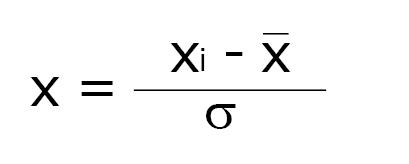Published online Jan 27, 2018. doi: 10.4254/wjh.v10.i1.105
Peer-review started: September 23, 2017
First decision: October 30, 2017
Revised: November 16, 2017
Accepted: January 15, 2018
Article in press: January 15, 2018
Published online: January 27, 2018
Processing time: 126 Days and 5.9 Hours
To develop metabonomic models (MMs), using 1H nuclear magnetic resonance (NMR) spectra of serum, to predict significant liver fibrosis (SF: Metavir ≥ F2), advanced liver fibrosis (AF: METAVIR ≥ F3) and cirrhosis (C: METAVIR = F4 or clinical cirrhosis) in chronic hepatitis C (CHC) patients. Additionally, to compare the accuracy of the MMs with the aspartate aminotransferase to platelet ratio index (APRI) and fibrosis index based on four factors (FIB-4).
Sixty-nine patients who had undergone biopsy in the previous 12 mo or had clinical cirrhosis were included. The presence of any other liver disease was a criterion for exclusion. The MMs, constructed using partial least squares discriminant analysis and linear discriminant analysis formalisms, were tested by cross-validation, considering SF, AF and C.
Results showed that forty-two patients (61%) presented SF, 28 (40%) AF and 18 (26%) C. The MMs showed sensitivity and specificity of 97.6% and 92.6% to predict SF; 96.4% and 95.1% to predict AF; and 100% and 98.0% to predict C. Besides that, the MMs correctly classified all 27 (39.7%) and 25 (38.8%) patients with intermediate values of APRI and FIB-4, respectively.
The metabonomic strategy performed excellently in predicting significant and advanced liver fibrosis in CHC patients, including those in the gray zone of APRI and FIB-4, which may contribute to reducing the need for these patients to undergo liver biopsy.
Core tip: The assessment of liver fibrosis in chronic hepatitis C patients is important to make therapeutic decisions and predict clinical outcomes. Due to various drawbacks related to the use of liver biopsy, individual markers and scores have been validated with feeble accuracy to assess intermediate stages of fibrosis. Our study showed promising results for the metabonomics strategy as a non-invasive tool to distinguish patients with significant fibrosis, advanced fibrosis, and cirrhosis, with sensitivity and specificity values above 95% and high accuracy in the gray zone of aspartate aminotransferase to platelet ratio index and fibrosis index based on four factors, which could avoid a large number of biopsies in these patients.
- Citation: Batista AD, Barros CJP, Costa TBBC, Godoy MMG, Silva RD, Santos JC, de Melo Lira MM, Jucá NT, Lopes EPA, Silva RO. Proton nuclear magnetic resonance-based metabonomic models for non-invasive diagnosis of liver fibrosis in chronic hepatitis C: Optimizing the classification of intermediate fibrosis. World J Hepatol 2018; 10(1): 105-115
- URL: https://www.wjgnet.com/1948-5182/full/v10/i1/105.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i1.105
Approximately 130-170 million people worldwide are chronically infected with the hepatitis C virus (HCV)[1] and an estimated 500000 individuals died in 2010 from virus-related illnesses[2]. In Brazil, the estimated prevalence of hepatitis C is 1450000 cases[3] and about 8000 deaths were due to HCV-related diseases in 2013[4].
The accurate diagnosis of significant fibrosis (SF), advanced fibrosis (AF) and cirrhosis (C) in the liver is important to determine the urgency of treatment, and to monitor complications of the disease. Hepatic histopathology assessment by liver biopsy is still considered the gold standard for evaluating liver fibrosis, but the procedure is invasive and may lead to major complications and death[5]. Sampling error, intra- and inter-observer variability leads to discordant results in 33% of cases[6], thus making biopsy an imperfect reference standard. For these reasons, several non-invasive serological biomarkers of fibrosis have been evaluated. Among them, aminotransferase to platelet ratio index (APRI) and fibrosis index based on four factors (FIB-4) stand out because they are based on readily available laboratory tests in clinical practice[7,8]. These surrogate markers have good accuracy as to excluding significant fibrosis and confirming cirrhosis. However they fail to diagnose intermediate fibrosis[9].
Metabonomics is defined as "a quantitative measure of the dynamic and multiparametric metabolic response of living organisms to pathological or genetic modifications[10]". As a result of homeostasis, the presence of a pathological condition changes the profile of endogenous metabolites, which can be monitored by 1H nuclear magnetic resonance (NMR) spectroscopy[11]. This method seeks to discriminate the samples in groups according to their biochemical status, thereby associating this status to a given condition[11-12]. The method has been studied in various liver diseases[13], and has been shown to be useful in distinguishing between patients with viral hepatitis and healthy volunteers[14-15] and performing well at identifying complications of liver cirrhosis[16-18]. Recently, our group showed that a partial least squares discriminant analysis (PLS-DA) metabonomic model (MM), based on the H-1 NMR spectroscopy of serum samples, presented a clear separation between 18 patients coinfected with schistosomiasis mansoni and hepatitis B virus (HBV) or HCV and 22 HBV or HCV mono-infected patients, with an accuracy, a predictive ability (Q2) and a coefficient of determination (R2) of 100%, 98.1% and 97.5%, respectively[19]. Therefore, the aim of this study was to develop and evaluate MMs, using 1H NMR spectrum of serum samples, as non-invasive markers of significant liver fibrosis, advanced liver fibrosis and cirrhosis in patients with chronic hepatitis C (CHC), and to compare their performance with the APRI and FIB-4.
This was a cross-sectional phase II validation diagnostic study[20], with prospective inclusion, by spontaneous demand, of CHC adult outpatients (anti-HCV and HCV-RNA detectable in serum) attended to at the hepatology clinic of the Hospital das Clínicas/ Universidade Federal de Pernambuco (HC/UFPE) between October/2012 and December/2015. Patients who had undergone a percutaneous liver biopsy in the previous 12 mo or had been clinically diagnosed with liver cirrhosis were included.
The clinical diagnosis of cirrhosis was based on characteristic symptoms and signals and/or according to evidence of chronic liver disease and/or portal hypertension on ultrasound (US), such as liver parenchymal heterogeneity, straight borders, reduced liver size, enhanced portal vein dimensions, presence of collateral vessels, splenomegaly, and/or signals of portal hypertension observed on upper gastrointestinal endoscopy, such as the presence of esophageal/gastric varices and/or hypertensive gastropathy.
Those undergoing antiviral treatment or diagnosed with periportal fibrosis induced by schistosomiasis, metabolic, autoimmune or cholestatic liver disease, HBV or HIV co-infection, neoplasia, or with ethanol ingestion > 20 g/d for women and > 30 g/d for men were excluded. All patients signed an informed consent form and the study was approved by the Ethics Committee of the Institution.
Fasting blood samples were collected from all patients by peripheral vein puncture. The laboratory tests were performed by an automated method. The HCV RNA was detected and the genotype was determined by real-time polymerase chain reaction, using COBAS® AmpliPrep/COBAS® TaqMan® (version 2, Roche, Pleasanton, CA, United States) with a detection limit of 15 IU/mL. The APRI and FIB-4 were calculated as described by Wai et al[7] (2003) and Sterling et al[8] (2006).
Serum samples, stored at -20 °C, were thawed at room temperature and prepared by adding 200 μL of D2O to 400 μL of serum. 1H NMR spectra were obtained using a Varian Unity Plus 300 spectrometer, operating at 299.95 MHz, at 300 K. The samples were analyzed using a pulse sequence with suppression of the resonance of water and T2 filter (PRESAT-CPMG), as follows: Pre-saturation time of 2.0 s, acquisition time of 1.704 s, 128 repetitions and spectral width of 4.8 kHz. Spectra were processed using line broadening equal to 0.3 Hz. The signal attributed to the methyl group of the lactate, in δ 1.33 ppm, was used as the internal reference of chemical shift. The baseline of the spectra was corrected manually.
The ultrasonography-guided percutaneous liver biopsies were performed using a 16 G x 90 mm Menghini needle in, at most, 2 punctures. Fragments with at least 15 mm and /or 6 complete portal tracts were included in the analysis. Fibrosis was classified as F0 to F4, in accordance with METAVIR[21], by two experienced pathologists, blinded to the clinical and serological results. The patients were allocated into three groups: SF (METAVIR F ≥ 2), AF (METAVIR F ≥ 3) and C (METAVIR F = 4 or clinical cirrhosis).
All spectra were processed using MestreNova software (version 9.0.1, MestreLab Research). The spectra were divided into 250 regions of 0.04 ppm, called bins, used to construct a dataset. The region containing the bins centered between δ 4.52 - 5.12 ppm was excluded so as to eliminate the residual signal of water. The spectra were normalized using the following expression:
Math 1
Where xi is the intensity in each bin, while (x1 + x2 + …… xn)/n is the arithmetic mean of the intensities observed in the bins and σ is the standard deviation.
Principal components analysis (PCA) formalism was initially applied to the dataset, to explore inherent clusters and to identify the presence of outliers. PCA did not present natural grouping in the classes of interest. Then, PLS-DA and linear discriminant analysis (LDA) supervised formalisms were used. Three PLS-DA models were constructed to predict FS, FA and C, respectively, using the MetaboAnalyst 3.0 platform[22]. The models were validated by leave-one-out-cross-validation (LOOCV) and by permutations tests. In addition, three LDA models were constructed, using the PCA matrix as input data, so as to predict SF, with five principal components (PCs); AF, with four PCs; and C, with five PCs. LDA models were validated by LOOCV, using STATISTICA software (version 10.0, Quest software).
For each LDA MM, and for APRI and FIB-4, a 2 x 2 contingency matrix was used to calculate sensitivity (SN), specificity (SP), the positive likelihood ratio (LR+), the negative likelihood ratio (LR-) and accuracy (A). A Receiver Operating Characteristic (ROC) curve was also constructed for APRI and FIB-4.
The descriptive and comparative analysis of the data was carried out using STATA (version 12.0, StataCorp, College Station, Texas) and GraphPad Prism software (version 5.0 for Windows, GraphPad Software, La Jolla, California). The qualitative variables were presented as absolute and relative frequencies, and the quantitative variables as means and standard deviation or medians and 25th and 75th percentile. Categorical variables were compared using the χ2 test, applying Fisher’s exact test, when necessary. The Mann-Whitney and Student’s t-test were used to compare non-parametric and parametric continuous measurements, respectively. All tests were applied with 95% confidence (P value ≤ 0.05).
The group studied consisted of 80 CHC patients initially selected. Eleven of the subjects (14%) were excluded due to: diagnosis of periportal fibrosis induced by schistosomiasis in 5 patients, hepatocellular carcinoma in 1, abuse of ethanol in 1 and inadequate liver fragment in 4. Therefore, 69 patients were evaluated, of whom 59.4% were female, with a mean age of 57 ± 12 years. The HCV genotype was determined in 67 patients, while 1b was the most frequent genotype in 36 (53.7%) patients, followed by genotype 3 in 16 (23.9%), genotype 1/1a in 13 (19.4%), genotype 2 in 1 (1.5%) and genotype 4 in 1 (1.5%) patients. The main characteristics of the casuistry are described in Table 1.
| Characteristics | Total (n = 69) | |
| Gender (n, %) | ||
| Male | 28 | 40.60% |
| Female | 41 | 59.40% |
| Age (yr)1 | 57.5 | ± 11.9 |
| BMI (kg/m2)1 | 27.7 | ± 4.7 |
| AST (U/L)2 | 51.5 | (33.7-88) |
| ALT (U/L)2 | 54 | (32.6-106) |
| AST/ALT1 | 1.03 | ± 0.44 |
| GGT (U/L)2 | 81.8 | (46.5-155) |
| Platelets (109/mm3)1 | 191 | ± 78 |
| Albumin (g/dL)1 | 4.08 | ± 0.56 |
| Total bilirubin (mg/dL)2 | 0.7 | (0.50-1.09) |
| Alkaline phosphatase (U/L)1 | 94.6 | ± 49.2 |
| INR1 | 1.09 | ± 0.14 |
| APRI2 | 0.8 | (0.44-2.18) |
| FIB-42 | 2.18 | (1.29-4.72) |
| Liver fragment length (cm)2 | 1.5 | (1.30-1.80) |
| Number of portal tracts2 | 15 | (12.0-20.0) |
| Stage of fibrosis (n, %) | ||
| F0 | 2 | 2.90% |
| F1 | 25 | 36.20% |
| F2 | 14 | 20.30% |
| F3 | 10 | 14.50% |
| F4 | 18 | 26.10% |
Liver biopsy was performed on 54 (78%) patients. There were no serious complications or deaths related to the procedure. The median fragment was 15 mm in length (P25: 13; P75: 18 mm), and there were 15 portal tracts (P25: 12; P75: 20 mm). The METAVIR fibrosis stage was distributed as follows: F0 in 2; F1 in 25; F2 in 14; F3 in 10 and F4 in 3 patients. The diagnosis of cirrhosis was clinically established in 15 (21.7%), thus classified according to the Child-Pugh score: 10 patients Child-Pugh A and 5 Child-Pugh B.
Therefore, 42 (60.9%) patients were classified as SF, 28 (40.6%) as AF and 18 (26.1%) as C. Patients with SF and AF presented a higher mean age, a higher mean value of INR and a higher median value of bilirubin, gamma-glutamyl transferase, APRI and FIB-4, as well as a lower mean platelet count and albumin serum level, when compared with the groups of a lower stage of fibrosis (Table 2).
| Fibrosis group | P value | ||||
| Significant (≥ F2) n = 42 | Non-significant (< F2) n = 27 | ||||
| Gender (M/F, %) | 16/26 | (40.6/59.4) | 12/15 | (38.1/61.9) | 0.600a |
| Age (yr)1 | 62.16 | ± 9.46 | 50.22 | ± 11.86 | < 0.0001b |
| BMI (kg/m2)1 | 28.6 | ± 4.9 | 26.3 | ± 3.8 | 0.051b |
| AST (U/L)2 | 61.5 | (45.5-103) | 34 | (29.6-47) | < 0 .0001c |
| ALT (U/L)2 | 57.5 | (41.7-108) | 39.1 | (26-81) | 0.04c |
| AST/ALT1 | 1.12 | ± 0.47 | 0.91 | ± 0.36 | < 0.0001b |
| GGT (U/L)2 | 107.5 | (65.8-168) | 56 | (32.8-82.9) | 0.001c |
| Platelets (109/mm3)1 | 162 | ± 72 | 239 | ± 61 | < 0.0001b |
| Albumin (g/dL)1 | 3.91 | ± 0.62 | 4.35 | ± 0.30 | 0.001b |
| Total bilirrubin (mg/dL)2 | 0.72 | (0.60-1.30) | 0.5 | (0.40-0.80) | 0.005c |
| Alkaline phosphatase (U/L)1 | 103.3 | ± 55.6 | 79.2 | ± 30.3 | 0.059b |
| INR1 | 1.13 | ± 0.17 | 1.03 | ± 0.06 | 0.002b |
| APRI2 | 1.14 | (0.75-2.84) | 0.48 | (0.27-0.79) | < 0.0001c |
| FIB-42 | 3.45 | (2.01-6.19) | 1.37 | (0.84-1.89) | < 0.0001c |
| Advanced (≥ F3) n = 28 | Non-advanced (< F3) n = 41 | ||||
| Gender (M/F, %) | 14/14 | (50.0/50.0) | 14/27 | (34.1-65.9) | 0.188a |
| Age (yr)1 | 62.23 | ± 9.42 | 54.25 | ± 12.46 | 0.004b |
| BMI (kg/m2)1 | 27.6 | ± 3.9 | 27.9 | ± 5.2 | 0.802b |
| AST (U/L)2 | 79 | (52.4-130) | 40 | (31.4-63) | 0.001c |
| ALT (U/L)2 | 62 | (42.5-113.3) | 43 | (29-84.8) | 0.078c |
| AST/ALT1 | 1.19 | ± 0.54 | 0.93 | ± 0.33 | 0.031b |
| GGT (U/L)2 | 110.5 | (76.1-159.3) | 66.5 | (36.2-137.8) | 0.005c |
| Platelets (109/mm3)1 | 138 | ± 65 | 228 | ± 63 | < 0.0001b |
| Albumin (g/dL)1 | 3.75 | ± 0.66 | 4.33 | ± 0.29 | < 0.0001b |
| Total bilirrubin (mg/dL)2 | 0.94 | (0.70-1.48) | 0.5 | (0.44-0.70) | < 0.0001c |
| Alkaline phosphatase (U/L)1 | 99.5 | ± 38.9 | 90.9 | ± 56.1 | 0.490b |
| INR1 | 1.18 | ± 0.17 | 1.03 | ± 0.07 | < 0.0001b |
| APRI2 | 2.13 | (0.99-3.75) | 0.59 | (0.39-0.92) | < 0.0001c |
| FIB-42 | 4.8 | (2.56-9.30) | 1.72 | (1.10-2.21) | < 0.0001c |
| Cirrhosis (F4) n = 18 | Non-cirrhosis (< F4) n = 51 | ||||
| Gender (M/F, %) | 7/11 | (38.9/61.1) | 21/30 | (41.2/58.8) | 0.865a |
| Age (yr)1 | 63.7 | ± 11.11 | 55.29 | ± 11.51 | 0.009b |
| BMI (kg/m2)1 | 27.7 | ± 3.4 | 27.7 | ± 5.1 | 0.983b |
| AST (U/L)2 | 69 | (54.1-101.5) | 43.2 | (32.5-80) | 0.059c |
| ALT (U/L)2 | 54 | (36.4-71) | 50 | (31-114) | 0.637c |
| AST/ALT1 | 1.38 | ± 0.56 | 0.91 | ± 0.31 | 0.003b |
| GGT (U/L)2 | 108.5 | (73.8-156.3) | 74.5 | (41.3-151.2) | 0.068c |
| Platelets (109/mm3)1 | 123 | ± 72 | 216 | ± 64 | < 0.0001b |
| Albumin (g/dL)1 | 3.41 | ± 0.63 | 4.31 | ± 0.29 | < 0.0001b |
| Total bilirrubin (mg/dL)2 | 1.27 | (0.70-2.88) | 0.6 | (0.47-0.80) | < 0.0001c |
| Alkaline phosphatase (U/L)1 | 107.2 | ± 41.3 | 89.7 | ± 51.5 | 0.202b |
| INR1 | 1.21 | ± 0.20 | 1.05 | ± 0.08 | 0.003b |
| APRI2 | 2.35 | (1.04-4.36) | 0.69 | (0.40-1.11) | < 0.0001c |
| FIB-42 | 5.63 | (4.37-11.27) | 1.85 | (1.17-2.38) | 0.001c |
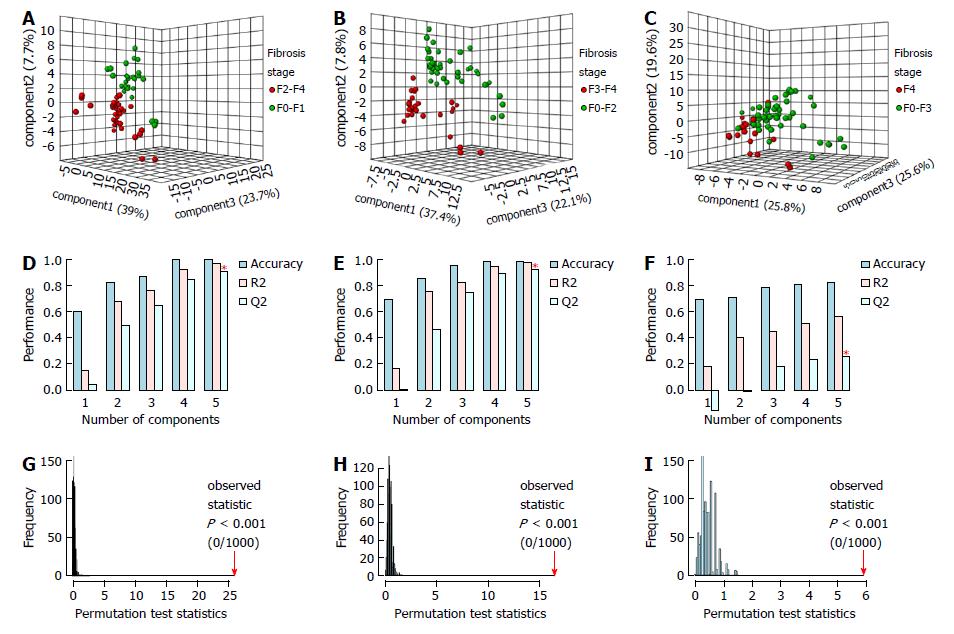
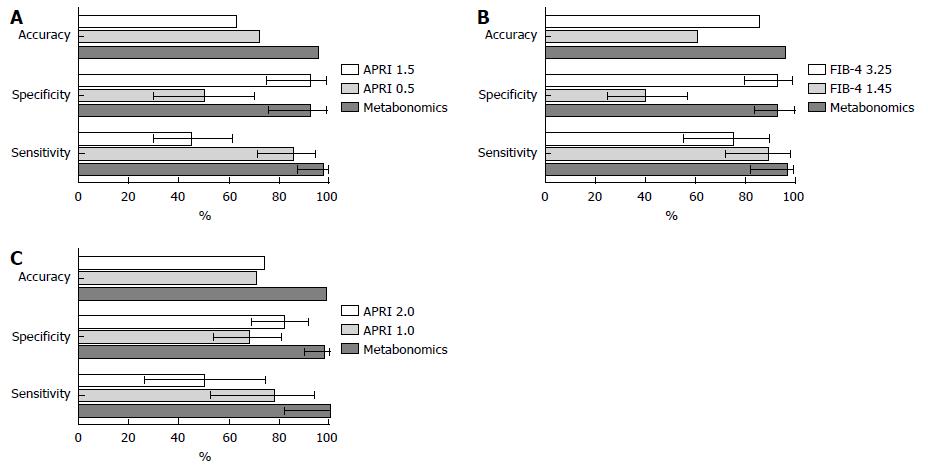
The PLS-DA MM for SF showed a clear discrimination between the samples with three latent components (Figure 1A). The model presented 100% accuracy, R2 and Q2 of 0.98 and 0.91, respectively, when five latent components were used (Figure 1D). The permutation tests, using up to 1000 permutations of the model, indicated that there was no permuted model better than the original one, with observed statistics at P < 0.001 (Figure 1G). The results of the LDA MM for SF are presented in Table 3 and were compared with APRI, with a lower and upper cut-off point of 0.5 and 1.5. The MM showed SN of 97.6% (95%CI: 87.4%-99.9%), similar to APRI cut-off of 0.5, which presented SN of 85.7% (95%CI: 71.5%-94.6%). The LR- of the MM was 0.03 (95%CI: 0.004-0.2), whereas the LR- of APRI values ≤ 0.5 was 0.3 (95%CI: 0.1-0.7). The MM presented SP of 92.6% (95%CI: 75.7%-99.1%) and LR+ 13.2 (95%CI: 3.5-50.1), similar to APRI cut-off of 1.5, which showed SP of 92.3% (95%CI: 74.9%-99.9%), with LR+ of 5.9 (95%CI: 1.5-23.2) for values > 1.5.
| Biopsy | P value | Sensitivity | Specificity | LR+ | LR- | A (%) | ||||||
| Significant fibrosis | ||||||||||||
| Model (n = 69) | F2-F4 | F0-F1 | (%) | 95%CI | (%) | 95%CI | 95%CI | 95%CI | ||||
| ≥ F2 | 41 | 2 | < 0.0011 | 97.6 | 87.4-99.9 | 92.6 | 75.7-99.1 | 13.2 | 3.5-50.1 | 0.03 | 0.004-0.2 | 95.7 |
| < F2 | 1 | 25 | ||||||||||
| APRI (n = 68) | ||||||||||||
| > 0.5 | 36 | 13 | 0.0012 | 85.7 | 71.5-94.6 | 50 | 29.9-0.70 | 1.71 | 1.2-2.6 | 0.3 | 0.1-0.7 | 72 |
| ≤ 0.5 | 6 | 13 | ||||||||||
| > 1.5 | 19 | 2 | 0.0011 | 45.2 | 29.9-61.3 | 92.3 | 74.9-99.0 | 5.9 | 1.5-23.2 | 0.6 | 0.4-0.8 | 63.2 |
| ≤ 1.5 | 23 | 24 | ||||||||||
| Advanced fibrosis | ||||||||||||
| Model (n = 69) | F3-F4 | F0-F2 | (%) | 95%CI | (%) | 95%CI | 95%CI | 95%CI | ||||
| ≥ F3 | 27 | 2 | < 0.0011 | 96.4 | 81.7-99.1 | 95.1 | 83.5-99.4 | 19.8 | 5.1-76.5 | 0.04 | 0.005-0.3 | 95.7 |
| < F3 | 1 | 39 | ||||||||||
| FIB-4 (n = 68) | ||||||||||||
| > 1.45 | 25 | 24 | 0.0121 | 89.3 | 71.8-97.7 | 40 | 24.9-56.7 | 1.5 | 1.1-2.0 | 0.3 | 0.1-0.8 | 60.3 |
| ≤ 1.45 | 3 | 16 | ||||||||||
| > 3.25 | 21 | 3 | < 0.0011 | 75 | 55.1-89.3 | 92.5 | 79.6-98.4 | 10 | 3.3-30.3 | 0.3 | 0.1-0.5 | 85.3 |
| ≤ 3.25 | 7 | 37 | ||||||||||
| Cirrhosis | ||||||||||||
| Model (n = 69) | F4 | F0-F3 | (%) | 95%CI | (%) | 95%CI | 95%CI | 95%CI | ||||
| F4 | 18 | 1 | < 0.0011 | 100 | 81.5-100 | 98 | 89.6-99.9 | 33.83 | 6.9-163.7 | 0.033 | 0.002-0.4 | 98.6 |
| < F4 | 0 | 50 | ||||||||||
| APRI (n = 68) | ||||||||||||
| > 1.00 | 14 | 16 | 0.0021 | 77.8 | 52.4-93.6 | 68 | 53.3-80.5 | 2.4 | 1.5-3.9 | 0.3 | 0.1-0.8 | 70.6 |
| ≤ 1.00 | 4 | 34 | ||||||||||
| > 2.00 | 9 | 9 | 0.0082 | 50 | 26.0-74.0 | 82 | 68.6-91.4 | 2.8 | 1.3-5.9 | 0.6 | 0.4-1.0 | 73.5 |
| ≤ 2.00 | 9 | 41 | ||||||||||
The PLS-DA MM for AF, using three latent components, discriminated all the samples, with 100% accuracy, R2 of 0.98 and Q2 of 0.93, with five latent components. Permutation tests indicated that the permuted models are not better than the original model (Figure 1B, E and H). The LDA MM for AF showed SN of 96.4% (95%CI: 81.7%-99.1%) and LR- of 0.04 (95%CI: 0.005-0.3), similar to FIB-4 cut-off of 1.45, which showed SN of 89.3% (95%CI: 71.8%-97.7%) and LR- of 0.3 (95%CI: 0.1-0.8) for values ≤ 1.5. The two methods also presented high SP and LR+, there being observed SP of 95.1% (95%CI: 83.5%-99.4%) and LR+ of 19.8 (95%CI: 5.1-76.5) for MM, and SP of 92.5% (95%CI: 79.6%-98.4%) and LR+ of 10 (95%CI: 3.3-30.3) for FIB-4 cut-off of 3.25 (Table 3).
The PLS-DA MM for C, using three latent components, also adequately discriminated the samples, attaining an accuracy of 84.0% with five latent components. The permutation tests indicated that the original model was not exceeded by any of the permuted models (Figure 1C, F and I). The LDA MM for C showed SN of 100% (95%CI: 81.5%-100%) and LR- of 0.03 (95%CI: 0.002-0.4), similar to APRI cut-off of 1.0, which presented SN of 77.8% (95%CI: 52.4-93.6), with LR- of 0.3 (95%CI: 0.1-0.8) for values ≤ 1.0. The MM presented SP of 98% (95%CI: 89.6%-99.9%), similar to APRI cut-off of 2.0, which showed SP of 82% (95%CI: 68.6 - 91.4). MM showed LR+ of 33.8 (95%CI: 6.9-163.7), which was higher than the APRI values > 2.0, which presented LR+ of 2.8 (95%CI: 1.3-5.9). This result indicates that, by MM, the likelihood of a positive test in the presence of cirrhosis is 33 times more likely than a positive test in the absence of cirrhosis, whereas, by the APRI it is 2.8 times more likely (Table 3).
In general, the MM presented a high accuracy and performance, similar to the APRI to predict SF and C, and similar to FIB-4 to predict AF (Figure 2). The area under ROC curve (AUROC) of APRI to predict SF and C was 0.79 (95%CI: 0.68-0.90) and 0.76 (95%CI: 0.61-0.91), respectively, whereas the AUROC of FIB-4 to predict AF was 0.84 (95%CI: 0.74-0.95) (Supplementary Figure 1).
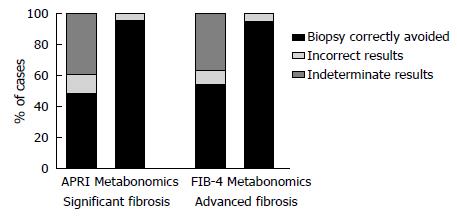
For the 68 patients with an APRI score, 27 (39.7%) had intermediate test values (> 0.5 and ≤ 1.5). Of these, 17 (63%) had SF by METAVIR. All these patients were correctly classified by the MM. Likewise, among 25 (36.8%) patients with FIB-4 values in the gray zone (> 1.45 and ≤ 3.25), the MM correctly identified 4 (16%) with AF. Figure 3 makes a comparison of the extent to which the MMs, APRI and FIB-4 would have correctly avoided the need for a biopsy.
In this study, it was observed that the higher the severity of liver fibrosis, the greater the mean age as well as the greater the impairment of liver function. In fact, this findings reflects the natural history of chronic hepatitis C, a fibrosing disease, which progresses to cirrhosis in about 30 years[23].
Our results suggest that the metabonomic strategy can discriminate F0-F1 from F2-F4 patients, F0-F2 from F3-F4 patients and F0-F3 from F4 patients. The MMs developed to predict SF, AF and C in CHC patients presented high performance, with values of SN, SP and an accuracy of above 90%. In practice, considering the confidence intervals, the results would be classified as similar to APRI and FIB-4. However, the MMs presented 100% accuracy for predicting SF and AF in the gray zone of APRI and FIB-4, when these indexes could not determine the absence or presence of significant and advanced fibrosis. If we consider the 39.7% of unclassified and 11.8% of incorrectly classified patients using APRI as a predictor of SF, liver biopsy would have been correctly avoided in 48.5% of cases. On the other hand, if the MM were used to this end, the biopsy would have been correctly avoided in 95.7% of the patients. Regarding FIB-4 analysis, considering the 36.8% of unclassified and 8.8% of incorrectly classified patients, using this index as the only predictor of AF, biopsy would have been correctly avoided in 54.4% of patients, while the MM would have prevented biopsy in 95.7% of patients (Figure 3).
The most widely indirect methods for the assessment of liver fibrosis in CHC patients in routine clinical practice are the non-commercial serological indexes APRI and FIB-4, and the physical methods, such as liver stiffness measurement, by elastography based on US (transient liver elastography, acoustic radiation force impulse elastography and 2D-shear wave elastography) or based on magnetic resonance imaging. In general, these methods do not distinguish well intermediate stages of fibrosis, although they are increasingly useful in the exclusion of significant fibrosis (< F2) and presence of advanced fibrosis (≥ F3). The serum levels of the extracellular matrix protein osteopontin are promising for the diagnosis of intermediate fibrosis, with increasing concentration in different stages of fibrosis groups from F0 to F4, progressively and significantly different between the groups, and with an AUROC of 0.977 for the discrimination of F1-F2 from F3-F4 patients[24]. Boursier et al[25] proposed the FibroMeter® + FibroScan® (FM+FS) algorithm, based on two fibrosis indexes (significant and advanced fibrosis indexes), from a combination of these two methods by logistic regression. Reliable diagnosis intervals of these two indexes were determined, resulting in a noninvasive classification of fibrosis in six classes. This classification showed an accuracy of 86.7% and using this algorithm, biopsy would be avoided in 100% of patients with significant and advanced fibrosis. However, these methods are based on high cost tests that are not always routinely available, especially in public health services in developing countries.
In fact, the NMR based metabonomics proved to be promising for evaluating the severity of liver disease, cirrhosis and its complications, which reflects the progression of fibrosis[26,27]. Amathieu et al[28] correlated the severity of hepatic impairment in 124 patients with chronic alcoholic liver disease, as measured by MELD, using the OPLS-DA model based on 1H NMR spectroscopy of serum samples. The same authors, using OPLS-DA model based on 1H NMR of serum samples, separated 93 patients with compensated alcoholic cirrhosis from 30 patients with acute on chronic liver failure, with good predictability[29]. Furthermore, Jiménez et al[30], using OPLS-DA MM, based on 1H NMR spectroscopy of serum samples from cirrhotic patients, were able to distinguish between 39 patients with minimal encephalopathy and 62 patients without encephalopathy, with R2 = 0.68 and Q2 = 0.63. A similar finding was described by Qi et al[31], who demonstrated that the OPLS-DA model, based on 1H NMR of serum samples, distinguished between 30 compensated HBV cirrhotic patients from 30 patients with decompensated cirrhosis, with 85% accuracy.
Regarding the evaluation of liver fibrosis, the metabonomic and metabolomic strategies have also shown promising results. In fact, Sands et al[32] on analyzing 1H NMR plasma spectroscopy of healthy controls and CHC patients, constructed OPLS-DA MMs, using METAVIR and the Enhanced Liver Fibrosis (ELF) score as reference standards. The models distinguished between 6 healthy controls and 34 CHC patients with moderate fibrosis (F1-F2) and 22 with advanced fibrosis (F3-F4) in the validation cohort. Embade et al[33] differentiated 27 cirrhotic patients (F4) from 30 patients without fibrosis (F0) using a PLS-DA model, through serum spectral analysis by 1H NMR, in CHC patients, with good predictive power (R2 = 0.8 and Q2 = 0.6). Sarfaraz et al[34] using serum 1H NMR spectroscopy of 45 CHC patients, showed that F0-F2 and F3-F4 patients were well discriminated with the OPLS-DA MM (R2= 0.67 e Q2 = 0.28). Moreover, Gabbani et al[35], constructed PLS-models with canonical analysis (PLS-CA), based on serial analysis of serum/plasma and urine NMR spectra in 33 CHC patients, 10 compensated cirrhotic patients and 23 with mild or moderate fibrosis, as measured by biopsy or transient hepatic elastography. The authors demonstrated that the models recognized compensated cirrhosis with better accuracy in plasma/serum samples (68% in plasma, 56% in serum and 50% in urine samples). Herein, the main finding of our study was the correct classification of patients in the gray zone of APRI and FIB-4, who had mainly intermediate fibrosis (METAVIR classification equal to F2).
In conclusion, the metabonomic strategy was able to distinguish between significant liver fibrosis, advanced liver fibrosis and cirrhosis in CHC patients, showing promising results as a non-invasive tool to evaluate liver fibrosis. Furthermore, the method presented high accuracy in the gray zone of APRI and FIB-4, which could avoid large numbers of biopsies in CHC patients.
Due to the small sample size, it was not possible to use one group for external validation. Therefore, further studies with a larger number of patients tested and external validation of the models are necessary, in order to confirm the performance of the MMs, for later incorporation into clinical practice.
Liver fibrosis is the most important prognostic factor in chronic hepatitis C (CHC). The gold standard method for liver fibrosis evaluation is the liver biopsy, which is invasive and subject to complications and misclassification.
The assessment of significant liver fibrosis is important to make therapeutic decisions and predict clinical outcomes. The serological and physical surrogate methods in hepatic fibrosis evaluation, such as APRI, FIB-4 and hepatic elastography, are good in excluding significant fibrosis and confirming advanced fibrosis, but they fail in diagnosing intermediate fibrosis. The metabonomics strategy is a method that seeks to discriminate biological samples, such as serum, through 1H nuclear magnetic resonance (NMR) spectroscopy, according to their biochemical status, thereby associating this status to a given condition. The method has been studied in various liver diseases, showing to be useful in identifying liver cirrhosis and its complications and with promising results in hepatic fibrosis evaluation in chronic liver diseases.
The aim of this study was to develop and evaluate metabonomic models (MMs), using 1H NMR spectrum of serum samples, as non-invasive tool for assessment of significant liver fibrosis, advanced liver fibrosis and cirrhosis in patients with CHC. Additionally, to compare the performance of the MMs with the serological surrogate indexes APRI and FIB-4 and to investigate the performance of MMs in the gray zone of these serological indexes.
This was a cross-sectional phase II validation diagnostic study, with prospective inclusion of CHC adult outpatients who had undergone percutaneous liver biopsy in the previous 12 mo or had been clinically diagnosed with liver cirrhosis. The clinical diagnosis of cirrhosis was based on characteristic symptoms and signals and/or according to evidence of chronic liver disease and/or portal hypertension on ultrasound and/or signals of portal hypertension observed on upper gastrointestinal endoscopy. Those undergoing antiviral treatment or diagnosed with other liver disease were excluded. All patients signed an informed consent form and the study was approved by the Ethics Committee of the Institution. Fasting blood samples were collected from all patients by peripheral vein puncture. The laboratory tests were performed by an automated method. The HCV RNA was detected by real-time polymerase chain reaction. APRI and FIB-4 were calculated as described by Wai et al (2003) and Sterling et al (2006). Serum samples, stored at -20 °C, were thawed at room temperature and prepared by adding 200 μL of D2O to 400 μL of serum. 1H NMR spectra were obtained using a Varian Unity Plus 300 spectrometer. The samples were analyzed using a pulse sequence with suppression of the resonance of water and T2 filter (PRESAT-CPMG). Spectra were processed using line broadening equal to 0.3 Hz. The signal attributed to the methyl group of the lactate was used as the internal reference of chemical shift. The baseline of the spectra was corrected manually. The ultrasonography-guided percutaneous liver biopsies were performed using a 16 G x 90 mm Menghini needle in, at most, 2 punctures. Fragments with at least 15 mm and /or 6 complete portal tracts were included in the analysis. Fibrosis was classified as F0 to F4, in accordance with METAVIR, by two experienced pathologists, blinded to the clinical and serological results. The patients were allocated into three groups: SF (METAVIR F ≥ 2), AF (METAVIR F ≥ 3) and C (METAVIR F = 4 or clinical cirrhosis). The descriptive and comparative analysis of the data was carried out using STATA and GraphPad Prism softwares. Categorical variables were compared using the Chi-square test, applying Fisher's exact test, when necessary. The Mann-Whitney and Student's t-test were used to compare non-parametric and parametric continuous measurements, respectively. All tests were applied with 95% confidence (P value ≤ 0.05). All spectra were processed using MestreNova software. The spectra were divided into 250 regions of 0.04 ppm, called bins, used to construct a dataset, eliminating the residual signal of water. The spectra were normalized and PLS-DA and LDA supervised formalisms were used. Three PLS-DA models were constructed to predict FS, FA and C, respectively, using the MetaboAnalyst 3.0 platform. The models were validated by leave-one-out-cross-validation (LOOCV) and by permutations tests. In addition, three LDA models were constructed, using the PCA matrix as input data, to predict SF; AF and CCs, and validated by LOOCV, using STATISTICA software. For each LDA MM, and for APRI and FIB-4, a 2 x 2 contingency matrix was used to calculate sensitivity (SN), specificity (SP), the positive likelihood ratio (LR+), the negative likelihood ratio (LR-) and accuracy (A), using liver biopsy as reference standard.
Sixty-nine patients were evaluated, of whom 59.4% were female, with a mean age of 57 ± 12 years. Liver biopsy was performed on 54 (78%) patients. The median fragment was 15 mm in length (P25: 13; P75: 18 mm), and there were 15 portal tracts (P25: 12; P75: 20). The METAVIR fibrosis stage was distributed as follows: F0 in 2; F1 in 25; F2 in 14; F3 in 10 and F4 in 3 patients. The diagnosis of cirrhosis was clinically established in 15 (21.7%), thus classified according to the Child-Pugh score: 10 patients Child-Pugh A and 5 Child-Pugh B. Therefore, 42 (60.9%) patients were classified as SF, 28 (40.6%) as AF and 18 (26.1%) as C. The PLS-DA MM for SF presented 100% accuracy, R2 and Q2 of 0.98 and 0.91, respectively. The results of the LDA MM for SF were compared to APRI and showed SN of 97.6% (95%CI: 87.4%-99.9%) and LR- of 0.03 (95%CI: 0.004-0.2), similar to APRI cut-off of 0.5, which presented SN of 85.7% (95%CI: 71.5%-94.6%) and LR- of 0.3 (95%CI: 0.1-0.7). The MM presented SP of 92.6% (95%CI: 75.7%-99.1%) and LR+ of 13.2 (95%CI: 3.5-50.1), similar to APRI cut-off of 1.5, which showed SP of 92.3% (95%CI: 74.9%-99.9%), with LR+ of 5.9 (95%CI: 1.5-23.2). The PLS-DA MM for AF discriminated all the samples, with 100% accuracy, R2 of 0.98 and Q2 of 0.93. LDA MM for AF showed SN of 96.4% (95%CI: 81.7%-99.1%) and LR- of 0.04 (95%CI: 0.005-0.3), similar to FIB-4 cut-off of 1.45, which showed SN of 89.3% (95%CI: 71.8%-97.7%) and LR- of 0.3 (95%CI: 0.1-0.8). The two methods also presented high SP and LR+, there being observed SP of 95.1% (95%CI: 83.5%-99.4%) and LR+ of 19.8 (95%CI: 5.1-76.5) for MM, and SP of 92.5% (95%CI: 79.6%-98.4%) and LR+ of 10 (95%CI: 3.3-30.3) for FIB- 4 cut-off of 3.25. The PLS-DA MM for C also adequately discriminated the samples, attaining an accuracy of 84.0%. The LDA MM for C showed SN of 100% (95%CI: 81.5%-100%) and LR- of 0.03 (95%CI: 0.002-0.4), similar to APRI cut-off of 1.0, which presented SN of 77.8% (95%CI: 52.4 - 93.6), with LR- of 0.3 (95%CI: 0.1-0.8). The MM presented SP of 98% (95%CI: 89.6%-99.9%), similar to APRI cut-off of 2.0, which showed SP of 82% (95%CI: 68.6 - 91.4), with higher LR+ of 33.8 (95%CI: 6.9-163.7), comparing to APRI cut-off of 2.0, which presented LR+ of 2.8 (95%CI: 1.3-5.9). In general, the MMs presented similar performance to APRI e FIB-4. However, their accuracy for predicting SF and AF in the gray zone of APRI and FIB-4 was 100%. Considering the 39.7% of unclassified and 11.8% of incorrectly classified patients using APRI as a predictor of SF, liver biopsy would have been correctly avoided in 48.5% of cases. On the other hand, if the MM were used to this end, the biopsy would have been correctly avoided in 95.7% of the patients. Regarding FIB-4 analysis, considering the 36.8% of unclassified and 8.8% of incorrectly classified patients, using this index as the only predictor of AF, biopsy would have been correctly avoided in 54.4% of patients, while the MM would have prevented biopsy in 95.7% of patients.
The metabonomic strategy was able to distinguish between significant liver fibrosis, advanced liver fibrosis and cirrhosis in CHC patients, showing promising results as a non-invasive tool to evaluate liver fibrosis. The main finding of our study was the correct classification of patients in the gray zone of APRI and FIB-4, who had mainly intermediate fibrosis (METAVIR classification equal to F2), which could avoid a large number of biopsies in CHC patients.
It is necessary to perform further studies testing a larger number of patients and with external validation of the models, in order to confirm the performance of the MMs, for later incorporation into clinical practice.
We are grateful to the Brazilian funding agencies CNPq and FACEPE, which are fomenters of research in our region, for encouraging research in this important field.
Manuscript source: Invited Manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Ferreira CN, Toshikuni N, Yamasaki T S- Editor: Cui LJ L- Editor: A E- Editor: Li RF
| 1. | Global Burden of Hepatitis C Working Group. Global burden of disease (GBD) for hepatitis C. J Clin Pharmacol. 2004;44:20-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 357] [Cited by in F6Publishing: 389] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 2. | Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-2128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9500] [Cited by in F6Publishing: 9261] [Article Influence: 771.8] [Reference Citation Analysis (0)] |
| 3. | Brasil. Ministério da Saúde. Boletim Epidemiológico-Hepatites Virais. 2015;1-29 Available from: http://www.aids.gov.br/es/node/90. [Cited in This Article: ] |
| 4. | Ferreira PR, Brandão-Mello CE, Estes C, Gonçales Júnior FL, Coelho HS, Razavi H, Cheinquer H, Wolff FH, Ferraz ML, Pessoa MG. Disease burden of chronic hepatitis C in Brazil. Braz J Infect Dis. 2015;19:363-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD; American Association for the Study of Liver Diseases. Liver biopsy. Hepatology. 2009;49:1017-1044. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1449] [Cited by in F6Publishing: 1446] [Article Influence: 96.4] [Reference Citation Analysis (1)] |
| 6. | Regev A, Berho M, Jeffers LJ, Milikowski C, Molina EG, Pyrsopoulos NT, Feng ZZ, Reddy KR, Schiff ER. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am J Gastroenterol. 2002;97:2614-2618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1504] [Cited by in F6Publishing: 1522] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 7. | Wai CT, Greenson JK, Fontana RJ, Kalbfleisch JD, Marrero JA, Conjeevaram HS, Lok AS. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38:518-526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2762] [Cited by in F6Publishing: 3062] [Article Influence: 145.8] [Reference Citation Analysis (0)] |
| 8. | Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, S Sulkowski M, Torriani FJ, Dieterich DT, Thomas DL. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317-1325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2633] [Cited by in F6Publishing: 3157] [Article Influence: 175.4] [Reference Citation Analysis (0)] |
| 9. | European Association for Study of Liver. Asociacion Latinoamericana para el Estudio del Higado. EASL-ALEH Clinical Practice Guidelines: Non-invasive tests for evaluation of liver disease severity and prognosis. J Hepatol. 2015;63:237-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1177] [Cited by in F6Publishing: 1234] [Article Influence: 137.1] [Reference Citation Analysis (0)] |
| 10. | Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181-1189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2918] [Cited by in F6Publishing: 2637] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 11. | Dunn WB, Ellis DI. Metabolomics: Current analytical platforms and methodologies. Trends Anal Chem. 2005;24:285–294. [DOI] [Cited in This Article: ] [Cited by in Crossref: 813] [Cited by in F6Publishing: 459] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 12. | Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054-1056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1368] [Cited by in F6Publishing: 1330] [Article Influence: 83.1] [Reference Citation Analysis (0)] |
| 13. | Amathieu R, Triba MN, Goossens C, Bouchemal N, Nahon P, Savarin P, Le Moyec L. Nuclear magnetic resonance based metabolomics and liver diseases: Recent advances and future clinical applications. World J Gastroenterol. 2016;22:417-426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 32] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Godoy MM, Lopes EP, Silva RO, Hallwass F, Koury LC, Moura IM, Gonçalves SM, Simas AM. Hepatitis C virus infection diagnosis using metabonomics. J Viral Hepat. 2010;17:854-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Munshi SU, Taneja S, Bhavesh NS, Shastri J, Aggarwal R, Jameel S. Metabonomic analysis of hepatitis e patients shows deregulated metabolic cycles and abnormalities in amino acid metabolism. J Viral Hepat. 2011;18:e591-e602. [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Shariff MI, Ladep NG, Cox IJ, Williams HR, Okeke E, Malu A, Thillainayagam AV, Crossey MM, Khan SA, Thomas HC. Characterization of urinary biomarkers of hepatocellular carcinoma using magnetic resonance spectroscopy in a Nigerian population. J Proteome Res. 2010;9:1096-1103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Shariff MIF, Gomaa AI, Cox IJ, Patel M, Williams HRT, Crossey MME. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: A validation study. J Proteome Res. 2011;10:1828-1836. [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 18. | Gao H, Lu Q, Liu X, Cong H, Zhao L, Wang H, Lin D. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782-785. [PubMed] [Cited in This Article: ] |
| 19. | Gouveia LR, Santos JC, Silva RD, Batista AD, Domingues ALC, Lopes EPA, Silva RO. Diagnosis of coinfection by schistosomiasis and viral hepatitis B or C using 1H NMR-based metabonomics. PLoS One. 2017;12:e0182196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 20. | Sackett DL, Haynes RB. The architecture of diagnostic research. BMJ. 2002;324:539-541. [PubMed] [Cited in This Article: ] |
| 21. | Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2860] [Cited by in F6Publishing: 2969] [Article Influence: 106.0] [Reference Citation Analysis (0)] |
| 22. | Xia J Sinelnikov I V. Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251-W257. [DOI] [Cited in This Article: ] [Cited by in Crossref: 2063] [Cited by in F6Publishing: 2072] [Article Influence: 230.2] [Reference Citation Analysis (0)] |
| 23. | Poynard T, Ratziu V, Benmanov Y, Di Martino V, Bedossa P, Opolon P. Fibrosis in patients with chronic hepatitis C: detection and significance. Semin Liver Dis. 2000;20:47-55. [PubMed] [Cited in This Article: ] |
| 24. | Matsue Y, Tsutsumi M, Hayashi N, Saito T, Tsuchishima M, Toshikuni N. Serum Osteopontin predicts degree of hepatic fibrosis and serves as a biomarker in patients with hepatitis C virus infection. PLoS One. 2015;10:e0118744. [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 25. | Boursier J, de Ledinghen V, Zarski JP, Fouchard-Hubert I, Gallois Y, Oberti F, Calès P; multicentric groups from SNIFF 32, VINDIAG 7, and ANRS/HC/EP23 FIBROSTAR studies. Comparison of eight diagnostic algorithms for liver fibrosis in hepatitis C: new algorithms are more precise and entirely noninvasive. Hepatology. 2012;55:58-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 26. | Le Moyec L, Triba M, Nahon P, Bouchemal N, Hantz E, Goossens C. Nuclear magnetic resonance metabolomics and human liver diseases: The principles and evidence associated with protein and carbohydrate metabolism (Review). Biomed Reports. 2017;6:387-395. [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Beyoğlu D, Idle , JR . The metabolomic window into hepatobiliary disease. J Hepatol. 2013;59:842-858. [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 28. | Amathieu R, Nahon P, Triba M, Bouchemal N, Trinchet JC, Beaugrand M, Dhonneur G, Le Moyec L. Metabolomic approach by 1H NMR spectroscopy of serum for the assessment of chronic liver failure in patients with cirrhosis. J Proteome Res. 2011;10:3239-3245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 29. | Amathieu R, Triba MN, Nahon P, Bouchemal N, Kamoun W, Haouache H, Trinchet JC, Savarin P, Le Moyec L, Dhonneur G. Serum 1H-NMR metabolomic fingerprints of acute-on-chronic liver failure in intensive care unit patients with alcoholic cirrhosis. PLoS One. 2014;9:e89230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 30. | Jiménez B, Montoliu C, MacIntyre DA, Serra MA, Wassel A, Jover M, Romero-Gomez M, Rodrigo JM, Pineda-Lucena A, Felipo V. Serum metabolic signature of minimal hepatic encephalopathy by (1)H-nuclear magnetic resonance. J Proteome Res. 2010;9:5180-5187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 31. | Qi SW, Tu ZG, Peng WJ, Wang LX, Ou-Yang X, Cai AJ, Dai Y. ¹H NMR-based serum metabolic profiling in compensated and decompensated cirrhosis. World J Gastroenterol. 2012;18:285-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 28] [Cited by in F6Publishing: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 32. | Sands CJ, Guha IN, Kyriakides M, Wright M, Beckonert O, Holmes E, Rosenberg WM, Coen M. Metabolic phenotyping for enhanced mechanistic stratification of chronic hepatitis C-induced liver fibrosis. Am J Gastroenterol. 2015;110:159-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 33. | Embade N, Mariño Z, Diercks T, Cano A, Lens S, Cabrera D. Metabolic characterization of advanced liver fibrosis in HCV patients as studied by serum 1H-NMR spectroscopy. PLoS One. 2016;11:e0155094. [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 34. | Sarfaraz MO, Myers RP, Coffin CS, Gao ZH, Shaheen AA, Crotty PM, Zhang P, Vogel HJ, Weljie AM. A quantitative metabolomics profiling approach for the noninvasive assessment of liver histology in patients with chronic hepatitis C. Clin Transl Med. 2016;5:33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 35. | Gabbani T, Marsico M, Bernini P, Lorefice E, Grappone C, Biagini MR, Milani S, Annese V. Metabolomic analysis with 1H-NMR for non-invasive diagnosis of hepatic fibrosis degree in patients with chronic hepatitis C. Dig Liver Dis. 2017;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |