppressor in cancer. Biochim Biophys Acta. 2015;1853:254-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 31. | Wang YL, Lee CC, Shen YC, Lin PL, Wu WR, Lin YZ, Cheng WC, Chang H, Hung Y, Cho YC, Liu LC, Xia WY, Ji JH, Liang JA, Chiang SF, Liu CG, Yao J, Hung MC, Wang SC. Evading immune surveillance via tyrosine phosphorylation of nuclear PCNA. Cell Rep. 2021;36:109537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Rosental B, Hadad U, Brusilovsky M, Campbell KS, Porgador A. A novel mechanism for cancer cells to evade immune attack by NK cells: The interaction between NKp44 and proliferating cell nuclear antigen. Oncoimmunology. 2012;1:572-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 33. | Liu Z, Jiang Z, Gao Y, Wang L, Chen C, Wang X. TP53 Mutations Promote Immunogenic Activity in Breast Cancer. J Oncol. 2019;2019:5952836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 34. | Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, Khan N, Ubellacker JM, Xie S, Metzger-Filho O, Hoog J, Ellis MJ, Ma CX, Ramm S, Krop IE, Winer EP, Roberts TM, Kim HJ, McAllister SS, Zhao JJ. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548:471-475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 700] [Cited by in RCA: 1072] [Article Influence: 134.0] [Reference Citation Analysis (0)] |
| 35. | Wang C, Yan J, Yin P, Gui L, Ji L, Ma B, Gao WQ. β-Catenin inhibition shapes tumor immunity and synergizes with immunotherapy in colorectal cancer. Oncoimmunology. 2020;9:1809947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 36. | Lyu H, Li M, Jiang Z, Liu Z, Wang X. Correlate the TP53 Mutation and the HRAS Mutation with Immune Signatures in Head and Neck Squamous Cell Cancer. Comput Struct Biotechnol J. 2019;17:1020-1030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 37. | He Y, Jiang Z, Chen C, Wang X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J Exp Clin Cancer Res. 2018;37:327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 374] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 38. | Jiang Z, Liu Z, Li M, Chen C, Wang X. Immunogenomics Analysis Reveals that TP53 Mutations Inhibit Tumor Immunity in Gastric Cancer. Transl Oncol. 2018;11:1171-1187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 39. | Yang YM, Kim SY, Seki E. Inflammation and Liver Cancer: Molecular Mechanisms and Therapeutic Targets. Semin Liver Dis. 2019;39:26-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 304] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 40. | Pombo Antunes AR, Scheyltjens I, Duerinck J, Neyns B, Movahedi K, Van Ginderachter JA. Understanding the glioblastoma immune microenvironment as basis for the development of new immunotherapeutic strategies. Elife. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 181] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 41. | Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS; Cancer Genome Atlas Research Network, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The Immune Landscape of Cancer. Immunity. 2018;48:812-830.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4007] [Cited by in RCA: 3808] [Article Influence: 544.0] [Reference Citation Analysis (0)] |
| 42. | Li L, Chen C, Wang X. DITHER: an algorithm for Defining IntraTumor Heterogeneity based on EntRopy. Brief Bioinform. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 43. | Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A, Bruun J, Micke P, de Reynies A, Nelson BH. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci U S A. 2019;116:9020-9029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 396] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 44. | Feng Q, Song D, Wang X. Pan-cancer analysis reveals that neurotrophin signaling correlates positively with anti-tumor immunity, clinical outcomes, and response to targeted therapies and immunotherapies in cancer. Life Sci. 2021;282:119848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 45. | Karanth SD, Katsumata Y, Nelson PT, Fardo DW, McDowell JK, Schmitt FA, Kryscio RJ, Browning SR, Braithwaite D, Arnold SM, Abner EL. Cancer diagnosis is associated with a lower burden of dementia and less Alzheimer's-type neuropathology. Brain. 2022;145:2518-2527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 46. | Kinney JW, Bemiller SM, Murtishaw AS, Leisgang AM, Salazar AM, Lamb BT. Inflammation as a central mechanism in Alzheimer's disease. Alzheimers Dement (N Y). 2018;4:575-590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1254] [Cited by in RCA: 1436] [Article Influence: 205.1] [Reference Citation Analysis (0)] |
| 47. | Meier-Stephenson FS, Meier-Stephenson VC, Carter MD, Meek AR, Wang Y, Pan L, Chen Q, Jacobo S, Wu F, Lu E, Simms GA, Fisher L, McGrath AJ, Fermo V, Barden CJ, Clair HDS, Galloway TN, Yadav A, Campágna-Slater V, Hadden M, Reed M, Taylor M, Kelly B, Diez-Cecilia E, Kolaj I, Santos C, Liyanage I, Sweeting B, Stafford P, Boudreau R, Reid GA, Noyce RS, Stevens L, Staniszewski A, Zhang H, Murty MRVS, Lemaire P, Chardonnet S, Richardson CD, Gabelica V, DePauw E, Brown R, Darvesh S, Arancio O, Weaver DF. Alzheimer's disease as an autoimmune disorder of innate immunity endogenously modulated by tryptophan metabolites. Alzheimers Dement (N Y). 2022;8:e12283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 48. | Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14:847-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1250] [Cited by in RCA: 1750] [Article Influence: 175.0] [Reference Citation Analysis (1)] |