Published online Mar 27, 2021. doi: 10.4254/wjh.v13.i3.343
Peer-review started: December 12, 2020
First decision: January 7, 2021
Revised: January 15, 2021
Accepted: March 8, 2021
Article in press: March 8, 2021
Published online: March 27, 2021
Processing time: 98 Days and 1.3 Hours
BIR repeat-containing ubiquitin conjugating enzyme (BRUCE) is a liver tumor suppressor, which is downregulated in a large number of patients with liver diseases. BRUCE facilitates DNA damage repair to protect the mouse liver against the hepatocarcinogen diethylnitrosamine (DEN)-dependent acute liver injury and carcinogenesis. While there exists an established pathologic connection between fibrosis and hepatocellular carcinoma (HCC), DEN exposure alone does not induce robust hepatic fibrosis. Further studies are warranted to identify new suppressive mechanisms contributing to DEN-induced fibrosis and HCC.
To investigate the suppressive mechanisms of BRUCE in hepatic fibrosis and HCC development.
Male C57/BL6/J control mice [loxp/Loxp; albumin-cre (Alb-cre)-] and BRUCE Alb-Cre KO mice (loxp/Loxp; Alb-Cre+) were injected with a single dose of DEN at postnatal day 15 and sacrificed at different time points to examine liver disease progression.
By using a liver-specific BRUCE knockout (LKO) mouse model, we found that BRUCE deficiency, in conjunction with DEN exposure, induced hepatic fibrosis in both premalignant as well as malignant stages, thus recapitulating the chronic fibrosis background often observed in HCC patients. Activated in fibrosis and HCC, β-catenin activity depends on its stabilization and subsequent translocation to the nucleus. Interestingly, we observed that livers from BRUCE KO mice demonstrated an increased nuclear accumulation and elevated activity of β-catenin in the three stages of carcinogenesis: Pre-malignancy, tumor initiation, and HCC. This suggests that BRUCE negatively regulates β-catenin activity during liver disease progression. β-catenin can be activated by phosphorylation by protein kinases, such as protein kinase A (PKA), which phosphorylates it at Ser-675 (pSer-675-β-catenin). Mechanistically, BRUCE and PKA were colocalized in the cytoplasm of hepatocytes where PKA activity is maintained at the basal level. However, in BRUCE deficient mouse livers or a human liver cancer cell line, both PKA activity and pSer-675-β-catenin levels were observed to be elevated.
Our data support a “BRUCE-PKA-β-catenin” signaling axis in the mouse liver. The BRUCE interaction with PKA in hepatocytes suppresses PKA-dependent phosphorylation and activation of β-catenin. This study implicates BRUCE as a novel negative regulator of both PKA and β-catenin in chronic liver disease progression. Furthermore, BRUCE-liver specific KO mice serve as a promising model for understanding hepatic fibrosis and HCC in patients with aberrant activation of PKA and β-catenin.
Core Tip: Upon diethylnitrosamine (DEN) exposure, BIR repeat-containing ubiquitin conjugating enzyme (BRUCE) liver-deficiency accelerates chronic liver diseases such as fibrosis and hepatocellular carcinoma (HCC) in mice. Our previous study established the role of BRUCE in the protection of the liver against DEN-induced liver injury and subsequent disease progression. Here we report a chronic fibrosis background induced by hepatic BRUCE knockout in mice that recapitulates the fibrosis background in HCC patients. We also report a BRUCE-dependent suppression of β-catenin activity through the suppression of protein kinase A (PKA) activity. This study provides a therapeutic potential involving the inhibition of PKA and β-catenin activities in patients with liver disease that carry BRUCE inactivating mutations.
- Citation: Vilfranc CL, Che LX, Patra KC, Niu L, Olowokure O, Wang J, Shah SA, Du CY. BIR repeat-containing ubiquitin conjugating enzyme (BRUCE) regulation of β-catenin signaling in the progression of drug-induced hepatic fibrosis and carcinogenesis. World J Hepatol 2021; 13(3): 343-361
- URL: https://www.wjgnet.com/1948-5182/full/v13/i3/343.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i3.343
The liver is constantly exposed to a variety of viral and bacterial products, environmental toxins, as well as alcohol intake and food antigens. The liver can rapidly detect damaging agents and protects itself against the damage without generating widespread inflammation and fibrosis, which are leading causes for liver cancers[1]. Diethylnitrosamine (DEN) is one of the most potent hepatocarcinogens that induces carcinogenic liver injury and development of hepatocellular carcinoma (HCC) in rodents. The application of DEN in rodents has become an attractive experimental model to study the pathogenetic alterations underlying hepato-genotoxic injury and the formation of HCC[2,3]. HCC represents the primary form of liver malignancy and the fourth most common cause of cancer-related deaths worldwide[4]. It has been well documented that the time and incidence of HCCs initiated by DEN differ greatly among mouse strains. DEN-induced HCC development is delayed in the tumor-resistant C57/BL6/J strain as compared to the more sensitive C3H/HE strain[5]. In addition, to induce robust hepatic fibrosis, a hallmark of human HCC development[6,7], administration of a single agent of DEN is insufficient in the C57/BL6/J strain, but when coupled with additional chemicals such as carbon tetrachloride, a fibrogenic agent, hepatic fibrosis is accelerated[8].
BIR repeat-containing ubiquitin-conjugating enzyme (BRUCE) is a hybrid ubiquitin conjugase and ligase[9]. BRUCE has two major pro-survival functions in vitro: Promotion of DNA damage repair and suppression of apoptosis. In the cell nucleus, BRUCE promotes DNA damage repair by homologous recombination (HR) to preserve genomic stability[10-12]. To achieve this function, BRUCE is recruited to damaged chromatin adjacent to DNA breaks, where it facilitates chromatin relaxation and accessibility, allowing for HR factors to be loaded onto DNA breaks to facilitate HR repair[11,12]. In the cytoplasm, BRUCE acts as a member of the inhibitor of apoptosis protein (IAP) family of proteins[13-15]. It inhibits the intrinsic mitochondrial pathway of apoptosis by post-translational ubiquitination of pro-apoptotic proteins to promote their degradation by the ubiquitin-proteasome system (UPS)[13,15-18]. In addition to these in vitro functions, we and others have demonstrated an in vivo anti-apoptosis function of BRUCE in mice, where it suppresses the mitochondrial pathway of apoptosis[16,17]. Furthermore, we have reported a DNA repair function of BRUCE in the protection of the mouse liver[3]. Utilizing an albumin-cre (Alb-cre) mediated liver-specific BRUCE knockout (LKO) mouse model, we have demonstrated for the first time that the BRUCE-ATR (Ataxia Telangiectasia and Rad3-related) signaling axis protects against DEN-induced liver injury and DEN-initiated HCC. BRUCE LKO mice had increased hepatocellular DNA damage accumulation induced by DEN, downregulated ATR-mediated DNA damage response, and an exacerbated HCC development with a fibrotic background[3]. However, the mechanisms underlying the liver fibrosis and HCC have not yet been characterized in this model.
Liver fibrosis is characterized by an excessive accumulation of the extracellular matrix (ECM) resulting in scar tissue formation[19]. Hepatocyte damage and death is an initial consequence of liver injury that initiates several events leading to the recruitment of inflammatory cells and the activation of hepatic stellate cells (HSCs)[20]. Upon liver damage, apoptotic hepatocytes release damage associated molecular patterns (DAMPs) and proinflammatory factors to activate neighboring Kupffer cells and HSCs, thereby inducing persistent inflammatory responses and fibrosis, respectively[19-21]. Activated HSCs are the principal source for deposition of ECM as activated HSCs are responsible for producing an excessive amount of ECM components, mainly collagens[20]. Although liver fibrosis occurs as a wound healing response to chronic toxin-mediated liver injury, chronic liver fibrosis can eventually lead to cirrhosis and HCC[2,22,23]. It is highly likely that the prognosis of liver fibrosis and HCC depend on genetic variations among multiple genes and the interactions of these genes with environmental factors and each other[24].
β-catenin is expressed throughout the adult liver. It is well documented that Wnt/β-catenin signaling regulates liver homeostasis, injury and tumorigenesis[25]. The nuclear expression and accumulation of β-catenin is an indication of its activation. As a leading contributor to chronic liver disease progression, aberrant β-catenin activation is detected during the early stages of chronic inflammation, fibrosis, steatosis, steatohepatitis, and hepatoblastoma as well as late stage of HCC. Aberrant activation of the Wnt/β-catenin signaling pathway (overexpression, mutations, increased nuclear expression of β-catenin) is found in up to 50% of human HCCs and correlated with tumor progression and poor prognosis[26,27].
While a hepatic fibrosis background is a hallmark of human HCC, the regulators of this pathology remain largely unclear. The HCC developed in our BRUCE LKO mouse model is associated with fibrosis[3], suggesting that BRUCE is a regulator of this pathology. Therefore, the BRUCE liver-KO mouse model allows us to examine how BRUCE regulates fibrosis and HCC. In this study, we used our mouse model to examine the pro-fibrotic and pro-tumorigenic signaling pathways. Furthermore, we also investigated the stages of chronic liver disease to determine the point of fibrosis induction, as well as tumor initiation in the BRUCE liver-KO mice.
The BRUCE Alb-Cre KO mice (C57/BL6 mice) were previously described[3]. Genotypes were confirmed by PCR and ablation of BRUCE protein expression in mouse liver tissues confirmed by Western blot.
To initiate chronic liver disease pathogenesis, DEN (Sigma, #N0756) was delivered intraperitoneally (i.p.) into control and BRUCE liver KO mice of 14-day old male mice at 25 mg/kg of body weight. Control and KO mice were sacrificed at the following time points: 3-, 6-, 8-, and 14-mo post exposure to DEN and livers were collected for further studies.
Slides were first dewaxed by three xylene washes for 6 min each. Slides were placed into two washes of 100% ethanol for 15 s each followed by a single 95% and 70% ethanol wash, for 15 s each. Slides were then washed with tap water for 1 min then dipped into filtered hematoxylin for 10-12 min. Slides were then rinsed in several washes of tap water until the water was clear. Slides were then dipped twice into a 0.3% Acid Solution (made with ethanol and HCl) then rinsed with tap water for 2 min. Slides were placed into 0.3% ammonia water (made with ammonium hydroxide and distilled water) until the tissue acquired a blue jean color. Slides were rinsed for 2 min in tap water then incubated in 95% ethanol for 20 s. Slides were then placed into Eosin-Y solution for 30 s-1 min, then dehydrated. The dehydration process included: 95% ethanol incubation for 20 s, three 100% ethanol incubations for 20 s, and three xylene incubators for 15 s each. Finally, slides were mounted.
Dewaxed slides were hydrated in an ethanol series: 100% for 5 min, 100% for 5 min, 95% for 3 min, and 70% for 3 min. Slides were then incubated in pico-sirius red for one hour followed by two times of washes in acidified water (made with glacial acetic acid). Slides were dried using filter paper then dehydrated in 3 changes of 100% ethanol for 3 min each. Finally, slides were incubated in xylene for 3 min each then mounted.
To quantify Sirius red images, the scale bar was first measured using the straight-line tool, creating a line along the length of the scale bar. Following this, the scale bar was measured by selecting Analyze > Set Scale to set the scale to micrometers. The scale bar length in micrometers was entered into the “known distance” space and the “um” was entered into “unit of length.” To split the image into three channels, we selected Image > Type > RGB stack then select Image > Stacks > Make Montage to view all three channels at once. The green channel (middle) was selected using the square tool then we selected Image > Adjust > Threshold. The slider was moved lower until the collagen is highlighted in red, then “Set” was selected. The square tool was then used to delete the scale bar area, which was then painted white to prevent its inclusion from the calculated area. Finally, to set measurements, we selected Analyze > Set Measurements and selected “area”, “area fraction”, “limit to threshold”, and “display label”. Finally, to measure we selected Analyze > Measure. The average of the measurements was taken as well as the standard deviation and graphed to represent the quantification analysis.
Liver samples were placed in RNAlater® Solution (Ambion, #AM7020) and kept at 4 °C. Liver RNA isolation was performed using the mirVana™ miRNA Isolation Kit (Ambion, #AM1560) according to the manufacturer’s protocol. RNA was sent to University of Cincinnati Genomics, Epigenomics and Sequencing Core for sequencing analysis.
Paraffin-embedded (formalin-fixed) liver tissue was sectioned to 5-8 μm thickness. Slides were deparaffinized in a series of xylene treatments (5 min). Slides were then rehydrated in an ethanol series (100% for 5 min, 100% for 5 min, 95% for 3 min, and 70% for 3 min). Slides were rinsed with 1 × PBS for 5 min. Antigen retrieval was performed using a solution of 0.1 M Citric Acid and 0.1 M Sodium Citrate. Antigen retrieval solution was boiled for 10 min, then slides were placed in the solution in Coplin jars and boiled in the microwave, 5 min at 100% power, then 5 min at 60% power twice. During each boil, top off the antigen retrieval solution with distilled water. Endogenous peroxidase was blocked by incubating the slides in 30% H2O2 in Methanol. Slides were washed twice in PBS for 5 min. Slides are blocked in 5% normal Goat Serum (Vector Labs, #S-1000) in PBST (made with 0.1% Triton X-100) for one hour at room-temperature. Primary antibody incubation was done overnight at 4 °C. Slides are washed twice with PBS for 5 min. Then slides were incubated with a secondary antibody for one hour at room-temperature. Slides were then washed twice in PBS for 10 min and incubated for 30 min with a Vectastain Elite ABC solution according to the manufacturer’s instructions (Vector Labs, #PK-6100). Slides were then washed twice in PBST for 5 min. Slides were developed by DAB (Sigma, #D3939). Slides were rinsed in tap water followed by a counterstain with hematoxylin. Slides were rinsed with tap water until water is clear then incubated in an acid rinse for 1 min. Slides were rinsed again and incubated with a bluing solution for 1 min. Slides were rinsed then dehydrated in an ethanol series, followed by xylene washes. Slides were mounted and analyzed. Primary Antibodies used in this study include alpha-smooth muscle actin (α-SMA) (CST 19245T), β-catenin (CST 9582), and Ki67 (CST 12202). The secondary antibody used was a biotinylated goat anti-rabbit immunoglobulin G antibody (Vector Labs, #BA-1000).
Images were analyzed using the “Fiji” version of ImageJ software. Image was opened. Color Deconvolution was selected for images stained specifically in the nuclei. To decrease the interference of cytoplasmic staining, images that had nuclear and cytoplasmic staining, under the image pull down, RGB stack was selected under type. To decrease cytoplasmic signal, go to Image > Type > RGB stack. Once the RGB window appears, select Image > Stacks > Make Montagne then perform color deconvolution. For both nuclear-specific and other images, select the Vectors pulldown > “HDAB”. The “Colour_2” image window was selected and measured. The units of intensity derived in the results window were transferred to an excel spreadsheet. The optimal density (O.D.) was calculated using the formula, O.D. = log (max intensity/mean intensity), where the max intensity should be 255. The average optimal density and standard deviations were calculated and graphed.
Slides were examined and percent nuclear positive hepatocytes per field (under 20 × magnification) were counted per 100 cells.
Slides were examined under 20 × magnification and percent nuclear positive cells were calculated per field using the cell counter feature in Fiji.
Control and LKO livers of mice 3- and 8-mo post-DEN exposure were harvested. Cytoplasmic and nuclear fractions were prepared from these livers using the Thermo Scientific Subcellular Protein Fractionation Kit for Tissues (Cat. No. 87790), according to the manufacturer’s recommendations.
Protein extracts (40-100 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose filter. The filter was blocked with 5% dry milk in PBST for 1 h at room temperature, followed by incubation with primary antibody overnight at 4 °C or 3 h at room temperature. The filter was then washed in PBST 3 times for 5 min each, followed by incubation with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. After washing with PBST, the filter was developed with ECL for 1 min and exposed to X-ray film. Quantification of the polypeptide bands was performed with the Fiji software.
Liver cDNA was isolated from RNA templates as described previously[3]. The PCR mix was made using the 1/10 cDNA solution as the template, primers, and the DreamTaq PCR master mix (2 ×) (Thermo Fischer Scientific, #K1071). The PCR products were separated by electrophoresis on a 2% agarose gel. RT-PCR was setup according to the iQ™ Sybr® Green Supermix (BioRad, #170-8882) with the 1/10 cDNA as the template. PCR conditions for the semi-quantitative and RT-PCR are as follows: Initial denaturation-95 °C for 1 cycle; for 40 cycles: Denaturing-95 °C, annealing-Tm as indicated below, extension-72 °C; and an optional hold at 4 °C. Gene primers were obtained from Integrated DNA Technologies.
RNA from Control and LKO livers of mice exposed to DEN for 8 mo was isolated as described previously[3]. RNA samples were submitted to the Genomics, Epigenomics and Sequencing Core at the University of Cincinnati.
HepG2 and THLE2 cells were purchased from ATCC. HepG2 cells were cultured in DMEM high glucose medium with 10% fetal bovine serum and 1% penicillin/ streptomycin at 37 °C in a CO2 (5%) incubator. THLE2 cells were cultured in special medium as suggested by ATCC. The siRNA transfection of cells was mediated by lipofectamine RNAiMAX (Thermo, Cat. No. 13778030) following manufacturer’s instruction.
THLE2 cells were fixed and stained with primary antibodies against BRUCE and protein kinase A (PKA). After washes, cells were incubated with secondary antibodies coupled with Alexa Fluor 594 and Alexa Fluor 488, respectively. Samples were analyzed and photos acquired under Zeiss Fluorescence Microscope.
Cell pellets were lysed and sonicated to elute whole cell lysates in RIPA buffer with protease inhibitor tablets (Roche) and phosphatase inhibitors of 10 mmol/L NaF and 50 mmol/L β-glycerophosphate. The lysates were centrifuged at 15000 g for 20 min and the supernatant was collected.
Antibodies: The antibodies used in this study were: BRUCE from Novus (NB300-264); α-SMA (CST 19245T); Total β-catenin (CST 9582); Ki67 (CST 12202); phospho-β-catenin Ser-675 (CST 4176); Lamin A/C (CST 4777); Actin (CST 3700); Glyceraldehyde-3-phosphate dehydrogenase (CST 2118); phospho-PKA substrate (RRXS*/T*) (100G7E) (CST 9624)
Reagents and siRNAs: DEN (#N0756) from Sigma; BRUCE siRNA and control siRNA were synthesized by Dharmacon[16]. Control siRNA sequence is UUCUCCGAACGUGUCACGUdTdT. The BRUCE siRNA sequence is GGCACAGCAGCTCTTATCA.
Data analysis: The results are expressed as the means ± SD of the determinations. The statistical significance of the difference was determined by a two-tailed Student’s t-test.
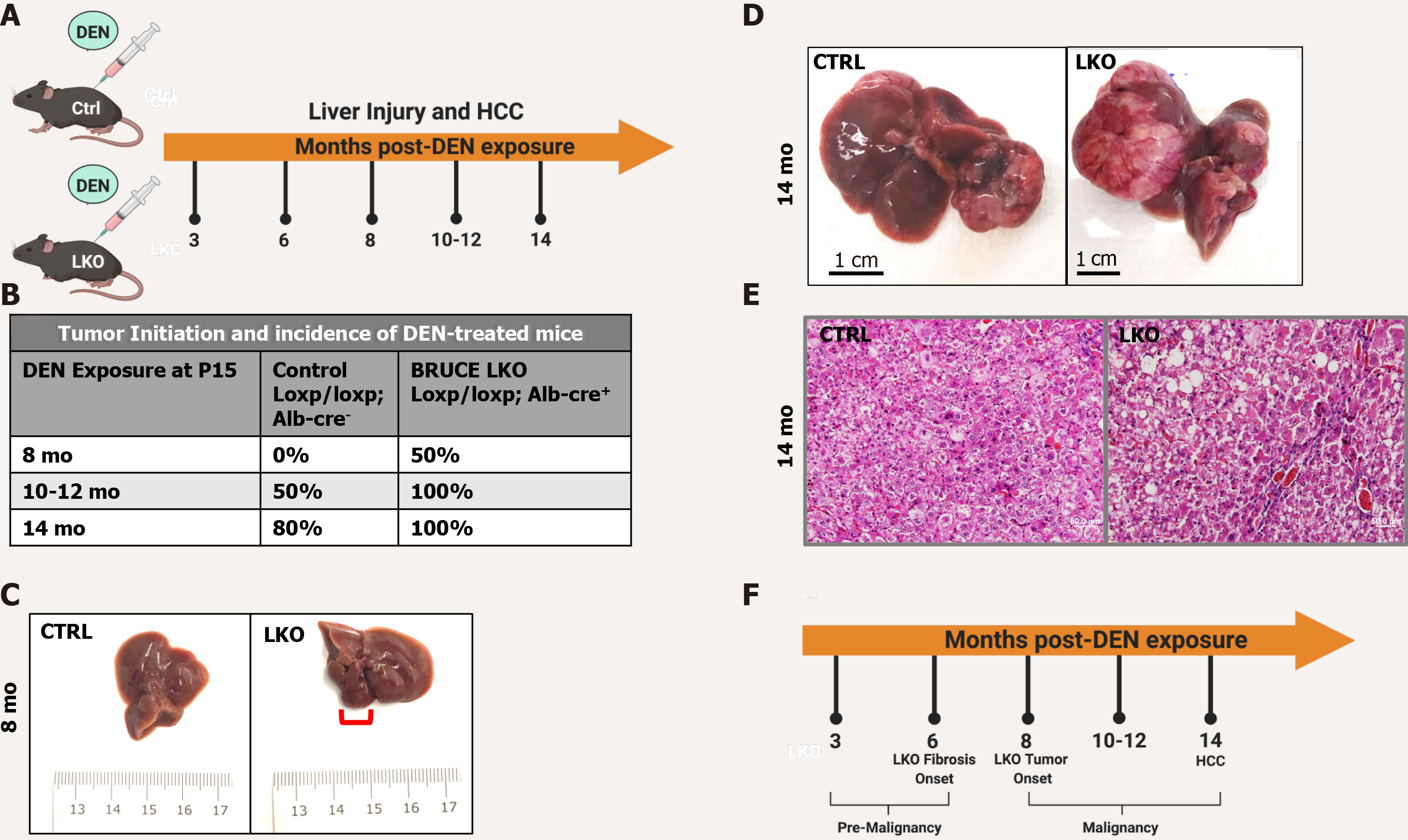
DEN administration in mice promotes chronic liver injury and HCC development[5]. Control (loxp/Loxp; Alb-Cre-) and LKO (loxp/Loxp; Alb-cre+) mice were exposed to DEN at postnatal day 15 to induce liver injury and malignant transformation to HCC. Mice with and without BRUCE expression in the liver were sacrificed at various time points for studies of liver disease progression (Figure 1A). Fifty percent of the LKO mice developed tumors 8 mo after DEN administration, while the control littermates did not begin to develop tumors until 10 mo post DEN administration (Figure 1B and C). By 14 mo, 100% of the LKO mice developed HCC (n = 17) whereas 80% of the control developed HCC (n = 10) (Figure 1B). More of the LKO mice developed an exacerbated HCC phenotype (Figure 1B and D). Histology of the HCC tumors revealed a trabecular architecture identical to the histologic patterns of HCC patients with poor prognosis (Figure 1E). The pathogenic highlights of this study were summarized into a pre-malignant and a malignant stage (Figure 1F). Together the data suggests that hepatic BRUCE deficiency accelerates and exacerbates DEN-induced HCC development in C57/BL6/J mice.
We have reported that DEN administration to BRUCE LKO mice induces more DNA damage accumulation in hepatocytes than that in control mice. We have also demonstrated that the repair of DEN induced hepatocellular DNA damage requires the BRUCE-ATR DNA repair axis[3]. Recently it has been reported that excessive hepatocyte apoptosis plays a tumor-promoting role in nonalcoholic steatohepatitis (NASH)-associated liver cancer in mice[28]. We and others have reported on the importance of BRUCE in the regulation of DNA damage response as well as inhibiting apoptosis[11,12,16], yet the connection of the aforementioned roles of BRUCE have not yet been examined for whether sustained DNA damage correlates with or triggers apoptosis in hepatocytes[29,30]. In addition, DEN exposure induces hepatocellular DNA damage and gene mutations in mice which is a carcinogenic mechanism underlying DEN-initiated liver injury and development of HCC[31]. However, the apoptotic regulators that are critical in the regulation of hepatic apoptosis induced by exposure to DEN have not been well established. We reasoned that BRUCE LKO mice have lost the IAP function of BRUCE in the liver, thus DEN exposure of LKO mice could induce more prominent hepatocyte apoptosis than in the control. Indeed, our RNA-seq analysis found that DEN administration results in a higher level of apoptotic gene expressions (Figure 2A), suggesting that hepatic BRUCE protects against DEN-induced hepatic apoptosis.
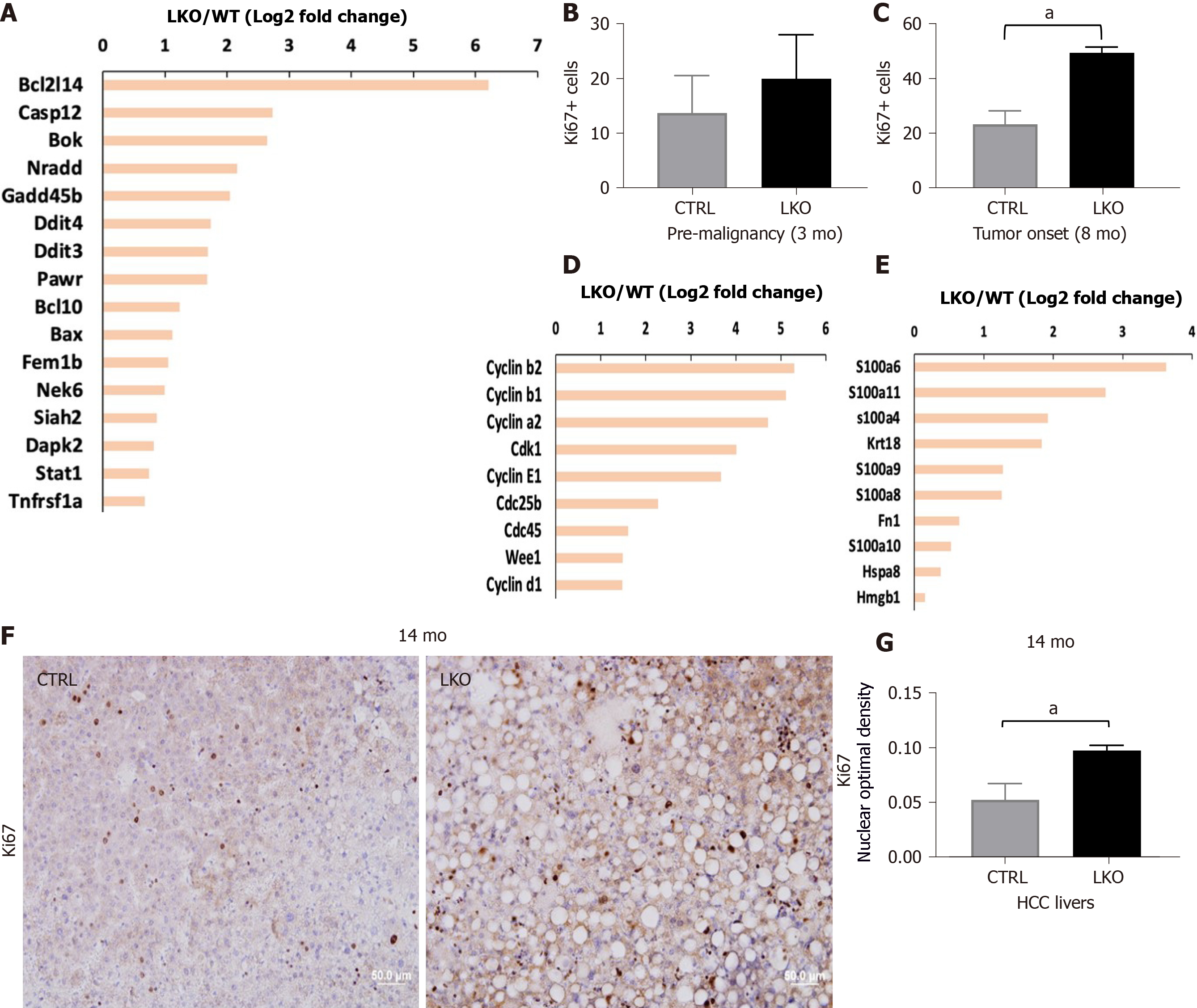
One of the pathological outcomes of hepatocyte apoptosis is the facilitation of hepatic inflammation and fibrosis through two major mechanisms: (1) Replenishment of lost hepatocytes via compensatory proliferation of quiescent hepatocytes through feed-forward apoptosis-proliferation circles[26,32]; and (2) Release of proinflammatory DAMPs which result in liver inflammation and fibrosis[33]. Our immunohistochemical staining of a proliferation marker, Ki67, showed enhanced hepatocyte compensatory proliferation in BRUCE-deficient mouse livers compared to the control, during the pre-malignant stage (3 mo post DEN administration) (Figure 2B). This proliferation continued to the time of tumor onset (8 mo post DEN exposure) (Figure 2C). This elevated hepatic proliferation is supported by increased expression of multiple critical cell-cycle regulatory genes in BRUCE-deficient livers by RNA-seq analysis (Figure 2D). In addition, gene expression of DAMP molecules including HMGB1 and S100 are also increased in KO livers (Figure 2E). Moreover, increased cellular proliferation in human HCC has been correlated with tumor progression and poor prognosis[34,35]. During the malignant stage of 14 mo post DEN exposure, the HCC tissues from LKO mice exhibited increased Ki67 expression compared to the HCCs from control mice (Figure 2F and G). Together our data demonstrate that hepatic BRUCE deficiency results in elevated hepatic cell apoptosis and proliferation. Moreover, the release of DAMPs would exacerbate liver inflammation.
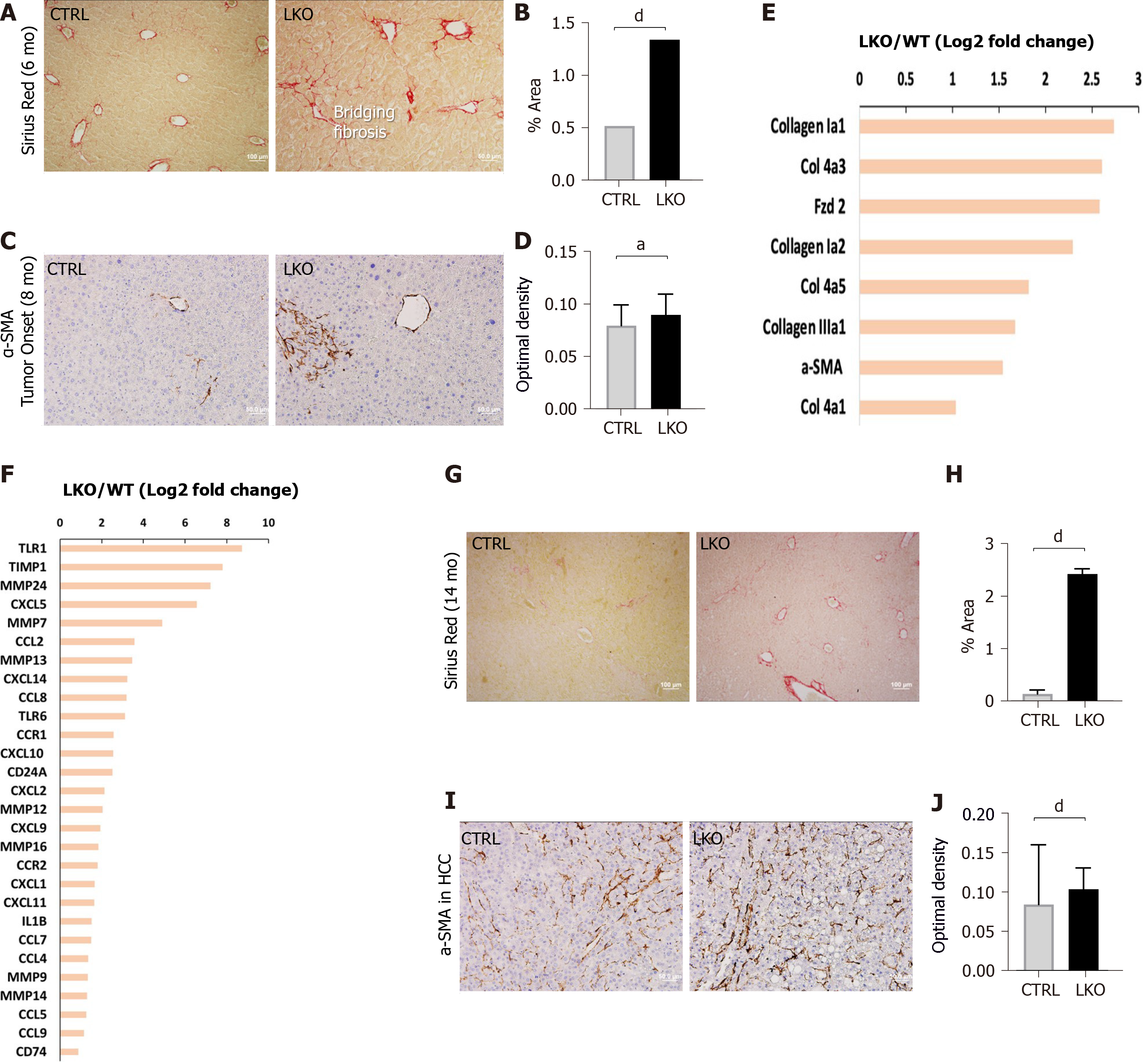
Hepatic fibrosis is characterized by an excessive accumulation of ECM in which collagen fibers are the major component produced by activated HSCs. A unique feature of liver cancer is its close association to liver fibrosis. More than 80% of HCCs develop in fibrotic or cirrhotic livers, suggesting an important role of liver fibrosis in the pre-malignant environment of the liver[6]. Although fibrosis is a feature of human HCC, it is not necessarily recapitulated in various murine liver disease models, which makes these models inferior in modeling the progression of fibrosis to HCC. Interestingly, the LKO mice developed significant fibrosis in the pre-malignant stage at 6 mo following DEN administration, as shown by the Sirius red staining of collagen fibers (Figure 3A and B). Notably the collagen fibers in BRUCE KO livers showed signs of the advanced stage of “bridging fibrosis”[36], as evidenced by the fibrotic spreading that extends between portal and central vein areas (Figure 3A). This bridging fibrosis is in sharp contrast to the control mice in which fibrosis was limited to portal or venular areas (Figure 3A). Upon chronic liver damage, injured hepatocytes undergo cell death which releases DAMP molecules and activate the normally quiescent HSCs[33]. To directly examine HSC activation, we analyzed the expression of α-SMA, which is expressed by HSCs and reflects their activation into a myofibroblast-like phenotype. The results revealed an increased α-SMA expression in HSCs which suggests an elevated activation of HSCs in LKO mice as compared to the control (Figure 3C and D). To validate these pro-fibrotic events at the gene expression level, RNA-seq analysis was conducted and multiple pro-fibrotic or fibrotic genes were found to have higher levels of expression in the LKO liver than that of the control (Figure 3E). Furthermore, elevated inflammatory gene expression was evident in LKO mice compared to control (Figure 3F). As liver fibrosis is characterized by the deposition of fibrillar collagens, the elevated gene expression of multiple types of collagens (Figure 3E) support the elevated fibrosis phenotype. In addition, HSCs contribute to the accumulation of ECM by producing excessive amounts of pro-fibrotic factors such as tissue inhibitor of metalloproteinases (TIMPs)[37]. Indeed, we observed an elevated level of TIMP1 gene expression in LKO mice (Figure 3F). Moreover, the turnover of ECM, controlled by matrix metalloproteinases (MMPs), also promotes fibrosis and therefore a number of MMPs are highly expressed in liver fibrosis[19]. We also found that the gene expression of a number of MMPs were increased in LKO mouse livers (Figure 3F). Altogether, the data indicate that DEN exposure of BRUCE LKO mice aggravates hepatic fibrosis at both the histological and gene expression levels.
To ascertain whether the hepatic fibrosis is sustained during the malignancy stage, HCC tissues from 14-mo exposed mice were analyzed for both Sirius red staining and α-SMA expression. The results from both analyses demonstrated an exacerbated fibrosis that concur with HCCs (Figure 3G-J), demonstrating that sustained and chronic fibrosis coexists with HCCs, which is a hallmark of human HCCs. Altogether, hepatic BRUCE deficiency in the DEN-induced HCC model is sufficient to drive fibrosis in both pre-malignant and malignant stages.
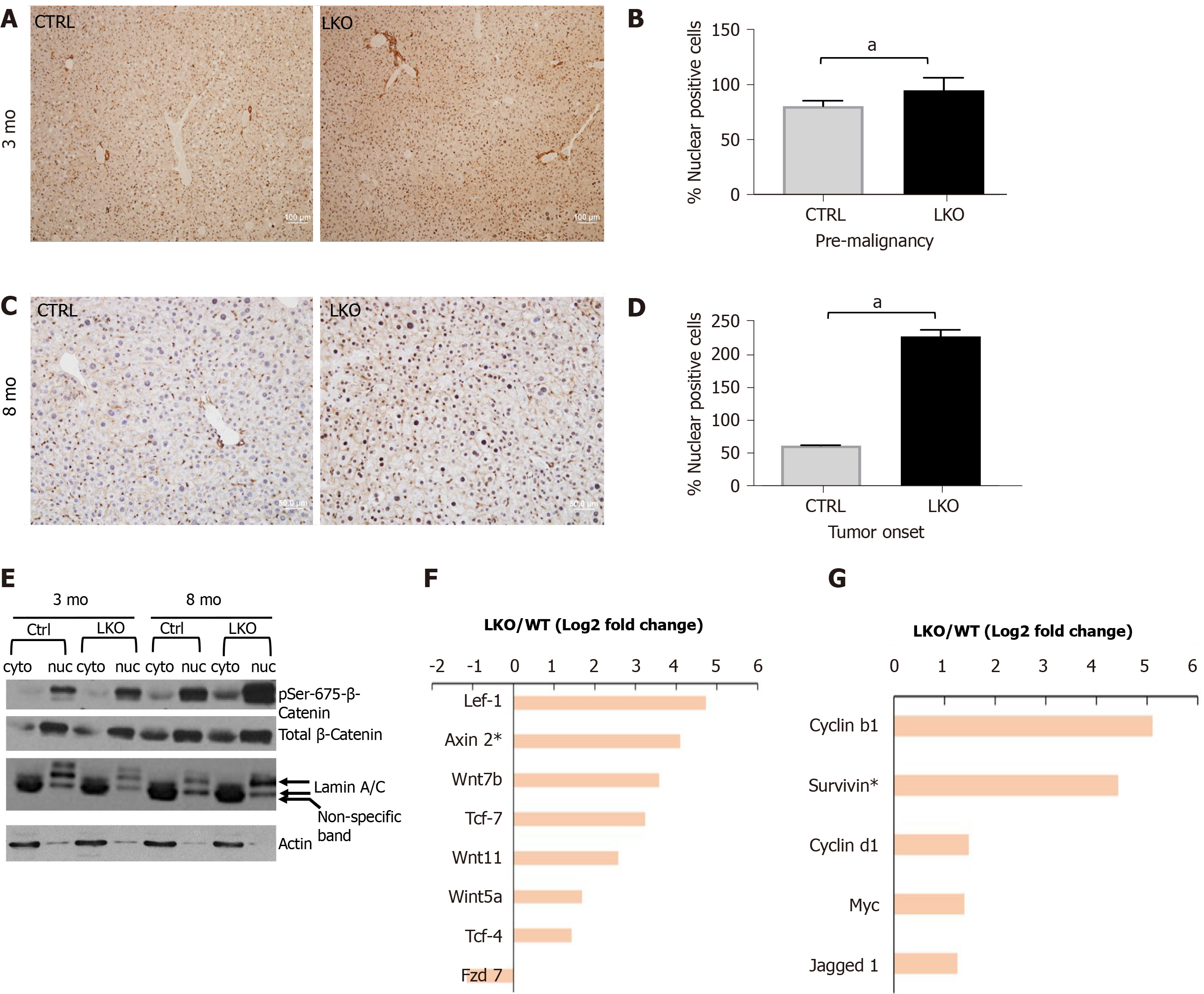
Activated Wnt/β-catenin pathway is indicative of the stabilization of β-catenin in the cytoplasm and its subsequent translocation to the cell nucleus, where it achieves its gene transcription function by activating gene expressions for promotion of hepatic inflammation and fibrosis[38-41]. Remarkably, there was a pronounced increase of nuclear localization of β-catenin in the liver sections from BRUCE LKO mice in the pre-malignant stage of 3 mo (Figure 4A and B) as well as at tumor onset of 8 mo post DEN exposure (Figure 4C and D). The nuclear localization of β-catenin in liver tissue sections assessed by immunohistochemistry (IHC) was further validated by a biochemical approach. Specifically, liver protein extracts from mice at the pre-malignant and tumor-onset stages were further fractionated into cytoplasmic and nuclear fractions and immunoblotted for β-catenin. There was a much higher level of total β-catenin in the nuclear fraction than in the cytosol of both control and LKO samples (Figure 4E). Promoted by the increase of nuclear β-catenin in liver tissue sections, we postulated that there could be an increase in β-catenin activity in the nuclear fraction of LKO liver samples. To test this possibility, we compared the levels of β-catenin phosphorylation at Ser-675, an activated form of β-catenin phosphorylated by cAMP-dependent PKA[42,43] in the control and LKO samples. Indeed, there was a dramatic increase of phospho-β-catenin at Ser-675 in the nuclear fractions of the LKO livers in both stages of pre-malignancy and tumor onset (Figure 4E), suggesting that β-catenin plays an important role in the promotion of hepatic inflammation and fibrosis in the early, pre-malignant stage. The RNA-seq analysis confirmed an upregulation in the expression of multiple Wnt ligands and regulators (Figure 4F), and β-catenin target genes (Figure 4G). Together the data demonstrate an aberrant activation of the Wnt/β-catenin pathway, which plays a pro-inflammatory and fibrotic role in LKO livers as shown in Figure 3. Collectively, these results indicate a new mechanism for an upregulated Wnt/β-catenin pathway resulting from hepatic BRUCE deficiency during the pre-malignant stage of hepatic inflammation and fibrosis in mice.
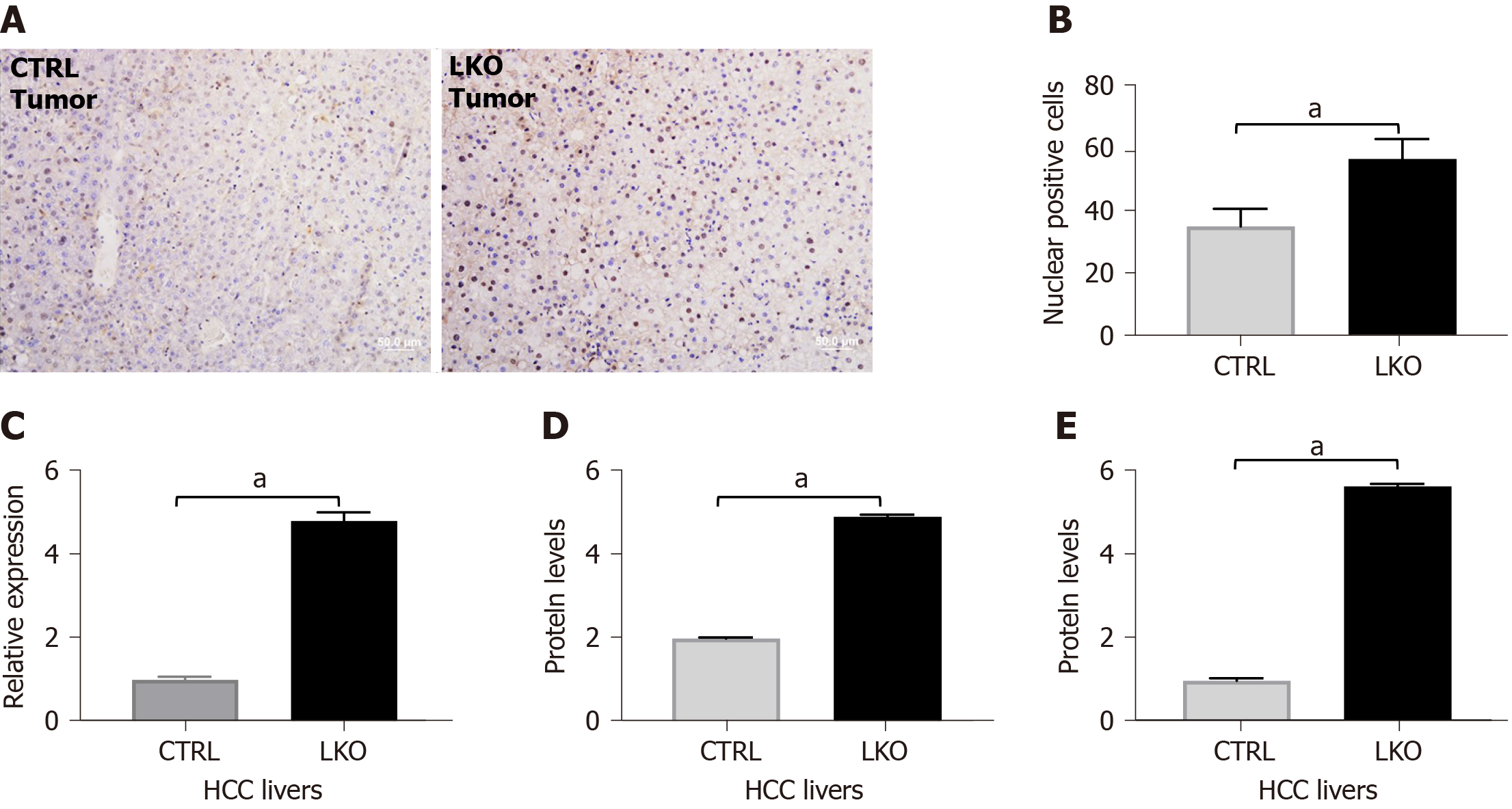
At age 14 mo, BRUCE LKO mice had a more exacerbated DEN-induced HCC phenotype (Figure 1D) consistent with a human HCC-like trabecular histological feature (Figure 1E). Livers from LKO mice maintained the nuclear localization of β-catenin (Figure 5A and B). Additionally, mRNA levels of β-catenin were measured by qRT-PCR and found to be increased in the LKO livers (Figure 5C). To confirm the IHC analysis of increased β-catenin, we analyzed β-catenin protein levels by Western blot analysis (Figure 5D) and noticed a concomitant increase in protein expression in LKO HCC livers. Additionally, cyclin D1, a downstream target of β-catenin, was increased at the protein level in LKO HCC tissues (Figure 5E). Together the data demonstrate that BRUCE deficiency increases β-catenin nuclear accumulation in DEN-induced HCC. As β-catenin activity plays a critical oncogenic role in the development of HCC, these data suggest that upregulated β-catenin activity induced by BRUCE deficiency contributes to the accelerated HCC development in mice.
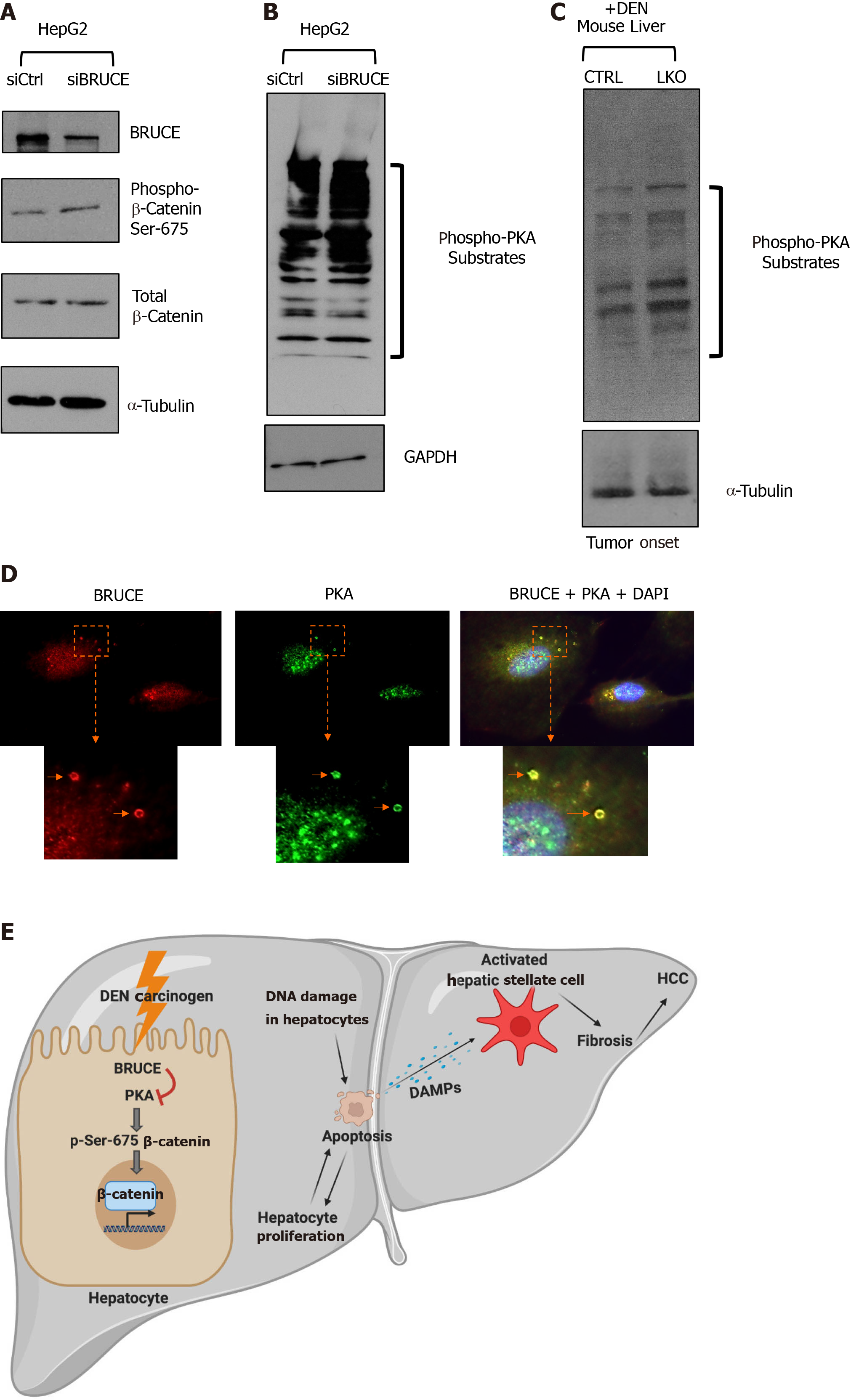
An increase of nuclear β-catenin in LKO mice at the stages of pre-malignant (3 mo), tumor onset (8 mo) and malignant (14 mo) (Figure 4) suggests that hepatic BRUCE regulates β-catenin activation. To investigate the underlying mechanisms, we utilized an in vitro cell culture system of the human liver cancer cell line HepG2 which allows for knockdown (KD) experiments. We first determined if an increase of phospho-β-catenin at Ser-675 can be induced by KD of BRUCE expression in HepG2 cells. HepG2 cells were transfected with either a control or a BRUCE siRNA followed by preparation of the whole cell protein lysates for Western blot analysis. Knockdown of BRUCE in HepG2 cells resulted in increased levels of both the total β-catenin protein and phospho-β-catenin at Ser-675 (Figure 6A), demonstrating that BRUCE negatively regulates β-catenin activation. Since phosphorylation of β-catenin at Ser-675 is PKA-dependent, we reasoned that loss of BRUCE expression might be linked to the activation of PKA activity. To test this possibility, we compared PKA activity in cells with and without BRUCE knockdown by examination of the levels of PKA phosphorylated substrates using an antibody specific to PKA substrates. The Western blotting results showed an increase in phospho-PKA substrates in BRUCE KD cells (Figure 6B), demonstrating that the loss of BRUCE expression induces activation of PKA, thereby resulting in a higher PKA activity. This link of BRUCE loss to PKA activity elevation was also reproduced in vivo in DEN-exposed mice, demonstrated by the increase of phospho-PKA substrates by Western blot analysis of liver protein extracts at the time of tumor onset in the LKO mice (Figure 6C). To further delineate the correlation of BRUCE loss with the upregulation of PKA activity, we performed co-immunofluorescence analysis of both proteins with a human normal hepatocyte line, THLE2. We found that BRUCE and PKA were co-localized in endosomes (Figure 6D). BRUCE is reported to be on endosomes in non-hepatocytes[44]; however, this is the initial report of BRUCE and PKA colocalization on endosomes in human hepatocytes. This colocalization suggests that in endosomes of hepatocytes, BRUCE interacts with PKA to restrain hyperactivation of PKA, whereas loss of BRUCE releases the restriction of PKA activation and thus PKA activity is elevated.
With the observation of PKA-dependent phospho-β-catenin at Ser-675 upon liver injury with DEN, we propose a new signaling axis of BRUCE-PKA-β-catenin in the regulation of liver function. In this axis, hepatic BRUCE suppresses hyperactivation of PKA activity, thereby preventing aberrant phosphorylation and activation of β-catenin as well as its subsequent profibrogenic and oncogenic functions. In livers devoid of BRUCE, there is a loss of the BRUCE-dependent negative regulation of PKA activation; therefore, PKA phosphorylates and activates β-catenin to aggravate hepatic fibrosis and accelerate HCC (schematic, Figure 6E).
We have previously reported the clinical relevance of BRUCE in liver diseases in which BRUCE downregulation is found in a large portion of liver disease patients, including fibrosis, hepatitis, NASH, and HCC[3]. Upon assessment of BRUCE protein expression levels in liver specimens (male and female patients), we found that BRUCE levels were reduced in 54.5% of hepatitis samples (n = 22), 46.7% of cirrhosis samples (n = 30), and 84% of HCC samples (n = 25)[3]. These findings suggest a correlation between BRUCE expression levels and various liver disease stages. Additionally, we previously reported a 6% rate of deleterious BRUCE mutations in HCC patients, as deduced through the Cancer Genome Atlas. This rate was comparable to the mutation rate of other key DNA damage response (DDR) genes such as, ATR, BRCA1 and BRCA2 in HCC patients. Furthermore, we delineated frameshift and nonsense mutations of BRUCE, particularly in BRUCE’s ubiquitin conjugating (UBC) domain[3]. Our group has previously established that the UBC domain of BRUCE is necessary for its DDR function[11,12]. Therefore, deleterious BRUCE mutations would inactivate BRUCE’s DDR function and could contribute to overall genomic instability leading to HCC development[3,12].
BRUCE has two major functions. It facilitates DDR to maintain genomic stability and as an IAP-family member, it suppresses apoptosis to maintain cell viability[15,16]. Liver KO of BRUCE abolishes both of these functions in the liver. While DDR inactivation and genomic instability promote HCC development in BRUCE-deficient settings[3], inflammation and fibrosis are well characterized risk factors in HCC pathogenesis[45,46]. Therefore, we focused on the progression of fibrosis in this study. The loss of hepatic BRUCE together with DEN administration contributes to an increase in DAMPs which lay the foundation for fibrosis as well as contribute to the progression of HCC. Loss of BRUCE’s anti-apoptotic function results in elevated hepatocyte apoptosis and the release of DAMPs, which promote compensatory hepatocyte proliferation (Figure 2). Upon release of DAMPs, the quiescent HSCs will become activated which will progressively trigger the onset of fibrosis (Figure 3). As previously reported, increased hepatic fibrosis and compensatory proliferation are contributors to both HCC and poor prognosis[6,33-35,47].
Nonetheless, there are a number of risk factors that predominate the development of HCC in humans. Infection with hepatitis B or C viruses, alcohol consumption and metabolic syndrome are also major risk factors. Since DNA damage and apoptosis are likely common to liver fibrosis and HCC induced by these risk factors, BRUCE is anticipated to also protect against liver diseases induced by these risk factors, which is our future direction for this research. In addition to DNA repair and anti-apoptosis, BRUCE also regulates autophagy and cellular energy levels as we previously published[48]. As autophagy is involved in the regulation of liver homeostasis and liver injury and because of the robust autophagic activity found in liver tissue[49-51], it is likely that BRUCE also regulates liver injury, fibrosis and carcinogenesis through autophagy. BRUCE likely coordinates multiple signaling pathways including autophagy, DNA repair and apoptosis to preserve liver homeostasis.
Liver diseases present a huge health threat and are on the rise. However, the molecular pathways leading to fibrosis and HCC are not fully defined, which have hampered the development of mechanism-based therapeutic intervention. Aberrant β-catenin activation and its nuclear localization in the promotion of liver disease is found in up to 50% of human HCCs[22,27]. It is believed that aberrant β-catenin activation is a key contributor to chronic liver disease progression. Finding upstream regulators of β-catenin pathogenic activation is necessary for identification of the right sub-group of chronic liver disease patients for considering mechanism-based therapeutic targeting.
This study provides new insights into the molecular pathways that contribute to liver fibrosis and HCC. We revealed a previously unknown hepatocellular “BRUCE-PKA-β-catenin signaling axis” involved in the regulation of fibrosis and HCC (Figure 6E). In this signaling axis, we have identified a novel role of hepatocellular BRUCE in the suppression of aberrant activation of β-catenin through preventing PKA-mediated phosphorylation and activation of β-catenin both in vivo and in vitro. Mechanistically, we have revealed a novel interaction between BRUCE and PKA in the hepatocyte cytoplasm at endosomes, which provides the support for a functional interaction of these two proteins in the regulation of liver functions. This is further supported by our observations that upon disruption of BRUCE function either by liver KO (animal) or KD (HepG2 cell line), the repression of PKA is derepressed and PKA-dependent phosphorylation of β-catenin at Ser-675 occurs. This β-catenin phosphorylation is associated with the early onset of fibrosis and accelerated HCC in our mouse model (Figure 6E).
How might BRUCE regulate PKA protein levels and its activation? BRUCE itself is a hybrid protein harboring ubiquitin conjugase and ligase activities[15]. During the intrinsic mitochondrial pathway of apoptosis, BRUCE catalyzes ubiquitination of pro-apoptotic proteins SMAC, Caspase-9 and others to reduce cellular apoptotic capacity to tip the balance of life and death towards cell death[17,51]. In addition, during DNA damage response induced by DNA double-strand and single-strand breaks, BRUCE ubiquitin ligase activity cooperates with the deubiquitinase USP8 to regulate ATM and ATR DNA damage responses to facilitate HR repair of DNA breaks[3,11,12]. Therefore, we propose that hepatic BRUCE-regulated protein ubiquitination signaling controls liver functions and conversely, lack of BRUCE expression results in dysregulation of ubiquitin signaling and accelerates liver disease development. BRUCE repression of PKA hyperactivation suggests a possible ubiquitination mechanism, in which BRUCE normally represses PKA activity through promotion of PKA ubiquitination and subsequent degradation through the UPS, thereby preventing β-catenin phosphorylation and activation. In the absence of BRUCE through genetic ablation of BRUCE in the mouse liver or gene knockdown in liver cancer cell lines, PKA becomes stabilized and β-catenin is activated. This possibility is currently under investigation in the lab.
This study has opened new avenues for focusing on BRUCE protection against liver injury, fibrosis and liver cancer. In addition to the “BRUCE-PKA-β-catenin signaling axis”, other functions of BRUCE can also impact liver disease progression. In this regard, we have shown that BRUCE’s function in the promotion of DNA damage repair is implicated in HCC development initiated by DEN[3]. Being an IAP in the suppression of mitochondrial pathway of apoptosis, BRUCE deficiency can make hepatocytes more susceptible to apoptosis under hepatic oxidative stress and detoxication, which are physiological processes inherent to livers. BRUCE also impacts autophagy and loss of BRUCE reduces cellular energy and increases autophagy flux[47]. Since autophagy regulates liver functions, the impact of BRUCE on liver autophagy and its connection with liver disease progression is under investigation in our lab. Future studies will focus on the interplay among these pathways in the maintenance of liver homeostasis and suppression of liver diseases in genetically modified murine models, including humanized murine models as it has shown promises to better understand human liver fibrosis.
The Wnt/β-catenin pathway has been regarded as a crucial mechanism involved in fibrosis and hepatocarcinogenesis. However, only a limited number of efficient targeted therapies are available for aberrant activation of this pathway in inhibiting chronic liver disease progression. Findings from this study provide the rationale to stratify the subset of liver disease patients with BRUCE mutant or deficiency and to test the therapeutic potential of targeting aberrant activation of the cAMP-PKA and Wnt/β-catenin pathways.
We previously reported the clinical relevance of somatic deleterious mutations in BRUCE or its downregulation in a large patient population with hepatitis, fibrosis and HCC[3]. In conclusion, this study identifies BRUCE as a suppressor of liver fibrosis in the premalignant and malignant stages in a DEN-induced hepatocarcinogenic murine model. Mechanistically, this study elucidates a previously unrecognized “BRUCE-PKA-β-catenin” signaling pathway contributing to hepatic proliferation, fibrosis and malignancy. Specifically, by using in vitro and in vivo approaches, we showed that hepatic BRUCE-deficiency releases its suppression of PKA kinase activity, leading to PKA-dependent phosphorylation and activation of β-catenin. In contrast to DEN exposure alone, which does not induce robust fibrosis, DEN treatment in a BRUCE null background accelerates fibrosis, which likely drives the early HCC development in BRUCE LKO mice. Considering the significant clinical relevance of BRUCE in patients with liver diseases, this study has demonstrated that our BRUCE LKO mouse model is a promising model for recapitulating human liver disease progression for dissecting the complicated pathological mechanisms underlying liver disease progression.
BIR repeat-containing ubiquitin-conjugating enzyme (BRUCE) is a known ubiquitin conjugase/Ligase hybrid that has been shown to inhibit apoptosis, regulate efficient DNA repair, and most recently promote tumor suppression in the liver. Our group previously showed that upon liver injury with diethylnitrosamine (DEN), loss of hepatic BRUCE promoted fibrosis and exacerbated hepatocellular carcinoma (HCC) development in mice.
About 80% of HCCs develop in fibrotic or cirrhotic livers, demonstrating the importance of understanding liver fibrosis as a factor contributing to hepatic malignancy. Identifying mechanisms that can regulate both fibrosis and HCC development simultaneously provides the possibility of opening therapeutic windows for treating fibrosis and HCC. Considering that over 50% of human HCCs have aberrant β-catenin mutations, targeting the Wnt/β-catenin has shown much promise. The key upstream regulators of this pathway that suppress fibrosis and HCC development remain elusive.
The objective of this study was to evaluate the mechanisms of BRUCE in inhibiting hepatic fibrosis and HCC upon liver injury induction.
Male C57/BL6/J control mice [loxp/Loxp; albumin-cre (Alb-cre)-] and BRUCE Alb-Cre KO mice (loxp/Loxp; Alb-Cre+) were injected with a single dose of DEN at postnatal day 15. Mice were sacrificed at various time points to examine liver disease progression and liver biopsies were used in the analyses of the proposed mechanism.
Based on the exacerbation of fibrosis and HCC phenotypes observed in the liver-specific BRUCE knockout (LKO) mice that we previously reported, we hypothesized that, “the onset of fibrosis and tumorigenesis are likely earlier events in LKO mice”. In the present study, we found that upon DEN-induction, BRUCE LKO livers developed fibrosis as early as after 6 mo of exposure. Additionally, the LKO mice developed tumors as early as 8-months after exposure compared to the WT tumor onset after 10 mo of DEN exposure. Furthermore, we observed increased accumulation of β-catenin, including its activity in LKO liver samples. The phosphorylation of β-catenin was determined by measuring nuclear levels of total β-catenin, and Ser-675 phosphorylated β-catenin. Additionally, the activity of protein kinase A (PKA), one of the upstream kinases that phosphorylates β-catenin at Ser-675, was found to be increased in both BRUCE-deficient mouse livers and a human liver cancer cell line. More importantly, BRUCE and PKA were found to be colocalized in the cytoplasm of hepatocytes.
In conclusion, this study further demonstrated BRUCE’s liver tumor suppressive function, by identifying the early onset of tumorigenesis in LKO mice. Furthermore, the current study elucidated a novel role of BRUCE in the negative regulation of PKA activity in order to negatively regulate β-catenin stabilization and activity. Together, BRUCE’s regulation of β-catenin through PKA, is a likely mechanism used to suppress hepatic diseases, such as fibrosis and HCC.
While further investigation is warranted, this study revealed the novel role of BRUCE in hepatic regulation of β-catenin upon liver injury. Further establishing BRUCE’s regulation of PKA activity can possibly provide more promising therapeutic approaches for treating liver disease patients with aberrant expression of BRUCE and β-catenin.
The authors would like to acknowledge the thesis committee for their support and guidance to Chrystelle L Vilfranc, and Susan Waltz and Carolyn Price for the support and training provided to Chrystelle L Vilfranc through the T32 training grant. Additionally, the authors are grateful to Dr. Sohaib Khan for additional review and editing of the manuscript.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yang L S-Editor: Fan JR L-Editor: A P-Editor: Wang LL
| 1. | Gao B. Basic liver immunology. Cell Mol Immunol. 2016;13:265-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 2. | Gao J, Wang GJ, Wang Z, Gao N, Li J, Zhang YF, Zhou J, Zhang HX, Wen Q, Jin H, Qiao HL. High CYP2E1 activity correlates with hepatofibrogenesis induced by nitrosamines. Oncotarget. 2017;8:112199-112210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Ge C, Vilfranc CL, Che L, Pandita RK, Hambarde S, Andreassen PR, Niu L, Olowokure O, Shah S, Waltz SE, Zou L, Wang J, Pandita TK, Du C. The BRUCE-ATR Signaling Axis Is Required for Accurate DNA Replication and Suppression of Liver Cancer Development. Hepatology. 2019;69:2608-2622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 2893] [Article Influence: 482.2] [Reference Citation Analysis (17)] |
| 5. | Heindryckx F, Colle I, Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. Int J Exp Pathol. 2009;90:367-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 293] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 6. | Affo S, Yu LX, Schwabe RF. The Role of Cancer-Associated Fibroblasts and Fibrosis in Liver Cancer. Annu Rev Pathol. 2017;12:153-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 511] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 7. | Yu LX, Ling Y, Wang HY. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. 2018;2:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 214] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 8. | Uehara T, Pogribny IP, Rusyn I. The DEN and CCl4 -Induced Mouse Model of Fibrosis and Inflammation-Associated Hepatocellular Carcinoma. Curr Protoc Pharmacol 2014; 66: 14.30.1-14. 3010;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 9. | Lotz K, Pyrowolakis G, Jentsch S. BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol. 2004;24:9339-9350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell. 2017;168:644-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 705] [Cited by in RCA: 980] [Article Influence: 122.5] [Reference Citation Analysis (0)] |
| 11. | Ge C, Che L, Du C. The UBC Domain Is Required for BRUCE to Promote BRIT1/MCPH1 Function in DSB Signaling and Repair Post Formation of BRUCE-USP8-BRIT1 Complex. PLoS One. 2015;10:e0144957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Ge C, Che L, Ren J, Pandita RK, Lu J, Li K, Pandita TK, Du C. BRUCE regulates DNA double-strand break response by promoting USP8 deubiquitination of BRIT1. Proc Natl Acad Sci USA. 2015;112:E1210-E1219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T. A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun. 1999;264:847-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 166] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 14. | Martin SJ. An Apollon vista of death and destruction. Nat Cell Biol. 2004;6:804-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 15. | Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol. 1998;141:1415-1422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 184] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 16. | Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, Katayama R, Hashimoto C, Zhang X, Noda T, Tsuruo T, Naito M. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 179] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 17. | Qiu XB, Markant SL, Yuan J, Goldberg AL. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 2004;23:800-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 111] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 741] [Article Influence: 74.1] [Reference Citation Analysis (0)] |
| 19. | Tu T, Calabro SR, Lee A, Maczurek AE, Budzinska MA, Warner FJ, McLennan SV, Shackel NA. Hepatocytes in liver injury: Victim, bystander, or accomplice in progressive fibrosis? J Gastroenterol Hepatol. 2015;30:1696-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 20. | Trauner M, Arrese M, Wagner M. Fatty liver and lipotoxicity. Biochim Biophys Acta. 2010;1801:299-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 216] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 21. | Khalaf AM, Fuentes D, Morshid AI, Burke MR, Kaseb AO, Hassan M, Hazle JD, Elsayes KM. Role of Wnt/β-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J Hepatocell Carcinoma. 2018;5:61-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 170] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 22. | Debebe A, Medina V, Chen CY, Mahajan IM, Jia C, Fu D, He L, Zeng N, Stiles BW, Chen CL, Wang M, Aggarwal KR, Peng Z, Huang J, Chen J, Li M, Dong T, Atkins S, Borok Z, Yuan W, Machida K, Ju C, Kahn M, Johnson D, Stiles BL. Wnt/β-catenin activation and macrophage induction during liver cancer development following steatosis. Oncogene. 2017;36:6020-6029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 23. | Li YS, Leng CL, Chen MT, Zhang WK, Li XJ, Tang HB, Shang HC, Zhu LH. Mouse hepatic neoplasm formation induced by trace level and low frequency exposure to diethylnitrosamine through β-catenin signaling pathway. Toxicol Res (Camb). 2016;5:210-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Monga SP. β-catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology. 2015;148:1294-1310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 398] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 25. | Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1568] [Article Influence: 82.5] [Reference Citation Analysis (0)] |
| 26. | Liu LJ, Xie SX, Chen YT, Xue JL, Zhang CJ, Zhu F. Aberrant regulation of Wnt signaling in hepatocellular carcinoma. World J Gastroenterol. 2016;22:7486-7499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 74] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 27. | Hirsova P, Bohm F, Dohnalkova E, Nozickova B, Heikenwalder M, Gores GJ, Weber A. Hepatocyte apoptosis is tumor promoting in murine nonalcoholic steatohepatitis. Cell Death Dis. 2020;11:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 558] [Cited by in RCA: 685] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 29. | Norbury CJ, Zhivotovsky B. DNA damage-induced apoptosis. Oncogene. 2004;23:2797-2808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 516] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 30. | Connor F, Rayner TF, Aitken SJ, Feig C, Lukk M, Santoyo-Lopez J, Odom DT. Mutational landscape of a chemically-induced mouse model of liver cancer. J Hepatol. 2018;69:840-850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 31. | Teoh NC, Dan YY, Swisshelm K, Lehman S, Wright JH, Haque J, Gu Y, Fausto N. Defective DNA strand break repair causes chromosomal instability and accelerates liver carcinogenesis in mice. Hepatology. 2008;47:2078-2088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 32. | Martin-Murphy BV, Holt MP, Ju C. The role of damage associated molecular pattern molecules in acetaminophen-induced liver injury in mice. Toxicol Lett. 2010;192:387-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 33. | Aithal GP, Guha N, Fallowfield J, Castera L, Jackson AP. Biomarkers in liver disease: emerging methods and potential applications. Int J Hepatol. 2012;2012:437508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Wu J, Shen SL, Chen B, Nie J, Peng BG. Numb promotes cell proliferation and correlates with poor prognosis in hepatocellular carcinoma. PLoS One. 2014;9:e95849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 35. | Manning DS, Afdhal NH. Diagnosis and quantitation of fibrosis. Gastroenterology. 2008;134:1670-1681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 309] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 36. | Geervliet E, Bansal R. Matrix Metalloproteinases as Potential Biomarkers and Therapeutic Targets in Liver Diseases. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 37. | Grünbaum A. "Meaning" connections and causal connections in the human sciences: the poverty of hermeneutic philosophy. J Am Psychoanal Assoc. 1990;38:559-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 38. | Chen Y, Chen X, Ji YR, Zhu S, Bu FT, Du XS, Meng XM, Huang C, Li J. PLK1 regulates hepatic stellate cell activation and liver fibrosis through Wnt/β-catenin signalling pathway. J Cell Mol Med. 2020;24:7405-7416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 39. | Nishikawa K, Osawa Y, Kimura K. Wnt/β-catenin Signaling as a Potential Target for the Treatment of Liver Cirrhosis Using Antifibrotic Drugs. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 40. | Roehlen N, Crouchet E, Baumert TF. Liver Fibrosis: Mechanistic Concepts and Therapeutic Perspectives. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 750] [Article Influence: 150.0] [Reference Citation Analysis (0)] |
| 41. | Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem. 2006;281:9971-9976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 354] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 42. | Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol Cell Biol. 2005;25:9063-9072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 343] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 43. | Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 44. | Herbst DA, Reddy KR. Risk factors for hepatocellular carcinoma. Clin Liver Dis (Hoboken). 2012;1:180-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 45. | Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68:526-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 523] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 46. | O'Rourke JM, Sagar VM, Shah T, Shetty S. Carcinogenesis on the background of liver fibrosis: Implications for the management of hepatocellular cancer. World J Gastroenterol. 2018;24:4436-4447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 89] [Cited by in RCA: 115] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 47. | Allaire M, Rautou PE, Codogno P, Lotersztajn S. Autophagy in liver diseases: Time for translation? J Hepatol. 2019;70:985-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 288] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 48. | Khambu B, Yan S, Huda N, Liu G, Yin XM. Homeostatic Role of Autophagy in Hepatocytes. Semin Liver Dis. 2018;38:308-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 49. | Ding WX. Role of autophagy in liver physiology and pathophysiology. World J Biol Chem. 2010;1:3-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 50. | Cui J, Gong Z, Shen HM. The role of autophagy in liver cancer: molecular mechanisms and potential therapeutic targets. Biochim Biophys Acta. 2013;1836:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 51. | Benten D, Kluwe J, Wirth JW, Thiele ND, Follenzi A, Bhargava KK, Palestro CJ, Koepke M, Tjandra R, Volz T, Lutgehetmann M, Gupta S. A humanized mouse model of liver fibrosis following expansion of transplanted hepatic stellate cells. Lab Invest. 2018;98:525-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |