Copyright
©The Author(s) 2019.
World J Hepatol. Feb 27, 2019; 11(2): 186-198
Published online Feb 27, 2019. doi: 10.4254/wjh.v11.i2.186
Published online Feb 27, 2019. doi: 10.4254/wjh.v11.i2.186
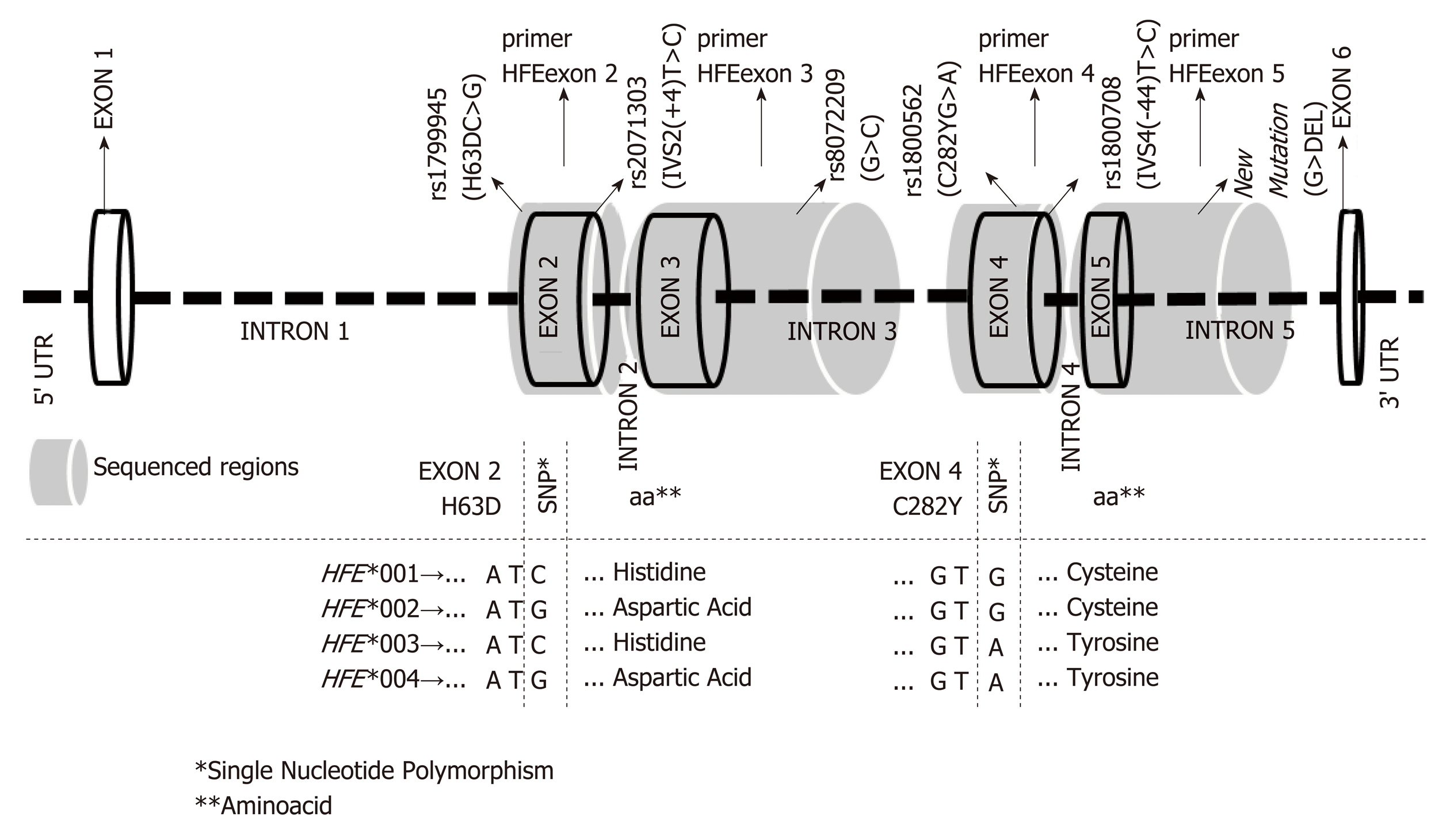
Figure 1 Structure of the HFE gene (ID# ENSG00000010704 - http://www.
ensembl.org) at chromosome region 6p21.3, showing the reference number (rs) of variation sites (NCBI Data base - http://www.ncbi.nlm.nih.gov/snp), previously associated with iron disorders. Shaded grey areas indicate the sequenced gene regions and the respective pairs of primers, as previously described[10]. The combination of these variation sites, translated into the official nomenclature for HFE alleles is also shown in the bottom chart; i.e., the combination of the triplet bases and respective encoded residues of the two most important mutations (H63DC>G and C282YG>A) that defined the four major HFE allele groups (mutated bases are shown in bold type). 1Single nucleotide polymorphism; 2Aminoacid.
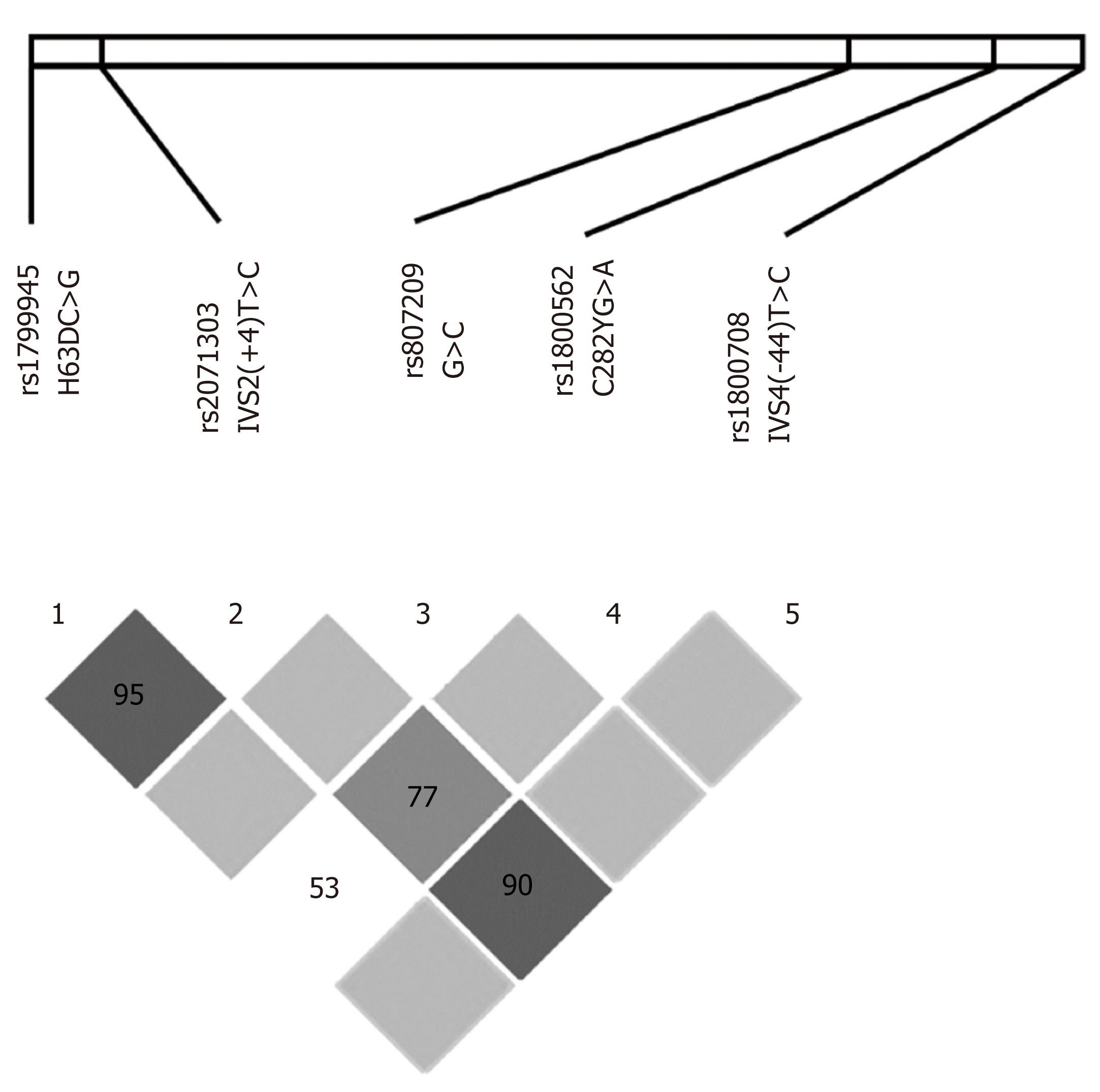
Figure 2 Linkage disequilibrium among single nucleotide polymorphisms observed along the coding region of the HFE gene.
Areas in dark gray represent strong linkage disequilibrium (LD) (LOD ≥ 2 and D' = 1), medium gray indicates moderate LD (LOD ≥ 2, D’ < 1), light gray indicates weak LD (LOD < 2, D' = 1); and white indicates no LD (LOD < 2, D’ < 1). Values of D' different from 1.00 are represented as a percentage within the square. LOD: Log of the odds; D': Linkage disequilibrium coefficient.
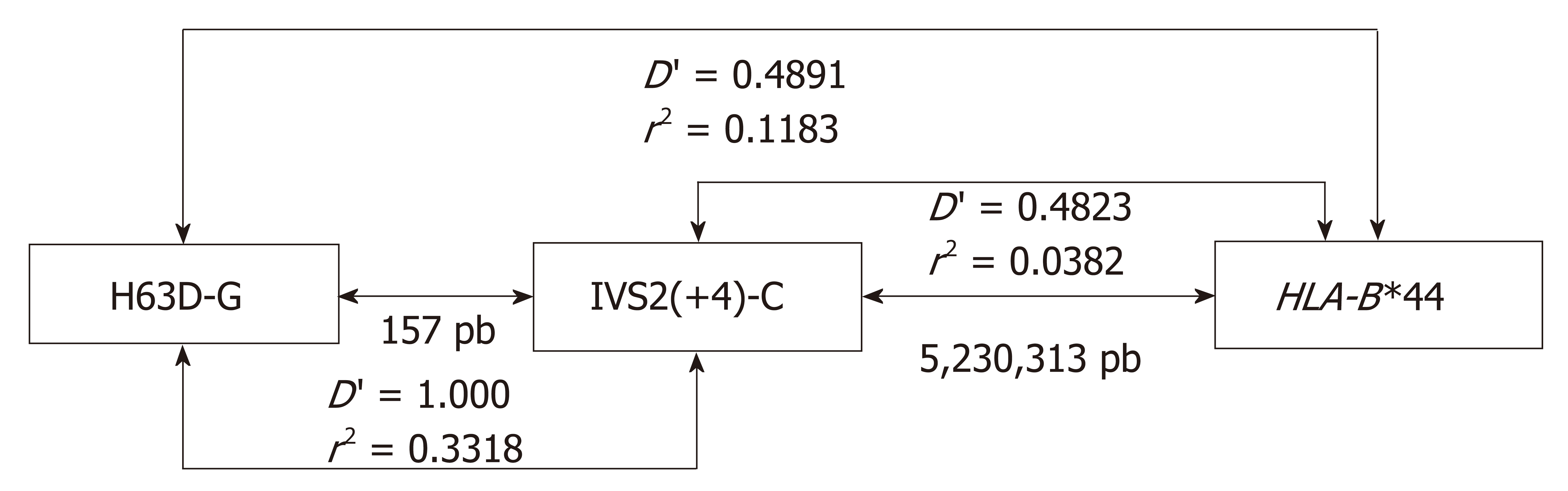
Figure 3 Linkage disequilibrium observed between two relevant HFE coding region [H63DC>G and IVS2(+4)T>C] single nucleotide polymorphism alleles and HLA-B alleles.
- Citation: de Campos WN, Massaro JD, Cançado ELR, Wiezel CEV, Simões AL, Teixeira AC, Souza FF, Mendes-Junior CT, Martinelli ALC, Donadi EA. Comprehensive analysis of HFE gene in hereditary hemochromatosis and in diseases associated with acquired iron overload. World J Hepatol 2019; 11(2): 186-198
- URL: https://www.wjgnet.com/1948-5182/full/v11/i2/186.htm
- DOI: https://dx.doi.org/10.4254/wjh.v11.i2.186