Published online Jun 26, 2022. doi: 10.4252/wjsc.v14.i6.393
Peer-review started: December 10, 2021
First decision: March 13, 2022
Revised: April 7, 2022
Accepted: May 28, 2022
Article in press: May 28, 2022
Published online: June 26, 2022
Processing time: 195 Days and 19.7 Hours
Over the past few decades, high-throughput screening (HTS) has made great contributions to new drug discovery. HTS technology is equipped with higher throughput, minimized platforms, more automated and computerized operating systems, more efficient and sensitive detection devices, and rapid data processing systems. At the same time, in vitro neurogenesis is gradually becoming important in establishing models to investigate the mechanisms of neural disease or deve
Core Tip: High-throughput screening (HTS) is a promising technology that can screen out targets from thousands of candidates. Here, we review the evidence that HTS could be beneficial in neurogenesis methods in various ways: The HTS method can screen out specific genes that induce neural induction, small molecules that facilitate neural differentiation, and three-dimensional microenvironments that could better modulate the microenvironments in vivo. We also focus on the application and prospects of HTS in in vitro neurogenesis, as organoid-based and microfluidic platforms are needed for future research.
- Citation: Zhang SY, Zhao J, Ni JJ, Li H, Quan ZZ, Qing H. Application and prospects of high-throughput screening for in vitro neurogenesis. World J Stem Cells 2022; 14(6): 393-419
- URL: https://www.wjgnet.com/1948-0210/full/v14/i6/393.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i6.393
High-throughput screening (HTS), also called large-scale cluster screening, first appeared in the 1980s and utilized microplates as a platform, an automated handling system as an operator, and a variety of highly sensitive detection instruments to screen out “hits” from thousands of drug candidates. As an integrated and multidisciplinary technology, HTS combines diverse fields, such as molecular biology, medicinal chemistry, mathematics, computer science and microelectronic technology. With its rapid, efficient, economic, microscale, highly automatic and computerized features, HTS technology has made great contributions to biomedical research, such as identifying new drug candidates for pharmacological research[1,2], enzyme engineering, including the directed evolution of enzymes[3], and genetic research[4].
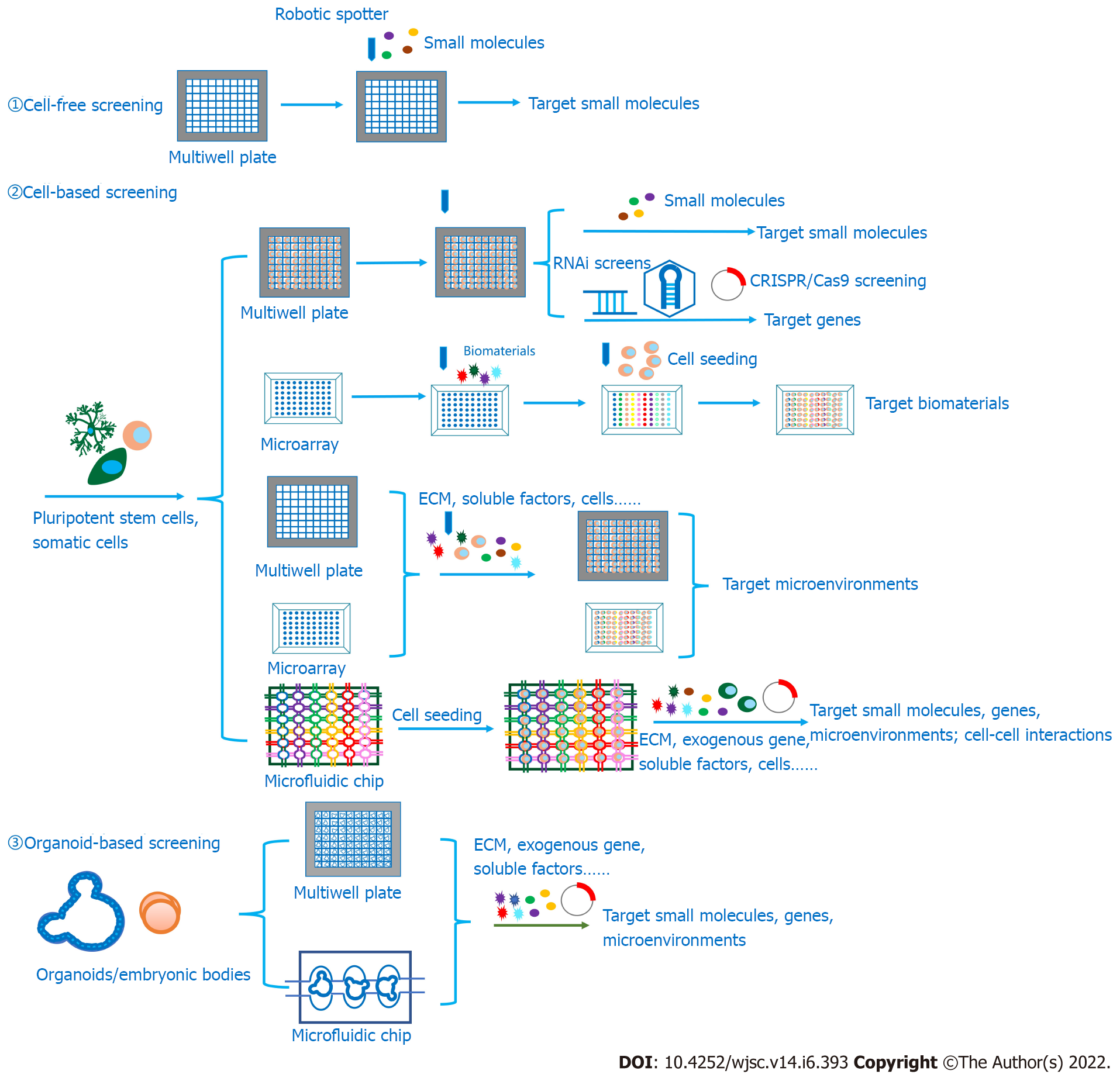
Based on ligand-target interactions, HTS can be performed between various candidates and targets, including substrates and enzymes, inhibitors and enzymes, ligands and receptors, proteins and proteins, and DNA and proteins[5]. According to the in vitro screening models, HTS can be mainly divided into cell-free assays, also called biochemical assays, and cell-based assays. While cell-free assays dominate in the early stage of HTS, cell-based assays are gradually gaining an essential role because some cellular processes, such as transmembrane transport, cytotoxicity effects or other off-target effects, can be tested in cellular models, and some screening targets are difficult to extract and purify from cells[6]. Significantly, in recent studies, screening targets have been extended from biochemical compounds such as enzymes, receptors, antibodies, nucleotides and living cells to tissues and even organoids to investigate protein–protein/DNA/RNA interactions, cell-protein interactions, cell–cell interactions and even protein-tissue interactions. Therefore, categories of testing candidates are also developing from biochemicals aiming at diverse targets such as enzymes and receptors in intracellular signaling pathways to microenvironments that are suitable for various cellular behaviors. Since the exploration of cell-extracellular matrix (ECM)-interactions is growing and three-dimensional (3D) cell culture technologies are developing, the HTS platform is evolving from two-dimensional (2D) to 3D. In previous research, hydrogel droplets and synthetic scaffolds could be attached to HTS platforms[7].
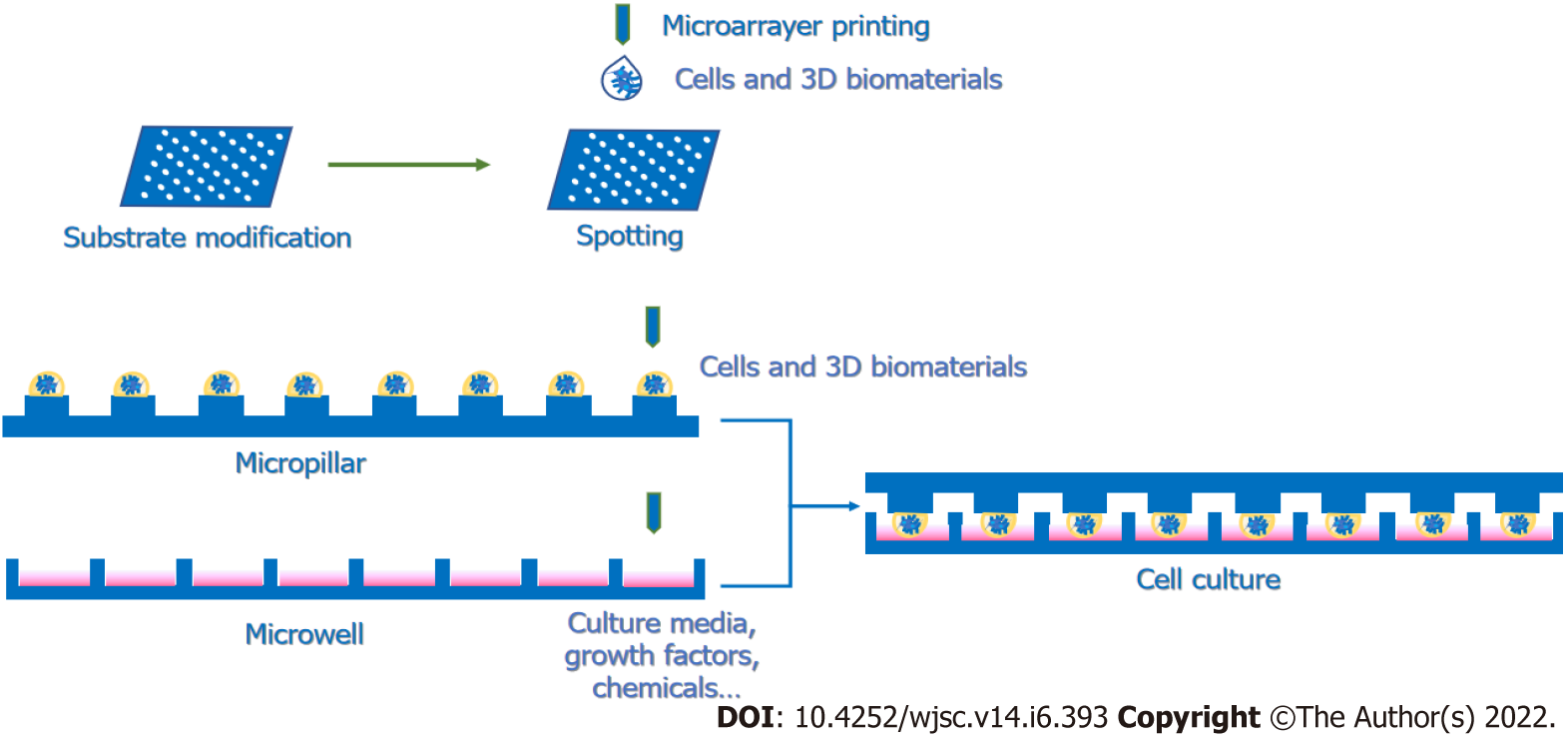
In addition to extending the variety of screening targets and candidates, researchers have also been working on improving the miniaturization, integration and automation of the screening platform to meet the increasing need for HTS applications in biomedical research. Specifically, the screening platform has developed from comprising microtiter plates with 96 wells to those with 384 wells and then to those with 1536 wells[8]. To further elevate the screening efficiency, microarrays are utilized to promote integration by immobilizing protein or DNA targets on glass chips. Then, to separate each spot, save reagents and create various cellular microenvironments, combinations of microwells and micropillars are applied for HTS[9,10].
Furthermore, microfluidic-based microarrays also play an important role in HTS because of their higher efficiency, improved automation, controlled microenvironments, adjustable flow parameters, achievement of microscale reaction volumes and the capacity for single-cell analysis. Methods based on microfluidic systems can be divided into two groups: Droplet-based microfluidics and array-based microfluidics[11]. Assay-based microfluidic devices have also been successfully utilized in HTS for drug screening[12,13], cell heterogeneity analysis[14], cell–cell interactions[15] and even cell–ECM interactions[15].
Compared with array-based microfluidic devices, droplet-based microfluidic devices are well suited for analyzing single-cell activities because biomolecules, particles or even single cells can be encapsulated in water-in-oil droplets that are emulsified when they are flowing through the droplet-producing devices to form the droplet library[16]. Every droplet contains a barcode that represents the elements encapsulated. The barcodes usually include nucleic acid sequences, which are capable of large screens, and fluorescent tags, which are suitable for real-time reading[17]. The droplets pooled in the library are then reinjected into the microfluidic device, usually merging with other cells or biomolecules to start the incubation, followed by a screening based on various characteristics, such as cell density[16] and fluorescence intensity[18]. In addition, droplets can also be sorted according to the variety of readouts. The strategies applied in droplet sorting are based on fluorescence-activated cell sorting (FACS), which is a mechanical actuation, also called reverse cell sorting, accomplished with the assistance of peristaltic pumps and valves[19] as well as dielectrophoresis[20].
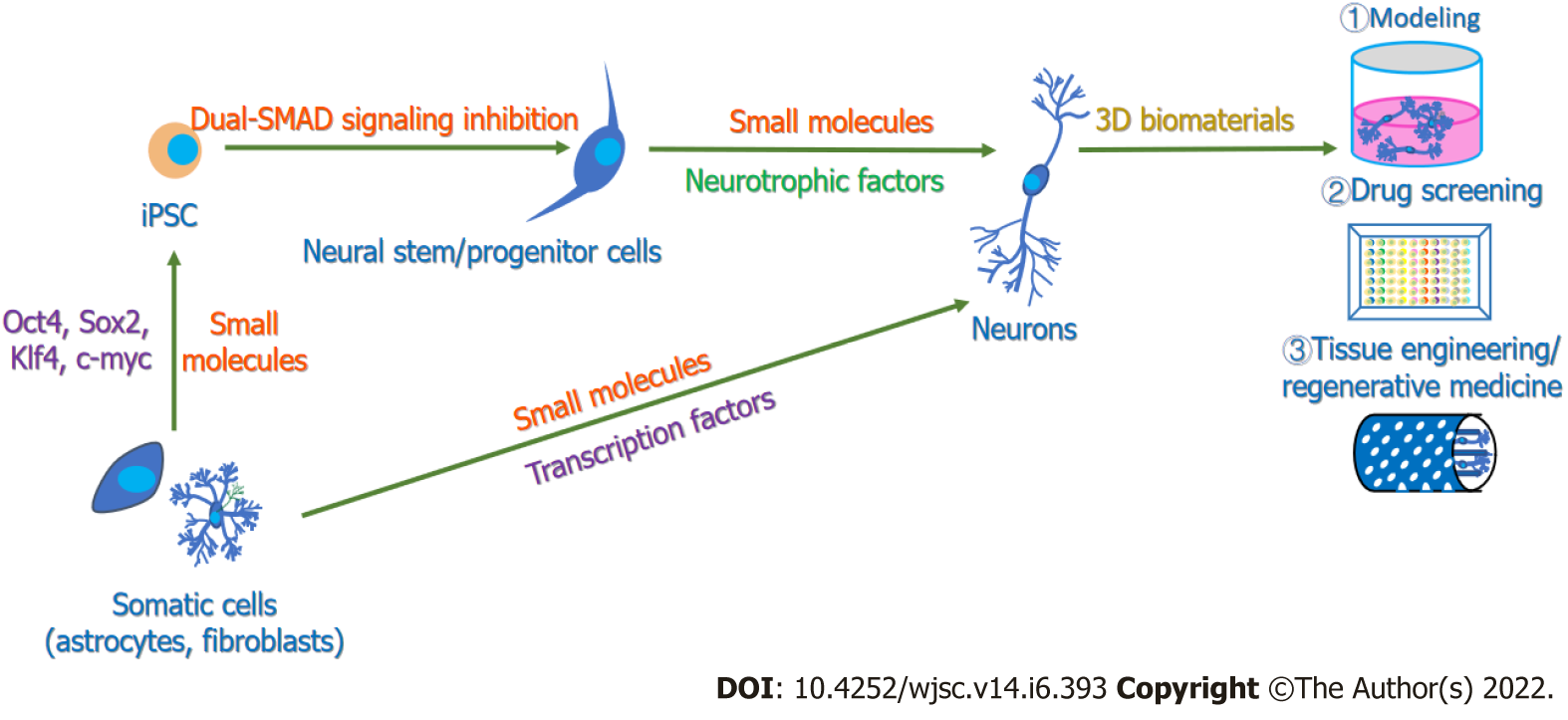
Over the years, the requirements of neurogenesis methods have grown with the increase in neurodegenerative diseases, and in vitro neurogenesis has been playing an important role in disease modeling, tissue engineering, drug screening and regenerative medicine[21-25]. However, the ways to generate mature and functional neural cells with high efficiency and cell purity remain a problem. Here, we discuss how HTS technology promotes the progression of in vitro neurogenesis in three sections, including screening out functional genes regulating neurogenesis, small molecules inducing neural lineage cells, and suitable microenvironments that facilitate in vitro neurogenesis. Furthermore, we review the applications of these obtained neural lineage cells using HTS technologies. Finally, with this review, we strengthen the connections between this promising and fast-developing technology and in vitro neurogenesis to raise awareness of generating more functional, mature and specific neural cells, as well as reproducible and standardized organoids with HTS technologies, for the sake of establishing robust neural developmental or disease models to better serve drug screening and regenerative medicine.
For the sake of designing various patterns for microarrays and microfluidic chips, depositing targets on the substrate surface is an essential step, which has been performed using a variety of methods, such as direct contact printing and noncontact printing, also known as ink-jet printing, photolithography, soft lithography, electron beam lithography, nanoimprint lithography, dip pen nanolithography, and laser-guided direct writing[26]. Direct contact printing can place desired biomolecules as ink from a stamp or a pin, linked to a high-precision robotic arm, to substrates with a reactive surface, which is usually accomplished by click reactions[27]. Noncontact printing can eject samples in the form of droplets to specific positions mainly by piezoelectric and thermal printers[26]. Photolithographic techniques can immobilize biomolecules on a substrate with photosensitive groups, for example, self-assembled monolayers such as alkanethiol[28], as linkers[29]. Patterns on the microarrays can be designed according to the patterns on the masks, which could selectively activate the photosensitive groups with UV light irradiation[29], and then the solubility of the photoresists will change, leaving the substrate in the development step. For soft lithographic techniques, the word “soft” can describe elastomeric stamps or channels, which are made of commonly used poly(dimethylsiloxane) (PDMS)[30]. PDMS stamps are utilized in microcontact printing, while channels are required in microfluidic channel flow patterning; these are the main methods used in soft lithographic techniques. The PDMS stamp can be fabricated using photolithographic techniques as the master is patterned with UV light and photoresists on the substrate, and then the liquid prepolymer is cast on the prepared master to form elastomeric stamps[30]. After that, these stamps can pattern the substrates through microcontact printing using molecules that can interact with biomolecules and cells.
To further enhance the resolution to the nanoscale for higher throughput, electron beam photolithography and dip pen nanolithography are applied for direct protein patterning on microarrays[31-35]. Nanoimprint lithography is also a nanostructure patterning technique that has been used to manipulate multiarchitectural chips with fields of topographies in nanometer dimensions to carry out high-throughput analysis for the screening of topographical structures that could promote stem cell differentiation[36,37]. In addition to using biomolecules as targets, cells can also be directly patterned into substrates, although attaching them to biomolecules that have been positioned to substrates is another matter. This technique is called laser-guided direct writing, in which the laser, focused by the lens, propels single cells with optical forces toward the substrate to form cell clusters[38]. This technique has been applied in tissue engineering through the reconstruction of tissues by micropatterning cells on soft matrices such as collagen or Matrigel to build cell–cell interactions that resemble those in the native microenvironment[39].
For neurobiological research, in vitro neurogenesis plays a significant role in conducting drug screening, establishing models for investigating mechanisms of neural development or diseases, and deepening research on regenerative medicine for cell therapy and tissue engineering[22,23,40]. Consequently, exploring more efficient methods for in vitro neurogenesis, including obtaining pure and functional neurons, building neural circuits, and forming neural tissue and even cerebral organoids, is of vital importance. To date, various methods have been used to manipulate in vitro neurogenesis (Figure 1). Embryonic stem cells (ESCs), pluripotent stem cells (PSCs) and mesenchymal stem cells (MSCs) have been induced to differentiate into functional neuronal cells through growth factors such as epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), and insulin-like growth factor 1 (IGF-1) as well as neurotrophic factors such as neurotrophin-3 (NT-3), brain-derived neurotrophic factor (BDNF), and glial cell line-derived neurotrophic factor (GDNF) (Supplementary Table 1)[41-43]. Sometimes, to obtain specific subtype neurons, key factors participating in subtype neuronal development are added; for example, sonic hedgehog (Shh) and fibroblast growth factor 8 (FGF8) are reported to be essential in the induction of dopaminergic neurons[44,45].
Furthermore, it has been found that the forced expression of distinct neurogenesis fate determinants, as well as the help of growth factors, can support the differentiation of ESCs and PSCs toward neural lineage cells (Supplementary Table 1)[46-49]. In fact, since induced PSCs (iPSCs) were developed by the integration of four transcription factors[50], neural transcription factors have been an efficient tool to reprogram fibroblasts[51,52], astrocytes[53,54] and other cell lines[55] to neurons in vitro. Recently, with the assistance of HTS, researchers have successfully explored some small molecules and their combinations to promote neural differentiation[56,57] and cell reprogramming[58-61] to obtain functional neurons (Supplementary Table 1). In these ways, functional and pure neurons with different neural subtypes, including dopaminergic neurons, cholinergic neurons, GABAergic neurons and glutaminergic neurons, have been generated.
Over the years, 3D culture systems have been developing rapidly, as they can provide cells with mechanical support, external stimuli and cell–ECM interactions to better mimic the architecture and functions of the in vivo microenvironment in living tissues and organs compared with conventional 2D cultures[62]. For many years, adding 3D matrices has become an attractive choice for the culture of neural cells and the generation of neural tissues or even neurospheres in vitro[63-65]. For cell culture, the three most commonly used models include the “on-gel model”, the “sandwich gel model” and the “in-gel model”. For even more complex 3D culture models, a study has cocultured predifferentiated human PSC (hPSC)-derived astrocytes with hPSC-derived, Ngn2-induced neurons on Matrigel-coated glass chips and observed more mature morphology of astrocytes, followed by the coculture of astrospheres and neurospheres on polytetrafluoroethylene membranes as organotypic-like cultures, and the astrocytes improved morphological complexity with fine leaflet-like structures[66].
There have also been other 3D culture systems containing spinner flasks and rotary bioreactors that can prevent cell attachment to the surface and aid in generating 3D spheroids[67], such as neural organoids. Over the past decade, organoid technologies have become a promising method to bridge the gap between cellular models and animal models and to better recapitulate the complexity of the cytoarchitecture at the tissue/organ level. The technology relies on the self-organization, self-renewal and differentiation capacity of ESCs or iPSCs and their potential to form cells from three germinal layers[68]. The protocol for generating cerebral organoids from hPSCs mainly includes the generation of embryonic bodies, the neural induction period, the neural differentiation period embedded in Matrigel, and the procedure for brain tissue growth and expansion in spinning bioreactors to provide enough oxygen[69,70]. Therefore, with the supplementation of 3D matrices and specific biomolecules, including growth factors and small molecules that precisely control the signaling pathways regulating neural lineage induction and neural differentiation, as well as devices providing sufficient oxygen for tissue growth, cerebral organoids containing discrete brain regions[69] and even organoids with specific brain regions, such as forebrain organoids[71,72], midbrain organoids[73,74], thalamic organoids[75] and cortical organoids[76], can also be generated. Specifically, to imitate morphogen concentration gradients in vivo, an inducible Shh-expressing hPSC line was constructed and embedded in one pore of developing organoids to create a Shh signaling gradient[72]. Treatment with LDN193189, SB431542 and XAV939 inhibited bone morphogenetic protein (BMP), transforming growth factor (TGF)-β, and Wnt signaling, respectively, to induce organoids toward forebrain identity[72,77]. Midbrain organoids were also generated based on the mechanism of regional patterning, as the addition of FGF8 and activation of Shh signaling have been proven to be significant elements in inducing midbrain dopaminergic identity[73,74,77,78]. Cortical organoids are induced through dual-SMAD signaling suppression following the induction of cortical identity patterns in vivo[76,77].
However, the self-organization ability of PSCs usually leads to an unpredictable arrangement of the internal structure of the organoids, which increases the uncertainty of their applications in in vitro modeling. Therefore, to improve the reproducibility between batches, a feasible method is to control the initial culture conditions, such as the starting cell types, cellular density, 3D matrices, and size and geometry of aggregates, because minimal deviations from the initial conditions will generally result in batch-to-batch controlled organoids[79]. To achieve this, trials have been performed to engineer 3D matrices and explore more suitable scaffolds, such as poly (lactic-co-glycolic acid) (PLGA) microfilaments, for guiding self-organization[71].
During neurogenesis, genes correlated with neural development are expressed in spatial and temporal order[80]. In previous studies, key genes have been identified[81] and have recently been the focus of further trials seeking to generate functional neural cells[48]. Significantly, basic helix loop helix transcription factors play important roles in neural specification and differentiation[82], and the forced expression of transcription factors has been manipulated to convert ESCs, iPSCs or nonpluripotent somatic cells into functional neurons even with specific subtypes, including midbrain dopaminergic neurons, spinal motor neurons, and forebrain GABAergic interneurons[21,51,83-86]. In addition to transcription factors, miRNAs are also effective tools to induce neural cell types; for example, Yoo et al[87] reported that miR-9/9* and miR-124, together with NeuroD2, could convert human fibroblasts into functional neurons by regulating SWI/SNF-like BAF chromatin-remodeling complexes.
For decades, functional gene identification has been carried out based on forward genetic approaches, mostly through whole-genome mutagenesis screening. Chemical mutagens, such as ethylmethanesulfonate and ethylnitrosourea, as well as polymerase chain reaction-based gene deletion strategies, are commonly applied to induce mutations, and phenotypes are evaluated to screen out functional genes[88-91]. However, for neurogenetic research, this method usually requires model organisms, including transgenic mice, Drosophila or zebrafish, which makes it time-consuming and difficult to manipulate screening on high-throughput platforms. Reverse genetic approaches are also becoming popular strategies to perform HTS to identify functional genes controlling various cellular behaviors. Since the discovery of RNA interference (RNAi), RNAi-based HTS has shown superiority in the identification of functional genes, the dissection of signal transduction pathways, and target exploration for drug development[92,93]. Compared with traditional forward genetic methods, RNAi screening fits more to cell-based screening, which is more suitable for conducting experiments at the cellular and molecular levels and is easier to utilize on HTS devices. Due to its advantages, this technique has been widely applied to identify genes that regulate neural development. Koizumi et al[94] injected double-stranded RNA into the preblastoderm embryos of Drosophila and found 22 genes that influence embryonic nervous system development, seven of which had unknown functions, nine of which had known functions, and the rest of which had known nervous system development phenotypes, such as dmt expressing a nuclear localization motif for peripheral nervous system development. Güneş et al[95] performed a selection-based screening and transduced CD34+ hematopoietic stem and progenitor cells and neural stem cells (NSCs) with shRNA. After next-generation sequencing and the comparison of shRNA representation in two cell types, they determined SMARCA4 to be the stemness regulator that controls NSC self-renewal by upregulating RE-1, a silencing transcription factor, and downregulating BAF53, suggesting its function in the repression of cell differentiation toward the neural lineage[95]. Other RNAi screening studies have also identified genes that play roles in neural outgrowth, axonal regeneration and neural cell death[88,96,97].
Over the years, with the discovery of the clustered regularly interspaced short palindromic repeats (CRISPR) system in prokaryotic organisms, CRISPR/Cas9 has opened new avenues toward HTS. The method of CRISPR-based HTS can be divided into arrayed/plate screens and pooled/barcode screens[4]. In arrayed screen format, the process is conducted in multiwell plates, and sgRNAs are individually delivered through viral transduction or transfection, followed by screen readout through high-throughput imaging, while for pooled screens, sgRNAs are synthesized and delivered as a pool, followed by viability screening through next-generation sequencing to evaluate the difference in the abundance of sgRNAs between samples for the identification of target genes or phenotypic screening, such as through FACS[4]. Compared with RNAi screening, CRISPR/Cas9-based screening has fewer off-target effects and is capable of investigating nontranscribed spacers and noncoding RNA, as RNAi can suppress gene expression only at the posttranscriptional level, whereas CRISPR/Cas9 can invalidate target genes at the genetic level[98]. Thus, the applications of CRISPR/Cas9 screening have grown in recent years, and genes related to bacterial toxicity[99], DNA mismatch repair pathways[100], new drug targets[101], cell viability and proliferation[102] have been identified in human cancer cells.
Significantly, CRISPR activation (CRISPR/a) and CRISPR interference (CRISPR/i) also play important roles in HTS in the identification of functional genes. These methods have been reported to serve as an effective tool in developmental biology studies. For instance, Genga et al[103] combined CRISPR/i screening and single-cell RNA-seq and identified FOXA2, the transcription factor that plays a significant role in endoderm development, as inhibition of FOXA2 impaired differentiation toward the foregut endoderm and the subsequent hepatic endoderm. In a neural differentiation study, Liu et al[104] utilized CRISPR/a screening through the establishment of a sgRNA library consisting of 55561 sgRNAs targeting all computationally predicted transcription factors and other DNA-binding factors and identified various transcription factors that could promote neural differentiation by sorting Tubb3-hCD8+ cells. They also studied the interactions between these hits through a combination of two sgRNAs and found that Ngn1, along with Brn2, Ezh2 or Foxo1, significantly improved the conversion efficiency of mouse embryonic fibroblasts (MEFs) into neurons[104]. The investigation of one of the hits, Ezh2, showed the downregulation of endodermal- and mesodermal-related genes in Ezh2-induced neurons, which indicates the possible mechanisms of enhanced neural differentiation of Ezh2[104].
Given that HTS possesses the ability to identify functional genes, particularly transcription factors related to neural development, future directions could still focus on the genetic networks of neurogenesis, especially the function of noncoding sequences, such as noncoding RNAs. For example, Zhu et al[105] designed a paired guide RNA CRISPR/Cas9 Library to delete approximately 700 long noncoding RNAs (lncRNAs) in the human liver cancer cell line Huh7.5OC. From a genome-scale lncRNA deletion screen, they found 9 lncRNAs positively or negatively correlated with the proliferation and survival of cancer cell lines[105]. Furthermore, CRISPR/a and CRISPR/i have also been proven to be useful in lncRNA screening. Liu et al[106] performed a genome-wide CRISPR/i screen with dCas9-KRAB targeting 16401 lncRNA loci in 6 transformed cell lines and an iPSC cell line, and the results showed that 499 lncRNAs participated in cell growth, such as LINC00263, the knockdown of which downregulated the proliferation of the U87 cell line and upregulated endoplasmic reticulum stress- and apoptosis-related genes. A CRISPR/a screen was also applied to identify lncRNAs related to the drug-resistance pathway in cancer cell lines[107]. In addition, CRISPR screening has been reported to discover functional miRNAs. Panganiban et al[108] conducted a genome-wide CRISPR/Cas9 screen and recently demonstrated that knockout of miR-124-3 led to upregulation of C/EBP homologous protein 10, the transcription factor associated with ER stress-mediated apoptosis, by regulating the IRE1 branch of the ER stress pathway. Moreover, screening out noncoding genes that serve as endogenous regulatory elements could help to further deepen our understanding of gene expression regulation. Klann et al[109] performed CRISPR/Cas9-based epigenomic regulatory element screening through dCas9-KRAB and dCas9-p300 to repress or activate the activity of the DNase I hypersensitive site and identified previously uncharacterized regulatory elements controlling the expression of b-globin and HER2. Baumann et al[110] strongly induced master transcription factor Sox1, as well as a neuroepithelial marker, through dCas9-VP64 to target the promoter of Sox1 and restored the neuronal differentiation potential of NPCs. Then, they transfected dCas9-Tet1 into NPCs that stably expressed dCas9-VP64 after transducing gRNA and found that the neuronal differentiation potency was increased with dCas9-Tet1 decreased DNA methylation levels around the transcription start of Sox1[110]. Thus, it is helpful to further understand the intrinsic interaction between activation of transcription factors and the regulation of epigenetic editing and even chromatin modification in neural development. Consequently, HTS could be a leading method to identify key genes and pathways, including transcription factors and noncoding regions related to neural development, which would provide novel targets for in vitro neurogenesis.
Although the overexpression of transcription factors in initial cells has made great progress in the induction of functional terminal differentiated cells, it could lead to safety problems such as tumorigenesis when the viral vectors integrate into the genomes of receptor cells[111]. Compared with the forced expression of transcription factors, utilizing cell-permeable chemical small molecules is safer, more cost-effective, less time-consuming, and easier to standardize. Thus, developing small molecules that could replace the effect of transcription factor overexpression has great prospects. This assumption first came into reality when Hou et al[112] identified a combination of seven small molecules that could reprogram MEFs to ESC-like PSCs. Before that, when many studies focused on the identification of small molecules that are capable of replacing defined transcription factors, HTS technology made huge contributions to identifying those specific small molecules.
For HTS technology, cell-free assays have been of great support for the identification of small molecules based on their effects on activating or blocking signaling pathways that facilitate or inhibit neural differentiation, which usually act as agonists or antagonists of kinases belonging to those signaling pathways. For example, SB431542 is an inhibitor of the ALK5/TGF-β1 receptor, which was identified by a flashplate-based assay with the immobilization of GST-tagged Smad3, and GST-tagged ALK5 was used as a kinase[113]. Except for using the interaction between biomolecules, especially between candidates and kinases or receptors affiliated with specific signaling pathways, cell-based methods are more commonly applied in screening compounds that could replace essential transcription factors, including in the period when the chemical induction method was the only method available for exploration. Takahashi and Yamanaka[50] screened out iPSCs by integrating a bgeo cassette, a fusion of the β-galactosidase and neomycin resistance genes, into Fbx15, a downstream gene of Oct4, to conduct iPSC screening through drug resistance to G418. However, iPSCs isolated in this manner were different from ESCs in their gene expression patterns, and screening was performed based on the activation of endogenous Nanog or Oct4 with a drug-resistance marker or the green fluorescent protein (GFP) reporter gene[114-116]. Recently, optical screening methods using fluorescent proteins, luciferase or the lacZ gene were proven to be effective in cell-based screening. To screen small molecules that could replace Sox2 in reprogramming, an Oct4-GFP transgenic reporter was used to provide Oct4-GFP+ colony numbers so that the reprogramming efficiency could be represented after retroviral transduction of MEFs with Oct4, Klf4 and c-Myc[117]. Then, from screening 200 small molecules, it was found that Repsox could substitute Sox2 even without c-Myc or the histone deacetylase inhibitor valproic acid (VPA), which could greatly improve reprogramming efficiency[118] through the inhibition of the TGF-β signaling pathway[117]. In addition, the G9a histone methyltransferase inhibitor BIX-01294 was also demonstrated to substitute Sox2 in the presence of Oct4 and Klf4 (c-Myc is dispensable when generating iPSCs from mouse and human fibroblasts)[119,120]. Other transcription factors, including Klf4, could also be replaced during reprogramming by VPA[121] or the GSK-3β or CDK inhibitor kenpaullone[122], and Oct4 could be replaced by the inhibitor of the ALK5/TGF-β1 receptor, SB431542 or Repsox[123].
In addition, HTS has been generally used to screen small molecules that could generate functional neurons to eliminate the ectopic expression of transcription factors. Li et al[59] screened approximately 5000 small molecules using Ascl1-infected mouse fibroblasts and found that forskolin, SB4315342, ISX9 and CHIR99021 could improve the number of TauEGFP-/Tuj1-positive neural cells. Subsequently, I-BET151 was screened out from approximately 1500 candidates in the presence of the former four small molecules and in the absence of the transcription factor Ascl1[59]. In subsequent tests, it was found that ISX9 was capable of activating the master neural genes NeuroD1 and Ngn2[59]. In addition, different combinations of various small molecules can also be screened to obtain more efficient cocktails for neural differentiation. Chambers et al[124] screened approximately 400 different combinations according to the day the compounds were added, and they confirmed that CHIR99021, DAPT and SU5402, which were added at day two of differentiation, along with SB431542 and LDN193189, could direct the differentiation of human NPC (hNPC) into nociceptive sensory neurons. They evaluated the decrease in PAX6+ cells and the improvement of TUJ1+ cells as the screening phenotype[124]. In another study, Zhang et al[61] performed a screen containing hundreds of combinations among 20 small compounds with different concentrations in a stepwise protocol and identified a cocktail including LDN193189, SB431542, TTNPB, thiazovivin, CHIR99021, VPA, DAPT, SAG, and purmorphamine that could convert human astrocytes into functional neurons.
To date, small molecules have been widely utilized in inducing neural cell lineages from PSCs or somatic cells, as a number of experiments have successfully generated functional neurons from ESCs, fibroblasts or astrocytes by chemical cocktails (Supplementary Table 2)[56,58-61,125,126]. Among the chemical compounds, SB431542 and LDN193189 have been widely applied to induce neuroectodermal cell lines from PSCs since the inhibition of dual-SMAD signaling was proven to be capable of converting human ESCs (hESCs) to PAX6+ neuroepithelial cells by the SB431542/Noggin protocol[127]. LDN193189 is an inhibitor of ALK2/ALK3 that was used to replace noggin, a soluble BMP antagonist. Dorsomorphin, also known as Compound C, was able to inhibit the BMP type 1 receptors ALK-2/ALK3/ALK6 and subsequently repress the phosphorylation of Smad1/5/8[128]. However, dorsomorphin resulted in moderate inhibition and unstable metabolism; thus, LDN193189 was recently developed and exhibited a highly decreased IC50[129]. In addition, CHIR99021, also named CT99021, is an inhibitor of GSK3β and can activate Wnt signaling, which has been demonstrated to play an important role in maintaining neural stem/progenitor cell proliferation and differentiation[130]. Nevertheless, CHIR99021 has been reported to induce the neural crest lineage through activation of the Wnt signaling pathway[124], the inhibition of which by XAV939 could help to generate cortical neurons[131]. Of note, CHIR99021 was added in the final differentiation step, as it can function in the promotion of axonal outgrowth and synapse formation[131]. Significantly, Kirkeby et al[57] tested CHIR99021 in a dose-dependent manner and found that hESCs differentiated into neural progenitors with all regions from the telencephalon to the posterior hindbrain along the rostrocaudal axis following increasing concentrations of CHIR99021. Except for the rostrocaudal axis, dorsoventral axis patterning could also be induced by small molecules, as ventralization can be regulated by Shh, and dorsalization can be controlled by the Wnt canonical pathway and the BMP pathway[77]. Purmorphamine is a Smoothened receptor agonist that can activate the Shh signaling pathway and has a similar effect as another small molecule called SAG, which is a potent Smoothened receptor agonist. Thus, neural subtypes could be enriched through the coordination of Shh signaling, Wnt canonical signaling and BMP signaling[132]. Forskolin is also a commonly used small molecule that functions as a diterpene adenylate cyclase activator, and the addition of forskolin could increase the level of intracellular cyclic AMP (cAMP). Significantly, the activation of cAMP/PKA-cAMP-responsive element binding (CREB) signaling, for example, by treatment with dibutyryl-cAMP, can phosphorylate CREB protein, which is an essential transcription factor regulating many target genes related to the survival, proliferation and differentiation of neurogenic cells, such as Bcl-2, BDNF, tyrosine hydroxylase and somatostatin[133]. Furthermore, SU5402 and PD0325901 are inhibitors of FGFR1 and mitogen-activated extracellular activated signal-regulated kinase (MEK), respectively, which all serve as inhibitors of FGF/MEK/ERK signaling. DAPT, a g-secretase inhibitor, is likewise a small molecule commonly applied as a Notch signaling inhibitor. There have been a number of studies using the inhibition of FGF and Notch signaling to suppress cell proliferation and thus lead to differentiation[124,126,131].
To date, with the support of the HTS method, many types of small molecules have been developed to convert pluripotent cells such as ESCs and NPCs or nonneural somatic cells such as fibroblasts into neural lineage cells. However, there is still great demand to generate mature neurons with specific neural subtypes and positional cues of different brain regions. To meet this requirement, more selective chemical compounds with optimum concentrations and combinations with different addition orders are desired. Maury et al[134] utilized the automated 384-well plate format to treat hNPCs with various concentrations, durations, and combinations of small molecules and directed NPCs to spinal motor neurons and cranial motor neurons with specific regional identities. Additionally, developing diverse types of small molecules that regulate gene transcription through different mechanisms could be another perspective to expand the collection of small molecules for neural lineage conversion. In addition to small molecules that work as signaling pathway modulators, other chemical compounds that can repress epigenetic-related enzymes, such as histone-modifying enzymes, DNA methylation-associated enzymes, and modulate nuclear receptors remain to be further explored[135]. For example, VPA, an inhibitor of histone deacetylase; RG108, an inhibitor of DNA methyltransferases; and TTNPB, a retinoic acid (RA) analog and a nuclear receptor RAR agonist, are applied in the conversion of neural cells through transdifferentiation methods[58,61,126,136]. Therefore, further investigations of epigenetic mechanisms and the orchestrated signaling processes underlying neural lineage specification are required to develop targeted small molecules.
The cell microenvironment consists of ECM, soluble molecules such as cytokines and hormones, and interactions with adjacent cells. To better facilitate neurogenesis in vitro, screening and reconstituting suitable microenvironments similar to those in vivo are important. However, traditional culture systems cannot be used to assess various microenvironmental factors at the same time. With the HTS platform, it is much more convenient and efficient to evaluate various parameters, including the type, topography, and stiffness of 3D materials and the types and concentration gradients of soluble biomolecules on a highly integrated chip.
Screening for 3D scaffolds: ECM molecules, mainly containing laminin, fibronectin, collagen Ⅳ, entactin, elastin, heparan sulfate proteoglycans, hyaluronan, chondroitin sulfate proteoglycans and tenascin-R in the CNS[137,138], are essential components of the microenvironment. In the natural matrices above, the widely applied 3D matrices are collagens[63,64,139,140], hyaluronan[141], and another commonly applied biomaterial called Matrigel[22,142,143] due to their superior contributions to the in vitro neural proliferation, differentiation and outgrowth of NSCs, NPCs or ESCs, some of which also build neural circuits and recapitulate CNS neural development.
Since these natural matrices are largely extracted from animals or cultured cells, it is difficult to control the biochemical and mechanical cues of batches[62]. Thus, to increase the reproducibility and reliability in further applications, synthetic scaffolds such as synthetic polymer hydrogels, which mainly contain self-assembling peptide hydrogels[144,145], poly(ethylene glycol) (PEG)[146-148], poly(lactic acid)[149,150], PLGA[151,152] and electrically conductive polymers including poly(pyrrole)[153-155], and carbon nanotubes[156,157] have become attractive tools for 3D in vitro neurogenesis (Supplemen
Microarrays based on glass slides on 2D platforms have been typically used for biomaterial screening, usually including polymer microarrays for screening synthetic polymer scaffolds and ECM or tissue microarrays for screening naturally sourced matrices. As mentioned before, biomaterials are patterned on slides through contact printing, injection printing and photolithography[159]. For polymer microarray screening, Anderson et al[160] fabricated a nanoliter-scale polymer array on which there were 576 different acrylate-based polymers in triplicate, synthesized by diverse combinations of 25 kinds of monomers through a light-activated radical initiator and UV light, attached to a poly (hyd
In addition to polymer-based arrays, ECM microarrays are also commonly utilized for the investigation and dissection of functional elements for regulating cell behavior. Nakajima et al[163] displayed ECM-based biomaterials, including collagen I, collagen IV, fibronectin and laminin, as well as artificial biomaterials containing acidic gelatins, basic gelatins, ProNectinTM F plus and ProNectinTM L, poly(L-lysine), and poly(ethyleneimine) with weight-averaged molecular weights of 800, 10000, 25000, and 750000 on gold-coated glass plates. The results showed that fibronectin, laminin, Pro-F, Pro-L and PEI-0.8 could support NSC adhesion, while collagen and gelatins had no effect on NSC adhesion[163]. The probable reason for these results could be that NSCs adhere fibronectin and laminin through b1 integrin, and an electrostatic interaction might have promoted NSC’s adhesion to PEI-0.8[163]. Ahmed et al[164] screened 190 combinations of 19 ECM proteins that were selected according to their expression in the ventral midbrain during dopaminergic neurogenesis and identified that Sparc, Sparc-like (Sparc-l1) and Nell2 could synergistically increase the number of TH+ neurons differentiated from long-term neuroepithelial stem cells.
In addition, tissue matrix-based microarrays were fabricated by removing soluble components and mechanically fragmented matrices from 11 different porcine tissues and organs, which could preserve the natural diversity and complexity of biomaterials[165]. In this way, studies could focus on naturally sourced ECM components from various tissues and organs and analyze the tissue/organ-specific differences that subsequently lead to cell lineage specification. While retaining the complexity of ECM proteins, disassembling functional domains could also be an effective approach. Lin et al[166] conducted a peptide microarray and seeded normal murine mammary gland cells and demonstrated that the peptides LTGKNFPMFHRN and MHRMPSFLPTTL could induce epithelial-to-mesenchymal transition and decrease E-cadherin levels.
Screening for surface topography and morphology: In addition to the type of biomaterial, surface topography and morphology can also support neural induction. Studies have been performed to investigate the impact that continuous, discontinuous and random topographies that biomaterials have on the guidance and outgrowth of axons and dendrites[167]. In particular, continuous topographies can impact the orientation and shape of NSCs through the regulation of cytoskeleton rearrangement and nucleus elongation, while discontinuous isotropic topographies are reported to induce NSCs to the glial lineage[168]. Thus, to explore more suitable biomaterials expected to support neural growth, precise surface topologies were screened to search for the topologies that promote axon and dendritic growth. Large-scale screening showed that anisotropic grating patterns could best facilitate axon growth, while dendrites showed almost no sensitivity to surface topologies[169]. Furthermore, matrix stiffness also plays an unignorable role in neural development, as distinct subtypes of neurons exhibit different neurite outgrowth rates when cultured in conditions of varying elasticity[170]. To investigate the mechanical properties suitable for controlling specific cellular behavior, HTS has been utilized; for instance, Kumachev et al[171] constructed a droplet-based screening platform by encapsulating mESCs into agarose microgels with different elastic moduli. Kourouklis et al[172] arrayed various combinations of five ECM proteins on a poly(acrylamide) hydrogel substrate with three different elastic moduli to assess the effect of substrate stiffness on the differentiation of bipotential mouse embryonic liver progenitor cells.
Screening for combinations of growth factors: Soluble bioactive molecules such as growth factors, including bFGF and EGF; members of the neurotrophin family, including BDNF and GDNF; and members of the TGF family are important parts of the microenvironment during neurogenesis, regulating neural proliferation and differentiation[173-175]. The HTS method could be used to evaluate the best candidate or the best combination to promote in vitro neurogenesis. Konagaya et al[176] immobilized five growth factors, including bFGF, EGF, IGF-1, BDNF, and ciliary neurotrophic factor (CNTF), on a chip and displayed them either as a single component or as the combination of any of two factors to explore their function on NSCs. They found that either bFGF or EGF alone could facilitate the proliferation of NSCs, and that the combination of these two factors showed a synergistic effect. Both IGF-1 and BDNF could facilitate NSC differentiation toward the neural lineage, but CNTF promoted glial lineage differentiation[176]. Nakajima et al[163] used a cell-based assay to coimmobilize growth factors and natural or synthetic matrices, and they found that EGF promoted the maintenance of NSCs and that two nerve growth factor (NGF) family members, NGF and NT-3, could facilitate NSCs toward neuronal differentiation. In addition to for cell-based microarrays requiring immobilization of biomolecules, Muckom et al[177] applied a high-throughput microculture system consisting of complementary micropillars and microwells that could hold 532 independent microenvironments for cell culture. They seeded adult rodent NSCs and provided 6 soluble factors, BMP4, TGF-β, FGF-2, shh, Wnt-3a and Ephin-B2, and evaluated the extent to which their individual signals and double, tertiary and quaternary signal combinations could influence neural differentiation[177]. Their results indicated that Wnt-3a and Ephin-B2 synergistically facilitated neural differentiation and maturation, while TGF-β, FGF-2 and Wnt-3a affected NSC proliferation and differentiation antagonistically[177].
Screening for 3D microenvironments: Beyond exploring single variants such as the abovementioned ECM proteins, surface topography, matrix stiffness and soluble factors, it could be more effective to combine various elements together and screen the whole microenvironment (Supplementary Table 4). A typically used approach is to premix ECM proteins and soluble signaling molecules in different combinations within multiwell plates, such as 384-well plates, and then to codispense the mixtures on substrate slides of the microarray. Later, cells are seeded on each spot. Lin et al[178] designed a microarray capable of screening microenvironments, including substrate stiffness, ECM matrices, various growth factors and cytokines. The elastic modulus could be adjusted by altering the base/cure ratio of PDMS to mimic hard tissues such as cartilage, cornea, and arterial walls, while regulating the acrylamide/bis-acrylamide ratio of PA could mimic soft tissues, including the brain, liver, and prostate[178]. Soen et al[179] also constructed a microarray for screening out microenvironments from 44 combinations of ECM proteins and signaling factors that promote the neural differentiation and specification of primary human NSCs (hNSCs). Moreover, Brafman et al[180] designed a 3D microarray screening method called arrayed cellular microenvironments, which could hold 8000 spots to screen microenvironments containing ECM proteins, growth factors and small molecules for evaluating cell attachment, growth and proliferation of hPSCs. To avoid interference between each spot on microarrays and to make the microculture system more suitable for culturing nonadherent cells, Gobaa et al[181] constructed arrays of PEG hydrogel microwells that could hold 2016 microenvironments to evaluate the effects of modular stiffness, bioactive molecules and ECM proteins on stem cells. The microwells were fabricated by stamping a silicon substrate with previously spotted biomolecules on a hydrogel substrate with different PEG concentrations to alter stiffness[181]. From screening these artificial niches, they studied the impact of laminin-1 and Jagged-1, the Notch ligand, on NSC fate[181].
However, when the microenvironment-based arrays are carried on a 2D platform, the cell–ECM interactions are weakened. For the sake of recapitulating the microenvironments and simulating the cellular states in vivo, 3D-based HTS platforms are recommended to evaluate the microenvironments proper to activate the specific cellular activities. Various microscale 3D culture screening systems mainly contain hanging drop plates, cellular microarrays and microwell plates (Supplementary Table 4)[67]. Because hanging drop plates are not suitable for long-term culture, these platforms might be more suitable for neural differentiation or organoid formation. Cellular microarrays, as well as microwell plates, are more broadly used for 3D HTS protocols, as these two methods allow for longer culture periods and more stability than hanging drop plates. For cellular microarrays, cells could be premixed with gels and coprinted on the substrates with robotic arrayers (Figure 2). For instance, Fernandes et al[182] designed a dual-slide incubation method that included a methyltrimethoxysilane-coated glass slide with preprinted all-trans-RA and FGF-4 and another poly(styrene-co-maleic anhydride)- and a poly(L-lysine)-coated glass slide containing mESCs embedded in alginate spotted with a robotic spotter. Another common platform contains two complementary chips, a microwell and a microchip. The procedure involves spotting the mixture of cells and 3D matrices on the top of the micropillar and adding culture medium with screening candidates, such as small molecules and soluble factors, into the microwell. Then, by stamping and incubating two complementary slides, the small molecules and growth factors can diffuse into the cell spots and trigger biological reactions (Figure 2). In this way, multiple elements in microenvironments can be screened to study the regulation of stem cell fate[183-185]. Given that microarray-based screening can result in interference between spots, microwell plates are also commonly used in 3D screening methods. Ranga et al[186] performed a 3D niche microarray on 1536-well plates that could control five characteristics: Matrix mechanical properties, ECM proteins, cell–cell interaction proteins, soluble factors and proteolytic degradability (matrix metalloproteinase sensitivity). Researchers cross-linked branched PEG-based macromers with specific peptide sequences susceptible to cell-secreted matrix metalloproteinases[186]. Then, they encapsulated mESCs in 3D PEG gels to investigate mESC proliferation and self-renewal properties in different combinations of microenvironments. From the completed studies, we can learn that HTS platforms, especially 3D platforms, are tools with great potential for constructing microenvironments to discover combinations of elements that could well facilitate in vitro neurogenesis, such as NSC proliferation and differentiation, and even the internal signaling in charge of those cell behaviors.
Since HTS technology can identify small molecules, specific genes and physiological microenvironments that contribute to neurogenesis in vitro, it is also important to focus on the clinical backgrounds and applications of the generated functional neurons or neural lineage cells using this technology.
Over the past 20 years, stem cell therapy and regenerative medicine have received considerable attention and have been expected to be applied in the treatment of CNS diseases, especially neurodegenerative diseases. The potential of stem cell therapy for CNS disease treatment lies in the capacity of NSCs to compensate for lost neurons with differentiated functional neurons, rebuild neural networks, secrete neurotrophic factors and reduce neuroinflammation to increase the survival rates of transplanted cells and healthy neurons[187,188]. Before clinical trials, studies of stem cell trans
For clinical studies, NPCs were transplanted into the dorsal putamina of patients with moderate PD, and a four-year evaluation was performed, which found this transplantation surgery to be safe and lacking in immune response or adverse effects[221]. Motor improvement and enhanced midbrain dopaminergic activity were shown, although they decreased somewhat over four years[221]. A long-term phase Ⅰ clinical trial also proved the safety of hNSCs and found a transitory decrease in the progression of the ALS Functional Rating Scale Revised up to four months post-transplantation[222]. These results are promising, and in future studies, neural precursors derived from iPSCs or somatic cells, especially fibroblasts, can be applied to clinical studies to show safety and efficiency. For an allogeneic approach, developing an iPSC bank based on the human leukocyte antigen haplotype could provide more possibilities for stem cell therapy[223], and the genetic editing of patient-derived iPSCs is also a feasible approach for autologous stem cell transplantation[224]. Meanwhile, exploring other cell sources for stem cell therapy could help broaden the field for clinical studies. Recently, a research group used NSCs isolated from midbrain organoids, which are generated from hPSCs, and transplanted them into rat PD models[225]. The results showed midbrain dopaminergic neuron engraftment and reproducible behavioral restoration in those PD models[225].
Neural lineage cells generated in vitro are also cell sources for neural tissue engineering, as they are becoming an attractive option for CNS disease treatment, considering that the support of 3D scaffolds can mimic the microenvironments that help the engrafted cells survive, integrate and differentiate. For brain injury repair, the injection of NSCs with a hyaluronate collagen scaffold loaded with controlled release of bFGF can recover cognitive function through the promotion of survival, differentiation and synaptic formation of NSCs in traumatic brain injury (TBI) rats[226]. Chitosan scaffolds are also common options for the neural tissue engineering treatment of TBI in animal models[227-229]. In addition to hydrogel materials, researchers also use porous scaffolds to prevent the collapse of scaffolds and provide enough space for neural differentiation, metabolic exchange, and neurite extension of grafted cells[230,231]. For SCI repair, collagen microchannel scaffolds and gelatin sponge scaffolds carrying NSCs with drugs or neurotrophic factors have enhanced tissue repair efficiency in SCI animal models[232,233]. Importantly, with the capacity to construct complex 3D microstructures, 3D bio
Functional neurons can also be used for in vitro modeling to achieve a better comprehension of CNS diseases and neural development mechanisms. For neural disease modeling, AD models have been constructed utilizing hNPCs with familial AD (FAD) genes in Matrigel-based 3D culture systems, and aggregated p-tau proteins and amyloid-β deposits resembling AD pathology were observed[22]. Furthermore, they showed that a high amyloid-β42/40 ratio could drive Aβ accumulation and phosphorylated tau protein accumulation in this 3D AD model[238]. To recapitulate neuroinflammation in AD, they also conducted a 3D AD triculture model containing hNPC-derived AD neurons/astrocytes and subsequently plated microglia in the microfluidic platform[239]. The results showed that migrating microglia, the upregulation of AD-related proinflammatory factors and the toxic effects of microglia on neurons and astrocytes could be observed in the 3D AD triculture model[239]. In addition to AD models, PD models have also been established using 3D culture with in vitro neural differentiation. Taylor-Whiteley et al[240] first constructed a 3D PD model by differentiating human SH-SY5Y neuroblastoma cells into dopaminergic cells with RA and BDNF cultured in Matrigel[240]. Next, they treated cells with preformed a-synuclein (a-syn) oligomers and observed a-syn-positive inclusions that resemble in vivo Lewy bodies in morphology[240]. Organoids have also been proven to be effective tools for 3D modeling. Kim et al[241] utilized hiPSCs with leucine-rich repeat kinase 2 G2019S mutation, which is a well-known trigger of late-onset familial and sporadic PD, to generate 3D midbrain organoids. From the 3D organoid model, they identified the TXNIP gene, which can contribute to the generation of α-syn in LRRK2-associated PD[241]. Another research group built a 3D sporadic AD model by treating brain organoids with human serum to mimic the serum exposure caused by a blood–brain barrier breakdown in AD[242]. AD-like pathologies could be observed in serum-exposed brain organoids, with increases in Aβ aggregates, phosphorylated microtubule-associated tau protein (p-Tau) levels, synaptic loss, apoptosis, and impaired neural networks[242]. In addition, 3D models aimed at other neural system diseases, including HD[243], hypoxic brain injury of prematurity[244] and brain tumors[245], have also been established. These 3D disease models provide us with feasible and valid platforms for future studies of disease pathogenesis and drug screening.
In addition to disease modeling, in vitro 3D models can also be constructed to recapitulate neural development. To study neural tube morphogenesis in vitro through 3D culture, Ranga et al[23] first performed combinational HTS to screen out appropriate parameters of 3D matrices, based on which they investigated the effects of early developmental signaling molecules, including RA, Shh, Wnt-3a, BMP4 and FGF8, on dorsal-ventral (D-V) patterning with their 3D neural tube model. Another study cultured mESCs in Matrigel or defined 3D scaffolds containing laminin and entactin or PEG and induced floor plate formation and D-V pattering with RA[246]. Mariani et al[247] induced human iPSCs to serum-free, floating embryoid body-like, quick aggregates with embryonic dorsal telencephalon properties, which could be used as an in vitro 3D model for human cortical development. With the use of cerebral organoids, gene expression programs and epigenetic signatures during human brain development were recapitulated, as well as the interaction patterns between different brain regions[248-250]. These models mimicking neural development could be applied to explore mechanisms underlying organogenesis and cell–cell interactions during neurogenesis[66,251] and could also be an option for studying neural genetic disorders[252], as well as a platform for drug screening.
Conversion efficiency is often discussed in articles focusing on cellular reprogramming and neural differentiation, which refers to the ratio of the target cell types to the initial cell types. The improvement of neural conversion efficiency is an important subject to address to increase the purity and efficiency of generated neurons for future clinical use. To overcome these difficulties, small molecules have been screened out to replace transcription factors, as small molecules can improve the conversion efficiency of cellular reprogramming compared to the overexpression of transcription factors[253]. For instance, VPA can enhance the reprogramming efficiency of somatic cells to iPSCs[118,121], and CHIR99021, LDN193189, and A83-01 can further improve the neural induction rate[136]. Significantly, the application of CRISPRa is capable of greatly enhancing reprogramming efficiency by targeting the human embryo genome activation-enriched Alu motif, leading to more efficient activation of Nanog and Rex1[254]. Therefore, activating endogenous loci controlling cellular reprogramming and neural lineage induction can be an effective way to increase neural conversion efficiency. An alternative way is to selectively ablate proliferative cells and keep functional neurons for the sake of guaranteeing the safety and efficiency of stem cell transplantation. This research was performed via pharmacological activation of the suicide gene within weeks after transplantation, and the yield of dopaminergic neurons and the recovery of motor functions were not affected by diminishing the graft size in the PD rat model[255]. Thus, in future studies, HTS technology can still play an important role in screening small molecules and endogenous genes, which can aid in improving the conversion efficiency and generate more functional neurons.
Over the decades, testing probes of HTS methods have been developed from molecules to cells and even to tissues/microenvironments and organoids. Currently, molecular, cell- and tissue-based screening systems have come into use, and it is also quite important to investigate organoid-based HTS devices with increased producibility and reduced heterogeneity between batches, allowing for large-scale screening[256]. Jorfi et al[257] screened FAD-mutated hNSCs or iPSC-derived neurospheroids with a 96-well cell culture plate with 1536 microwells. They embedded the neurospheroids in Matrigel and screened several chemical compounds to assess their impact on neural differentiation[257]. The establishment of a high-throughput bioengineered human cardiac organoid in the 96-well format was also reported[258]. After that, 105 hit compounds from approximately 5000 candidates, which were screened from iPSC-derived cardiomyocytes in the 2D platform, were screened over a 3-log scale concentration range that requires approximately 1000 human cardiac organoids to develop compounds with the capacity for cardiomyocyte proliferation[259]. Another study generated kidney organoids from hiPSCs utilizing multiwell plates, and this HTS-compatible platform was used to screen out an inhibitor of nonmuscle myosin II ATPase activity as a specific activator of polycystic kidney disease cystogenesis in organoids[260]. Renner et al[261] also developed an automated workflow that could integrate midbrain organoid culture, immunostaining and high-content imaging for high-throughput chemical screening using a 96-well format, which could save manual operation and improve the compatibility of organoid culture and HTS. Although high-content imaging analysis has been a powerful tool to evaluate organoid generation, for brain organoids, it is probable that the evaluation of neural circuit dynamics, such as that through 3D microelectrode arrays, could become a standard in upcoming studies[262,263]. Furthermore, combinations with the automated workflow of organoid culture and artificial intelligence can shed light on CNS disease modeling and drug discovery for clinical trials[264].
Over the years, HTS devices have been developed, ranging from multiwell plates to microarrays; notably, microfluidic devices are gradually showing their features in HTS technology. The lab-on-chip method has contributed to this development. Schudel et al[265] designed a microfluidic chip to separate cell clusters by dividing the chip into one part for siRNA patterning and another for target screening to study virus–host interactions. Furthermore, to improve the screening efficiency after cell transduction, Wang et al[266] designed a droplet-based microfluidic platform compatible with single-cell screening to identify the yeast Saccharomyces cerevisiae with elevated protein production through RNAi screening and searched for genetic targets capable of improving protein secretion. Han et al[267] first utilized a CRISPR/Cas9 screen on a microfluidic platform, also called a microfluidic separation chip, on which cells transduced with the lenti-CRISPR kinase library were sorted to examine transport distances to evaluate cell deformability[267]. For chemical screening, Titmarsh et al[268] constructed a high-density microbioreactor array that could provide 8100 chambers for the proliferation of hPSCs or hPSC-derived cardiomyocytes. They found that CHIR99021 showed the best effect on human cardiomyocyte proliferation among purmorphamine, IGF-1 and FGF-2[268]. Although the microfluidic array could provide thousands of chambers as reactors, the numbers of candidates allowed for one screening are usually limited. Thus, exploiting microfluidic devices that are able to hold more isolated channels for screening more candidates at a time has great potential.
However, the HTS platforms currently available for cell/organoid-based screening are mainly well plates that lack automation and integration and commonly cause reagent waste. Therefore, in further research, the microfluidic platform shows great promise to achieve a higher throughput and autocontrolled and integrated properties. For instance, Schuster et al[269] designed an automated microfluidic 3D cellular and organoid culture platform for the culture of pancreatic ductal adenocarcinoma organoids generated from single cells from patients. The platform could contain 20 independent experimental conditions and 200 individual chambers that are large enough to hold growing organoids[269]. The researchers performed dynamic and combinational drug screening and recorded the incidences of cellular apoptosis and death to evaluate the treatment effect of the temporal drug combinations.
In addition, microfluidic devices could also be utilized to perform cell coculture using droplet-based microfluidic systems, which could function in studying the microenvironments of cell–cell interactions under high-throughput conditions[270]. Other researchers have also designed high-throughput 3D coculture systems on microfluidic chips[271,272]. With these methods, HTS could be performed on these platforms to screen out 3D microenvironments containing cellular interactions, such as synaptic connections between neurons and astrocytes. In addition, microfluidic chips have been applied in generating concentration gradients of biomolecules to study steepness-dependent neural chemotaxis on high-throughput 3D platforms[273]. Rifes et al[274] constructed a microfluidic platform to generate gradients of CHIR99021 to activate Wnt signaling, and they modeled neural tube development in this 3D microfluidic system. Therefore, in future studies, HTS will be performed on microfluidic systems due to their capacity to better recapitulate the microenvironments in vivo, which is a strategy that shows great promise.
HTS technologies are playing increasingly important roles in neurogenesis in vitro due to their ability to screen out crucial genes controlling neural lineage determination, small chemical molecules regulating cell fate, and microenvironments, including 3D matrices, soluble factors, physical parameters and interactions with other cell types (Figure 3). After screening out suitable microenvironments, these culture conditions could be applied in generating mature and functional neurons, neural tissues and organoids in vitro for further applications, such as 3D modeling and drug screening, to investigate neural diseases or developmental mechanisms and explore medical solutions. With the requirements of 3D models, 3D-based screening with tissues or organoids is developing to better evaluate screening outcomes from an overall perspective than molecular or cell-based screening can. Meanwhile, the screening devices are trending toward minimization, automation and integration, from multiwell plates to microarrays and microfluidic devices, to conduct the screening process in a high-throughput manner that requires less time and consumes fewer reagents. Today, the need for combinational screening is growing, as investigations of the interactions between different drugs or environmental factors are vital to developing combined therapies and novel culture conditions. In addition, it is notable that microfluidics makes it easier to perform high-throughput combinational screening with nanodroplets and microwell array plates that can hold only two nanodroplets in a well[275]. Overall, from past studies and due to the fast development of HTS devices, we anticipate that HTS technologies will be able to make great contributions to in vitro neurogenesis and solve other problems in regenerative medicine in future studies.
To conclude, HTS technology could help to dissect the mechanisms of genetic regulation during neurodevelopment, identify niche-targeted small molecules and secreted factors to promote en
We thank the Biological and Medical Engineering Core Facilities of Beijing Institute of Technology.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Nakeep S, Egypt; Papadopoulos K, Thailand; Song BW, South Korea A-Editor: Liu X, China S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Chen W, Chen M, Barak LS. Development of small molecules targeting the Wnt pathway for the treatment of colon cancer: a high-throughput screening approach. Am J Physiol Gastrointest Liver Physiol. 2010;299:G293-G300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 2. | Penchovsky R, Stoilova CC. Riboswitch-based antibacterial drug discovery using high-throughput screening methods. Expert Opin Drug Discov. 2013;8:65-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Tan Y, Zhang Y, Han Y, Liu H, Chen H, Ma F, Withers SG, Feng Y, Yang G. Directed evolution of an α1,3-fucosyltransferase using a single-cell ultrahigh-throughput screening method. Sci Adv. 2019;5:eaaw8451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 4. | Schuster A, Erasimus H, Fritah S, Nazarov PV, van Dyck E, Niclou SP, Golebiewska A. RNAi/CRISPR Screens: from a Pool to a Valid Hit. Trends Biotechnol. 2019;37:38-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 5. | Liu B, Li S, Hu J. Technological advances in high-throughput screening. Am J Pharmacogenomics. 2004;4:263-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 67] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | An WF, Tolliday N. Cell-based assays for high-throughput screening. Mol Biotechnol. 2010;45:180-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 127] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 7. | Jonczyk R, Kurth T, Lavrentieva A, Walter JG, Scheper T, Stahl F. Living Cell Microarrays: An Overview of Concepts. Microarrays (Basel). 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 419] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 9. | Lee SY, Doh I, Lee DW. A High Throughput Apoptosis Assay using 3D Cultured Cells. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Doh I, Kwon YJ, Ku B, Lee DW. Drug Efficacy Comparison of 3D Forming and Preforming Sphere Models with a Micropillar and Microwell Chip Platform. SLAS Discov. 2019;24:476-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Thorsen TA. Microfluidic tools for high-throughput screening. Biotechniques. 2004;36:197-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Qiang L, Guo J, Han Y, Jiang J, Su X, Liu H, Qi Q, Han L. A novel anti Candida albicans drug screening system based on high-throughput microfluidic chips. Sci Rep. 2019;9:8087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Mondal S, Hegarty E, Martin C, Gökçe SK, Ghorashian N, Ben-Yakar A. Large-scale microfluidics providing high-resolution and high-throughput screening of Caenorhabditis elegans poly-glutamine aggregation model. Nat Commun. 2016;7:13023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 14. | Lu Y, Chen JJ, Mu L, Xue Q, Wu Y, Wu PH, Li J, Vortmeyer AO, Miller-Jensen K, Wirtz D, Fan R. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal Chem. 2013;85:2548-2556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 15. | Toh YC, Zhang C, Zhang J, Khong YM, Chang S, Samper VD, van Noort D, Hutmacher DW, Yu H. A novel 3D mammalian cell perfusion-culture system in microfluidic channels. Lab Chip. 2007;7:302-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 291] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 16. | Guo MT, Rotem A, Heyman JA, Weitz DA. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12:2146-2155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 784] [Cited by in RCA: 663] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 17. | Sesen M, Alan T, Neild A. Droplet control technologies for microfluidic high throughput screening (μHTS). Lab Chip. 2017;17:2372-2394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 18. | Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA. 2009;106:14195-14200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 701] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 19. | Fu AY, Chou HP, Spence C, Arnold FH, Quake SR. An integrated microfabricated cell sorter. Anal Chem. 2002;74:2451-2457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 251] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 20. | Baret JC, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9:1850-1858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 713] [Cited by in RCA: 634] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 21. | Kim J, Su SC, Wang H, Cheng AW, Cassady JP, Lodato MA, Lengner CJ, Chung CY, Dawlaty MM, Tsai LH, Jaenisch R. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 214] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D'Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515:274-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 753] [Cited by in RCA: 888] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 23. | Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, Lutolf MP. Neural tube morphogenesis in synthetic 3D microenvironments. Proc Natl Acad Sci USA. 2016;113:E6831-E6839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 171] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 24. | Fantuzzo JA, Robles DA, Mirabella VR, Hart RP, Pang ZP, Zahn JD. Development of a high-throughput arrayed neural circuitry platform using human induced neurons for drug screening applications. Lab Chip. 2020;20:1140-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Zhao C, Tan A, Pastorin G, Ho HK. Nanomaterial scaffolds for stem cell proliferation and differentiation in tissue engineering. Biotechnol Adv. 2013;31:654-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 26. | Algahtani MS, Scurr DJ, Hook AL, Anderson DG, Langer RS, Burley JC, Alexander MR, Davies MC. High throughput screening for biomaterials discovery. J Control Release. 2014;190:115-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Voskuhl J, Brinkmann J, Jonkheijm P. Advances in contact printing technologies of carbohydrate, peptide and protein arrays. Curr Opin Chem Biol. 2014;18:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Ko IK, Kato K, Iwata H. Parallel analysis of multiple surface markers expressed on rat neural stem cells using antibody microarrays. Biomaterials. 2005;26:4882-4891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 38] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Roth EA, Xu T, Das M, Gregory C, Hickman JJ, Boland T. Inkjet printing for high-throughput cell patterning. Biomaterials. 2004;25:3707-3715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 555] [Cited by in RCA: 391] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 30. | Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363-2376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1224] [Cited by in RCA: 847] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 31. | Kim S, Marelli B, Brenckle MA, Mitropoulos AN, Gil ES, Tsioris K, Tao H, Kaplan DL, Omenetto FG. All-water-based electron-beam lithography using silk as a resist. Nat Nanotechnol. 2014;9:306-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 164] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 32. | Pal RK, Yadavalli VK. Silk protein nanowires patterned using electron beam lithography. Nanotechnology. 2018;29:335301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Bat E, Lee J, Lau UY, Maynard HD. Trehalose glycopolymer resists allow direct writing of protein patterns by electron-beam lithography. Nat Commun. 2015;6:6654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 34. | Ginger DS, Zhang H, Mirkin CA. The evolution of dip-pen nanolithography. Angew Chem Int Ed Engl. 2004;43:30-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 806] [Cited by in RCA: 516] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 35. | Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702-1705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 952] [Cited by in RCA: 656] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 36. | Moe AA, Suryana M, Marcy G, Lim SK, Ankam S, Goh JZ, Jin J, Teo BK, Law JB, Low HY, Goh EL, Sheetz MP, Yim EK. Microarray with micro- and nano-topographies enables identification of the optimal topography for directing the differentiation of primary murine neural progenitor cells. Small. 2012;8:3050-3061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 37. | Ankam S, Suryana M, Chan LY, Moe AA, Teo BK, Law JB, Sheetz MP, Low HY, Yim EK. Substrate topography and size determine the fate of human embryonic stem cells to neuronal or glial lineage. Acta Biomater. 2013;9:4535-4545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 38. | Odde DJ, Renn MJ. Laser-guided direct writing for applications in biotechnology. Trends Biotechnol. 1999;17:385-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 138] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Nahmias Y, Odde DJ. Micropatterning of living cells by laser-guided direct writing: application to fabrication of hepatic-endothelial sinusoid-like structures. Nat Protoc. 2006;1:2288-2296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 79] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 40. | Zhang T, Ke W, Zhou X, Qian Y, Feng S, Wang R, Cui G, Tao R, Guo W, Duan Y, Zhang X, Cao X, Shu Y, Yue C, Jing N. Human Neural Stem Cells Reinforce Hippocampal Synaptic Network and Rescue Cognitive Deficits in a Mouse Model of Alzheimer's Disease. Stem Cell Reports. 2019;13:1022-1037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 41. | Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, Harrison NL, Studer L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12543-12548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 746] [Cited by in RCA: 713] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 42. | Liu Y, Liu H, Sauvey C, Yao L, Zarnowska ED, Zhang SC. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc. 2013;8:1670-1679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 221] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 43. | Borkowska P, Fila-Danilow A, Paul-Samojedny M, Kowalczyk M, Hart J, Ryszawy J, Kowalski J. Differentiation of adult rat mesenchymal stem cells to GABAergic, dopaminergic and cholinergic neurons. Pharmacol Rep. 2015;67:179-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 44. | Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 966] [Cited by in RCA: 918] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 45. | Cho MS, Lee YE, Kim JY, Chung S, Cho YH, Kim DS, Kang SM, Lee H, Kim MH, Kim JH, Leem JW, Oh SK, Choi YM, Hwang DY, Chang JW, Kim DW. Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:3392-3397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 202] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 46. | Kim JH, Auerbach JM, Rodríguez-Gómez JA, Velasco I, Gavin D, Lumelsky N, Lee SH, Nguyen J, Sánchez-Pernaute R, Bankiewicz K, McKay R. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature. 2002;418:50-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1216] [Cited by in RCA: 1096] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 47. | Sun AX, Yuan Q, Tan S, Xiao Y, Wang D, Khoo AT, Sani L, Tran HD, Kim P, Chiew YS, Lee KJ, Yen YC, Ng HH, Lim B, Je HS. Direct Induction and Functional Maturation of Forebrain GABAergic Neurons from Human Pluripotent Stem Cells. Cell Rep. 2016;16:1942-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 48. | Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Südhof TC. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1337] [Cited by in RCA: 1124] [Article Influence: 93.7] [Reference Citation Analysis (0)] |
| 49. | Yang N, Chanda S, Marro S, Ng YH, Janas JA, Haag D, Ang CE, Tang Y, Flores Q, Mall M, Wapinski O, Li M, Ahlenius H, Rubenstein JL, Chang HY, Buylla AA, Südhof TC, Wernig M. Generation of pure GABAergic neurons by transcription factor programming. Nat Methods. 2017;14:621-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 244] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 50. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18186] [Article Influence: 957.2] [Reference Citation Analysis (0)] |
| 51. | Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035-1041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2235] [Cited by in RCA: 2274] [Article Influence: 151.6] [Reference Citation Analysis (0)] |
| 52. | Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 854] [Cited by in RCA: 777] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 53. | Corti S, Nizzardo M, Simone C, Falcone M, Donadoni C, Salani S, Rizzo F, Nardini M, Riboldi G, Magri F, Zanetta C, Faravelli I, Bresolin N, Comi GP. Direct reprogramming of human astrocytes into neural stem cells and neurons. Exp Cell Res. 2012;318:1528-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 54. | Heinrich C, Blum R, Gascón S, Masserdotti G, Tripathi P, Sánchez R, Tiedt S, Schroeder T, Götz M, Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 337] [Cited by in RCA: 385] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 55. | Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Südhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 279] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 56. | Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, Widmer HR, Eggan K, Goldstein PA, Anderson SA, Studer L. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 453] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 57. | Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, Lindvall O, Parmar M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1:703-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 506] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 58. | Hu W, Qiu B, Guan W, Wang Q, Wang M, Li W, Gao L, Shen L, Huang Y, Xie G, Zhao H, Jin Y, Tang B, Yu Y, Zhao J, Pei G. Direct Conversion of Normal and Alzheimer's Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell. 2015;17:204-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 372] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 59. | Li X, Zuo X, Jing J, Ma Y, Wang J, Liu D, Zhu J, Du X, Xiong L, Du Y, Xu J, Xiao X, Chai Z, Zhao Y, Deng H. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell. 2015;17:195-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 330] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 60. | Gao L, Guan W, Wang M, Wang H, Yu J, Liu Q, Qiu B, Yu Y, Ping Y, Bian X, Shen L, Pei G. Direct Generation of Human Neuronal Cells from Adult Astrocytes by Small Molecules. Stem Cell Reports. 2017;8:538-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 61. | Zhang L, Yin JC, Yeh H, Ma NX, Lee G, Chen XA, Wang Y, Lin L, Chen L, Jin P, Wu GY, Chen G. Small Molecules Efficiently Reprogram Human Astroglial Cells into Functional Neurons. Cell Stem Cell. 2015;17:735-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 238] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 62. | Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1902] [Cited by in RCA: 1961] [Article Influence: 108.9] [Reference Citation Analysis (4)] |
| 63. | O'Connor SM, Stenger DA, Shaffer KM, Maric D, Barker JL, Ma W. Primary neural precursor cell expansion, differentiation and cytosolic Ca(2+) response in three-dimensional collagen gel. J Neurosci Methods. 2000;102:187-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 66] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 64. | Ma W, Fitzgerald W, Liu QY, O'Shaughnessy TJ, Maric D, Lin HJ, Alkon DL, Barker JL. CNS stem and progenitor cell differentiation into functional neuronal circuits in three-dimensional collagen gels. Exp Neurol. 2004;190:276-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 164] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 65. | Distler T, Lauria I, Detsch R, Sauter CM, Bendt F, Kapr J, Rütten S, Boccaccini AR, Fritsche E. Neuronal Differentiation from Induced Pluripotent Stem Cell-Derived Neurospheres by the Application of Oxidized Alginate-Gelatin-Laminin Hydrogels. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 66. | Krencik R, Seo K, van Asperen JV, Basu N, Cvetkovic C, Barlas S, Chen R, Ludwig C, Wang C, Ward ME, Gan L, Horner PJ, Rowitch DH, Ullian EM. Systematic Three-Dimensional Coculture Rapidly Recapitulates Interactions between Human Neurons and Astrocytes. Stem Cell Reports. 2017;9:1745-1753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 67. | Montanez-Sauri SI, Beebe DJ, Sung KE. Microscale screening systems for 3D cellular microenvironments: platforms, advances, and challenges. Cell Mol Life Sci. 2015;72:237-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 68. | Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, Johansen LE, van Dijk RE, Scheefhals N, Berdenis van Berlekom A, Ribes Martínez E, Kling S, MacGillavry HD, van den Berg LH, Kahn RS, Hol EM, de Witte LD, Pasterkamp RJ. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9:4167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 422] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 69. | Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373-379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3083] [Cited by in RCA: 3521] [Article Influence: 293.4] [Reference Citation Analysis (0)] |
| 70. | Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329-2340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 1127] [Article Influence: 102.5] [Reference Citation Analysis (0)] |
| 71. | Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 549] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 72. | Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, Studer L. Specification of positional identity in forebrain organoids. Nat Biotechnol. 2019;37:436-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 230] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 73. | Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Göke J, Tan ZY, Saw TY, Tan CP, Lokman H, Lee Y, Kim D, Ko HS, Kim SO, Park JH, Cho NJ, Hyde TM, Kleinman JE, Shin JH, Weinberger DR, Tan EK, Je HS, Ng HH. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016;19:248-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 588] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 74. | Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, Jarazo J, Walter J, Brüggemann I, Boussaad I, Berger E, Fleming RMT, Bolognin S, Schwamborn JC. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports. 2017;8:1144-1154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 301] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 75. | Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, Lee SH, Weissman SM, Park IH. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell. 2019;24:487-497.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 326] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 76. | Schukking M, Miranda HC, Trujillo CA, Negraes PD, Muotri AR. Direct Generation of Human Cortical Organoids from Primary Cells. Stem Cells Dev. 2018;27:1549-1556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 77. | Tao Y, Zhang SC. Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell. 2016;19:573-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 210] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 78. | Roussa E, Krieglstein K. Induction and specification of midbrain dopaminergic cells: focus on SHH, FGF8, and TGF-beta. Cell Tissue Res. 2004;318:23-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 79. | Brassard JA, Lutolf MP. Engineering Stem Cell Self-organization to Build Better Organoids. Cell Stem Cell. 2019;24:860-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 227] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 80. | Vieira MS, Santos AK, Vasconcellos R, Goulart VAM, Parreira RC, Kihara AH, Ulrich H, Resende RR. Neural stem cell differentiation into mature neurons: Mechanisms of regulation and biotechnological applications. Biotechnol Adv. 2018;36:1946-1970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 81. | Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 665] [Cited by in RCA: 674] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 82. | El Wazan L, Urrutia-Cabrera D, Wong RC. Using transcription factors for direct reprogramming of neurons in vitro. World J Stem Cells. 2019;11:431-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (1)] |
| 83. | Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 992] [Article Influence: 70.9] [Reference Citation Analysis (0)] |
| 84. | Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Björklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108:10343-10348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 597] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 85. | Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 512] [Cited by in RCA: 519] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 86. | Colasante G, Lignani G, Rubio A, Medrihan L, Yekhlef L, Sessa A, Massimino L, Giannelli SG, Sacchetti S, Caiazzo M, Leo D, Alexopoulou D, Dell'Anno MT, Ciabatti E, Orlando M, Studer M, Dahl A, Gainetdinov RR, Taverna S, Benfenati F, Broccoli V. Rapid Conversion of Fibroblasts into Functional Forebrain GABAergic Interneurons by Direct Genetic Reprogramming. Cell Stem Cell. 2015;17:719-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 87. | Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 841] [Cited by in RCA: 784] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 88. | Sepp KJ, Hong P, Lizarraga SB, Liu JS, Mejia LA, Walsh CA, Perrimon N. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 2008;4:e1000111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 89. | Srivastava AK, Mohan S, Wergedal JE, Baylink DJ. A genomewide screening of N-ethyl-N-nitrosourea-mutagenized mice for musculoskeletal phenotypes. Bone. 2003;33:179-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 90. | Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci USA. 2003;100:11529-11534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 229] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 91. | Zohn IE, Anderson KV, Niswander L. Using genomewide mutagenesis screens to identify the genes required for neural tube closure in the mouse. Birth Defects Res A Clin Mol Teratol. 2005;73:583-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 92. | Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW, Lancini C, van Lohuizen M. In vivo RNAi screen for BMI1 targets identifies TGF-β/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell. 2013;23:660-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 93. | Yin H, Kassner M. In Vitro High-Throughput RNAi Screening to Accelerate the Process of Target Identification and Drug Development. Methods Mol Biol. 2016;1470:137-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 94. | Koizumi K, Higashida H, Yoo S, Islam MS, Ivanov AI, Guo V, Pozzi P, Yu SH, Rovescalli AC, Tang D, Nirenberg M. RNA interference screen to identify genes required for Drosophila embryonic nervous system development. Proc Natl Acad Sci USA. 2007;104:5626-5631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 95. | Güneş C, Paszkowski-Rogacz M, Rahmig S, Khattak S, Camgöz A, Wermke M, Dahl A, Bornhäuser M, Waskow C, Buchholz F. Comparative RNAi Screens in Isogenic Human Stem Cells Reveal SMARCA4 as a Differential Regulator. Stem Cell Reports. 2019;12:1084-1098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 96. | Sekine Y, Lin-Moore A, Chenette DM, Wang X, Jiang Z, Cafferty WB, Hammarlund M, Strittmatter SM. Functional Genome-wide Screen Identifies Pathways Restricting Central Nervous System Axonal Regeneration. Cell Rep. 2018;23:415-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 97. | Zhang J, Wang H, Sherbini O, Ling-Lin Pai E, Kang SU, Kwon JS, Yang J, He W, Eacker SM, Chi Z, Mao X, Xu J, Jiang H, Andrabi SA, Dawson TM, Dawson VL. High-Content Genome-Wide RNAi Screen Reveals CCR3 as a Key Mediator of Neuronal Cell Death. eNeuro. 2016;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 98. | Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 930] [Cited by in RCA: 878] [Article Influence: 87.8] [Reference Citation Analysis (0)] |
| 99. | Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 Library for functional genomics in human cells. Nature. 2014;509:487-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 547] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 100. | Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2015] [Cited by in RCA: 2109] [Article Influence: 175.8] [Reference Citation Analysis (0)] |
| 101. | Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3572] [Cited by in RCA: 3859] [Article Influence: 350.8] [Reference Citation Analysis (0)] |
| 102. | Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1328] [Cited by in RCA: 1189] [Article Influence: 118.9] [Reference Citation Analysis (0)] |
| 103. | Genga RMJ, Kernfeld EM, Parsi KM, Parsons TJ, Ziller MJ, Maehr R. Single-Cell RNA-Sequencing-Based CRISPRi Screening Resolves Molecular Drivers of Early Human Endoderm Development. Cell Rep. 2019;27:708-718.e10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 104. | Liu Y, Yu C, Daley TP, Wang F, Cao WS, Bhate S, Lin X, Still C 2nd, Liu H, Zhao D, Wang H, Xie XS, Ding S, Wong WH, Wernig M, Qi LS. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell. 2018;23:758-771.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 150] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 105. | Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, Yuan P, Brown M, Liu XS, Wei W. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 Library. Nat Biotechnol. 2016;34:1279-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 321] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 106. | Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 538] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 107. | Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, Victor J, Sauvageau M, Monteleone E, Rinn JL, Provero P, Church GM, Clohessy JG, Pandolfi PP. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell. 2018;173:649-664.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 237] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 108. | Panganiban RA, Park HR, Sun M, Shumyatcher M, Himes BE, Lu Q. Genome-wide CRISPR screen identifies suppressors of endoplasmic reticulum stress-induced apoptosis. Proc Natl Acad Sci USA. 2019;116:13384-13393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 109. | Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol. 2017;35:561-568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 287] [Cited by in RCA: 310] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 110. | Baumann V, Wiesbeck M, Breunig CT, Braun JM, Köferle A, Ninkovic J, Götz M, Stricker SH. Targeted removal of epigenetic barriers during transcriptional reprogramming. Nat Commun. 2019;10:2119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 111. | Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2012;13:713-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 332] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 112. | Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 938] [Cited by in RCA: 1024] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 113. | Callahan JF, Burgess JL, Fornwald JA, Gaster LM, Harling JD, Harrington FP, Heer J, Kwon C, Lehr R, Mathur A, Olson BA, Weinstock J, Laping NJ. Identification of novel inhibitors of the transforming growth factor beta1 (TGF-beta1) type 1 receptor (ALK5). J Med Chem. 2002;45:999-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 253] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 114. | Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1902] [Article Influence: 105.7] [Reference Citation Analysis (0)] |
| 115. | Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3332] [Cited by in RCA: 3081] [Article Influence: 171.2] [Reference Citation Analysis (0)] |
| 116. | Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 532] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 117. | Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 618] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 118. | Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1320] [Cited by in RCA: 1199] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 119. | Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2072] [Cited by in RCA: 1974] [Article Influence: 109.7] [Reference Citation Analysis (0)] |
| 120. | Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 640] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 121. | Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 994] [Cited by in RCA: 949] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 122. | Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, Bollong M, Kunick C, Brinker A, Cho CY, Schultz PG, Jaenisch R. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci USA. 2009;106:8912-8917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 282] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 123. | Tan F, Qian C, Tang K, Abd-Allah SM, Jing N. Inhibition of transforming growth factor β (TGF-β) signaling can substitute for Oct4 protein in reprogramming and maintain pluripotency. J Biol Chem. 2015;290:4500-4511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 124. | Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi SH, Studer L. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715-720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 413] [Cited by in RCA: 459] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 125. | Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547-551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1289] [Cited by in RCA: 1431] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 126. | Yang Y, Chen R, Wu X, Zhao Y, Fan Y, Xiao Z, Han J, Sun L, Wang X, Dai J. Rapid and Efficient Conversion of Human Fibroblasts into Functional Neurons by Small Molecules. Stem Cell Reports. 2019;13:862-876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 127. | Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3036] [Cited by in RCA: 2677] [Article Influence: 167.3] [Reference Citation Analysis (0)] |
| 128. | Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 878] [Cited by in RCA: 847] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 129. | Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18:4388-4392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 285] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 130. | Bengoa-Vergniory N, Kypta RM. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell Mol Life Sci. 2015;72:4157-4172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 131. | Qi Y, Zhang XJ, Renier N, Wu Z, Atkin T, Sun Z, Ozair MZ, Tchieu J, Zimmer B, Fattahi F, Ganat Y, Azevedo R, Zeltner N, Brivanlou AH, Karayiorgou M, Gogos J, Tomishima M, Tessier-Lavigne M, Shi SH, Studer L. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol. 2017;35:154-163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 132. | Du ZW, Chen H, Liu H, Lu J, Qian K, Huang CL, Zhong X, Fan F, Zhang SC. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun. 2015;6:6626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 284] [Cited by in RCA: 308] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 133. | Dworkin S, Mantamadiotis T. Targeting CREB signalling in neurogenesis. Expert Opin Ther Targets. 2010;14:869-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 134. | Maury Y, Côme J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, Peschanski M, Martinat C, Nedelec S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol. 2015;33:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 281] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 135. | Yu C, Liu K, Tang S, Ding S. Chemical approaches to cell reprogramming. Curr Opin Genet Dev. 2014;28:50-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 136. | Zhang M, Lin YH, Sun YJ, Zhu S, Zheng J, Liu K, Cao N, Li K, Huang Y, Ding S. Pharmacological Reprogramming of Fibroblasts into Neural Stem Cells by Signaling-Directed Transcriptional Activation. Cell Stem Cell. 2016;18:653-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 137. | Choi BH. Role of the basement membrane in neurogenesis and repair of injury in the central nervous system. Microsc Res Tech. 1994;28:193-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 138. | Sorg BA, Berretta S, Blacktop JM, Fawcett JW, Kitagawa H, Kwok JC, Miquel M. Casting a Wide Net: Role of Perineuronal Nets in Neural Plasticity. J Neurosci. 2016;36:11459-11468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 315] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 139. | Huang F, Shen Q, Zhao J. Growth and differentiation of neural stem cells in a three-dimensional collagen gel scaffold. Neural Regen Res. 2013;8:313-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 140. | Han J, Xiao Z, Chen L, Chen B, Li X, Han S, Zhao Y, Dai J. Maintenance of the self-renewal properties of neural progenitor cells cultured in three-dimensional collagen scaffolds by the REDD1-mTOR signal pathway. Biomaterials. 2013;34:1921-1928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 141. | Brännvall K, Bergman K, Wallenquist U, Svahn S, Bowden T, Hilborn J, Forsberg-Nilsson K. Enhanced neuronal differentiation in a three-dimensional collagen-hyaluronan matrix. J Neurosci Res. 2007;85:2138-2146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 118] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 142. | Kothapalli CR, Kamm RD. 3D matrix microenvironment for targeted differentiation of embryonic stem cells into neural and glial lineages. Biomaterials. 2013;34:5995-6007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 143. | Kim YH, Choi SH, D'Avanzo C, Hebisch M, Sliwinski C, Bylykbashi E, Washicosky KJ, Klee JB, Brüstle O, Tanzi RE, Kim DY. A 3D human neural cell culture system for modeling Alzheimer's disease. Nat Protoc. 2015;10:985-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 192] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 144. | Cheng TY, Chen MH, Chang WH, Huang MY, Wang TW. Neural stem cells encapsulated in a functionalized self-assembling peptide hydrogel for brain tissue engineering. Biomaterials. 2013;34:2005-2016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 277] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 145. | Sun Y, Li W, Wu X, Zhang N, Zhang Y, Ouyang S, Song X, Fang X, Seeram R, Xue W, He L, Wu W. Functional Self-Assembling Peptide Nanofiber Hydrogels Designed for Nerve Degeneration. ACS Appl Mater Interfaces. 2016;8:2348-2359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 163] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 146. | Freudenberg U, Hermann A, Welzel PB, Stirl K, Schwarz SC, Grimmer M, Zieris A, Panyanuwat W, Zschoche S, Meinhold D, Storch A, Werner C. A star-PEG-heparin hydrogel platform to aid cell replacement therapies for neurodegenerative diseases. Biomaterials. 2009;30:5049-5060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 215] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 147. | Naghdi P, Tiraihi T, Ganji F, Darabi S, Taheri T, Kazemi H. Survival, proliferation and differentiation enhancement of neural stem cells cultured in three-dimensional polyethylene glycol-RGD hydrogel with tenascin. J Tissue Eng Regen Med. 2016;10:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 148. | Mosley MC, Lim HJ, Chen J, Yang YH, Li S, Liu Y, Smith Callahan LA. Neurite extension and neuronal differentiation of human induced pluripotent stem cell derived neural stem cells on polyethylene glycol hydrogels containing a continuous Young's Modulus gradient. J Biomed Mater Res A. 2017;105:824-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 149. | Yang F, Murugan R, Wang S, Ramakrishna S. Electrospinning of nano/micro scale poly(L-lactic acid) aligned fibers and their potential in neural tissue engineering. Biomaterials. 2005;26:2603-2610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1408] [Cited by in RCA: 1092] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 150. | Barroca N, Marote A, Vieira SI, Almeida A, Fernandes MHV, Vilarinho PM, da Cruz E Silva OAB. Electrically polarized PLLA nanofibers as neural tissue engineering scaffolds with improved neuritogenesis. Colloids Surf B Biointerfaces. 2018;167:93-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 151. | Sperling LE, Reis KP, Pozzobon LG, Girardi CS, Pranke P. Influence of random and oriented electrospun fibrous poly(lactic-co-glycolic acid) scaffolds on neural differentiation of mouse embryonic stem cells. J Biomed Mater Res A. 2017;105:1333-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 152. | Olson HE, Rooney GE, Gross L, Nesbitt JJ, Galvin KE, Knight A, Chen B, Yaszemski MJ, Windebank AJ. Neural stem cell- and Schwann cell-loaded biodegradable polymer scaffolds support axonal regeneration in the transected spinal cord. Tissue Eng Part A. 2009;15:1797-1805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 153. | Lu Y, Li T, Zhao X, Li M, Cao Y, Yang H, Duan YY. Electrodeposited polypyrrole/carbon nanotubes composite films electrodes for neural interfaces. Biomaterials. 2010;31:5169-5181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 154. | Xu H, Holzwarth JM, Yan Y, Xu P, Zheng H, Yin Y, Li S, Ma PX. Conductive PPY/PDLLA conduit for peripheral nerve regeneration. Biomaterials. 2014;35:225-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 186] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 155. | Stewart E, Kobayashi NR, Higgins MJ, Quigley AF, Jamali S, Moulton SE, Kapsa RM, Wallace GG, Crook JM. Electrical stimulation using conductive polymer polypyrrole promotes differentiation of human neural stem cells: a biocompatible platform for translational neural tissue engineering. Tissue Eng Part C Methods. 2015;21:385-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 156. | Jan E, Kotov NA. Successful differentiation of mouse neural stem cells on layer-by-layer assembled single-walled carbon nanotube composite. Nano Lett. 2007;7:1123-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 181] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 157. | Serrano MC, Nardecchia S, García-Rama C, Ferrer ML, Collazos-Castro JE, del Monte F, Gutiérrez MC. Chondroitin sulphate-based 3D scaffolds containing MWCNTs for nervous tissue repair. Biomaterials. 2014;35:1543-1551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 158. | Dewitt DD, Kaszuba SN, Thompson DM, Stegemann JP. Collagen I-matrigel scaffolds for enhanced Schwann cell survival and control of three-dimensional cell morphology. Tissue Eng Part A. 2009;15:2785-2793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 159. | Zhang D, Lee J, Kilian KA. Synthetic Biomaterials to Rival Nature's Complexity-a Path Forward with Combinatorics, High-Throughput Discovery, and High-Content Analysis. Adv Healthc Mater. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 160. | Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 533] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 161. | Anderson DG, Putnam D, Lavik EB, Mahmood TA, Langer R. Biomaterial microarrays: rapid, microscale screening of polymer-cell interaction. Biomaterials. 2005;26:4892-4897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 170] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 162. | Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho SW, Mitalipova M, Pyzocha N, Rojas F, Van Vliet KJ, Davies MC, Alexander MR, Langer R, Jaenisch R, Anderson DG. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat Mater. 2010;9:768-778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 486] [Cited by in RCA: 412] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 163. | Nakajima M, Ishimuro T, Kato K, Ko IK, Hirata I, Arima Y, Iwata H. Combinatorial protein display for the cell-based screening of biomaterials that direct neural stem cell differentiation. Biomaterials. 2007;28:1048-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 122] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 164. | Ahmed M, Owens MJS, Toledo EM, Arenas E, Bradley M, Ffrench-Constant C. Combinatorial ECM Arrays Identify Cooperative Roles for Matricellular Proteins in Enhancing the Generation of TH+ Neurons From Human Pluripotent Cells. Front Cell Dev Biol. 2021;9:755406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 165. | Beachley VZ, Wolf MT, Sadtler K, Manda SS, Jacobs H, Blatchley MR, Bader JS, Pandey A, Pardoll D, Elisseeff JH. Tissue matrix arrays for high-throughput screening and systems analysis of cell function. Nat Methods. 2015;12:1197-1204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 134] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 166. | Lin E, Sikand A, Wickware J, Hao Y, Derda R. Peptide microarray patterning for controlling and monitoring cell growth. Acta Biomater. 2016;34:53-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 167. | Simitzi C, Ranella A, Stratakis E. Controlling the morphology and outgrowth of nerve and neuroglial cells: The effect of surface topography. Acta Biomater. 2017;51:21-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 168. | Simitzi C, Karali K, Ranella A, Stratakis E. Controlling the Outgrowth and Functions of Neural Stem Cells: The Effect of Surface Topography. Chemphyschem. 2018;19:1143-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 169. | Li W, Tang QY, Jadhav AD, Narang A, Qian WX, Shi P, Pang SW. Large-scale topographical screen for investigation of physical neural-guidance cues. Sci Rep. 2015;5:8644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 170. | Nichol RH 4th, Catlett TS, Onesto MM, Hollender D, Gómez TM. Environmental Elasticity Regulates Cell-type Specific RHOA Signaling and Neuritogenesis of Human Neurons. Stem Cell Reports. 2019;13:1006-1021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 171. | Kumachev A, Greener J, Tumarkin E, Eiser E, Zandstra PW, Kumacheva E. High-throughput generation of hydrogel microbeads with varying elasticity for cell encapsulation. Biomaterials. 2011;32:1477-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 172. | Kourouklis AP, Kaylan KB, Underhill GH. Substrate stiffness and matrix composition coordinately control the differentiation of liver progenitor cells. Biomaterials. 2016;99:82-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 173. | Wong RW, Guillaud L. The role of epidermal growth factor and its receptors in mammalian CNS. Cytokine Growth Factor Rev. 2004;15:147-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 182] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 174. | Chen BY, Wang X, Wang ZY, Wang YZ, Chen LW, Luo ZJ. Brain-derived neurotrophic factor stimulates proliferation and differentiation of neural stem cells, possibly by triggering the Wnt/β-catenin signaling pathway. J Neurosci Res. 2013;91:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 175. | Battista D, Ferrari CC, Gage FH, Pitossi FJ. Neurogenic niche modulation by activated microglia: transforming growth factor beta increases neurogenesis in the adult dentate gyrus. Eur J Neurosci. 2006;23:83-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 234] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 176. | Konagaya S, Kato K, Nakaji-Hirabayashi T, Arima Y, Iwata H. Array-based functional screening of growth factors toward optimizing neural stem cell microenvironments. Biomaterials. 2011;32:5015-5022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 177. | Muckom R, McFarland S, Yang C, Perea B, Gentes M, Murugappan A, Tran E, Dordick JS, Clark DS, Schaffer DV. High-throughput combinatorial screening reveals interactions between signaling molecules that regulate adult neural stem cell fate. Biotechnol Bioeng. 2019;116:193-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 178. | Lin CH, Lee JK, LaBarge MA. Fabrication and use of microenvironment microarrays (MEArrays). J Vis Exp. 2012;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 179. | Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 161] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 180. | Brafman DA, Chien S, Willert K. Arrayed cellular microenvironments for identifying culture and differentiation conditions for stem, primary and rare cell populations. Nat Protoc. 2012;7:703-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 181. | Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Artificial niche microarrays for probing single stem cell fate in high throughput. Nat Methods. 2011;8:949-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 305] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 182. | Fernandes TG, Kwon SJ, Bale SS, Lee MY, Diogo MM, Clark DS, Cabral JM, Dordick JS. Three-dimensional cell culture microarray for high-throughput studies of stem cell fate. Biotechnol Bioeng. 2010;106:106-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 183. | Nierode GJ, Gopal S, Kwon P, Clark DS, Schaffer DV, Dordick JS. High-throughput identification of factors promoting neuronal differentiation of human neural progenitor cells in microscale 3D cell culture. Biotechnol Bioeng. 2019;116:168-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 184. | Nierode GJ, Perea BC, McFarland SK, Pascoal JF, Clark DS, Schaffer DV, Dordick JS. High-Throughput Toxicity and Phenotypic Screening of 3D Human Neural Progenitor Cell Cultures on a Microarray Chip Platform. Stem Cell Reports. 2016;7:970-982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 185. | Joshi P, Yu KN, Kang SY, Kwon SJ, Kwon PS, Dordick JS, Kothapalli CR, Lee MY. 3D-cultured neural stem cell microarrays on a micropillar chip for high-throughput developmental neurotoxicology. Exp Cell Res. 2018;370:680-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 186. | Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. 3D niche microarrays for systems-level analyses of cell fate. Nat Commun. 2014;5:4324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 171] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 187. | Sivandzade F, Cucullo L. Regenerative Stem Cell Therapy for Neurodegenerative Diseases: An Overview. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 108] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 188. | De Gioia R, Biella F, Citterio G, Rizzo F, Abati E, Nizzardo M, Bresolin N, Comi GP, Corti S. Neural Stem Cell Transplantation for Neurodegenerative Diseases. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 159] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 189. | Younsi A, Zheng G, Riemann L, Scherer M, Zhang H, Tail M, Hatami M, Skutella T, Unterberg A, Zweckberger K. Long-Term Effects of Neural Precursor Cell Transplantation on Secondary Injury Processes and Functional Recovery after Severe Cervical Contusion-Compression Spinal Cord Injury. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 190. | Baklaushev VP, Durov OV, Kalsin VA, Gulaev EV, Kim SV, Gubskiy IL, Revkova VA, Samoilova EM, Melnikov PA, Karal-Ogly DD, Orlov SV, Troitskiy AV, Chekhonin VP, Averyanov AV, Ahlfors JE. Disease modifying treatment of spinal cord injury with directly reprogrammed neural precursor cells in non-human primates. World J Stem Cells. 2021;13:452-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 191. | Armijo E, Edwards G, Flores A, Vera J, Shahnawaz M, Moda F, Gonzalez C, Sanhueza M, Soto C. Induced Pluripotent Stem Cell-Derived Neural Precursors Improve Memory, Synaptic and Pathological Abnormalities in a Mouse Model of Alzheimer's Disease. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 192. | Zhang HA, Yuan CX, Liu KF, Yang QF, Zhao J, Li H, Yang QH, Song D, Quan ZZ, Qing H. Neural stem cell transplantation alleviates functional cognitive deficits in a mouse model of tauopathy. Neural Regen Res. 2022;17:152-162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 193. | Wianny F, Dzahini K, Fifel K, Wilson CRE, Bernat A, Dolmazon V, Misery P, Lamy C, Giroud P, Cooper HM, Knoblauch K, Procyk E, Kennedy H, Savatier P, Dehay C, Vezoli J. Induced Cognitive Impairments Reversed by Grafts of Neural Precursors: A Longitudinal Study in a Macaque Model of Parkinson's Disease. Adv Sci (Weinh). 2022;9:e2103827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 194. | Pereira MCL, Boese AC, Murad R, Yin J, Hamblin MH, Lee JP. Reduced dopaminergic neuron degeneration and global transcriptional changes in Parkinson's disease mouse brains engrafted with human neural stems during the early disease stage. Exp Neurol. 2022;352:114042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 195. | Nelke A, García-López S, Martínez-Serrano A, Pereira MP. Multifactoriality of Parkinson's Disease as Explored Through Human Neural Stem Cells and Their Transplantation in Middle-Aged Parkinsonian Mice. Front Pharmacol. 2021;12:773925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 196. | Park HJ, Jeon J, Choi J, Kim JY, Kim HS, Huh JY, Goldman SA, Song J. Human iPSC-derived neural precursor cells differentiate into multiple cell types to delay disease progression following transplantation into YAC128 Huntington's disease mouse model. Cell Prolif. 2021;54:e13082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 197. | Yoon Y, Kim HS, Jeon I, Noh JE, Park HJ, Lee S, Park IH, Stevanato L, Hicks C, Corteling R, Barker RA, Sinden JD, Song J. Implantation of the clinical-grade human neural stem cell line, CTX0E03, rescues the behavioral and pathological deficits in the quinolinic acid-lesioned rodent model of Huntington's disease. Stem Cells. 2020;38:936-947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 198. | Forostyak S, Forostyak O, Kwok JCF, Romanyuk N, Rehorova M, Kriska J, Dayanithi G, Raha-Chowdhury R, Jendelova P, Anderova M, Fawcett JW, Sykova E. Transplantation of Neural Precursors Derived from Induced Pluripotent Cells Preserve Perineuronal Nets and Stimulate Neural Plasticity in ALS Rats. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 199. | Kawai M, Imaizumi K, Ishikawa M, Shibata S, Shinozaki M, Shibata T, Hashimoto S, Kitagawa T, Ago K, Kajikawa K, Shibata R, Kamata Y, Ushiba J, Koga K, Furue H, Matsumoto M, Nakamura M, Nagoshi N, Okano H. Long-term selective stimulation of transplanted neural stem/progenitor cells for spinal cord injury improves locomotor function. Cell Rep. 2021;37:110019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 200. | Rodriguez-Pallares J, Garcia-Garrote M, Parga JA, Labandeira-Garcia JL. Dose-dependent effect of mesenchymal stromal cells co-grafted with dopaminergic neurons in a Parkinson's disease rat model. J Cell Mol Med. 2021;25:9884-9889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 201. | Yu Y, Gu S, Huang H, Wen T. Combination of bFGF, heparin and laminin induce the generation of dopaminergic neurons from rat neural stem cells both in vitro and in vivo. J Neurol Sci. 2007;255:81-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 202. | Matsuse D, Kitada M, Ogura F, Wakao S, Kohama M, Kira J, Tabata Y, Dezawa M. Combined transplantation of bone marrow stromal cell-derived neural progenitor cells with a collagen sponge and basic fibroblast growth factor releasing microspheres enhances recovery after cerebral ischemia in rats. Tissue Eng Part A. 2011;17:1993-2004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 203. | Redmond DE Jr, McEntire CR, Kingsbery JP, Leranth C, Elsworth JD, Bjugstad KB, Roth RH, Samulski RJ, Sladek JR Jr. Comparison of fetal mesencephalic grafts, AAV-delivered GDNF, and both combined in an MPTP-induced nonhuman primate Parkinson's model. Mol Ther. 2013;21:2160-2168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 204. | Zhong SJ, Gong YH, Lin YC. Combined intranasal nerve growth factor and ventricle neural stem cell grafts prolong survival and improve disease outcome in amyotrophic lateral sclerosis transgenic mice. Neurosci Lett. 2017;656:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 205. | Moriarty N, Gantner CW, Hunt CPJ, Ermine CM, Frausin S, Viventi S, Ovchinnikov DA, Kirik D, Parish CL, Thompson LH. A combined cell and gene therapy approach for homotopic reconstruction of midbrain dopamine pathways using human pluripotent stem cells. Cell Stem Cell. 2022;29:434-448.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 206. | Wakeman DR, Redmond DE Jr, Dodiya HB, Sladek JR Jr, Leranth C, Teng YD, Samulski RJ, Snyder EY. Human neural stem cells survive long term in the midbrain of dopamine-depleted monkeys after GDNF overexpression and project neurites toward an appropriate target. Stem Cells Transl Med. 2014;3:692-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 207. | Sharma R, McMillan CR, Niles LP. Neural stem cell transplantation and melatonin treatment in a 6-hydroxydopamine model of Parkinson's disease. J Pineal Res. 2007;43:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 208. | Yan J, Xu L, Welsh AM, Chen D, Hazel T, Johe K, Koliatsos VE. Combined immunosuppressive agents or CD4 antibodies prolong survival of human neural stem cell grafts and improve disease outcomes in amyotrophic lateral sclerosis transgenic mice. Stem Cells. 2006;24:1976-1985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 97] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 209. | Ma D, Zhao L, Zhang L, Li Y, Li L. Icariin Promotes Survival, Proliferation, and Differentiation of Neural Stem Cells In Vitro and in a Rat Model of Alzheimer's Disease. Stem Cells Int. 2021;2021:9974625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 210. | Rodriguez-Pallares J, Rodriguez-Perez AI, Muñoz A, Parga JA, Toledo-Aral JJ, Labandeira-Garcia JL. Effects of Rho Kinase Inhibitors on Grafts of Dopaminergic Cell Precursors in a Rat Model of Parkinson's Disease. Stem Cells Transl Med. 2016;5:804-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 211. | Alastrue-Agudo A, Rodriguez-Jimenez FJ, Mocholi EL, De Giorgio F, Erceg S, Moreno-Manzano V. FM19G11 and Ependymal Progenitor/Stem Cell Combinatory Treatment Enhances Neuronal Preservation and Oligodendrogenesis after Severe Spinal Cord Injury. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 212. | McGinley LM, Sims E, Lunn JS, Kashlan ON, Chen KS, Bruno ES, Pacut CM, Hazel T, Johe K, Sakowski SA, Feldman EL. Human Cortical Neural Stem Cells Expressing Insulin-Like Growth Factor-I: A Novel Cellular Therapy for Alzheimer's Disease. Stem Cells Transl Med. 2016;5:379-391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 213. | Khazaei M, Ahuja CS, Nakashima H, Nagoshi N, Li L, Wang J, Chio J, Badner A, Seligman D, Ichise A, Shibata S, Fehlings MG. GDNF rescues the fate of neural progenitor grafts by attenuating Notch signals in the injured spinal cord in rodents. Sci Transl Med. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 214. | Wu CC, Lien CC, Hou WH, Chiang PM, Tsai KJ. Gain of BDNF Function in Engrafted Neural Stem Cells Promotes the Therapeutic Potential for Alzheimer's Disease. Sci Rep. 2016;6:27358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 215. | Ma H, Yu B, Kong L, Zhang Y, Shi Y. Neural stem cells over-expressing brain-derived neurotrophic factor (BDNF) stimulate synaptic protein expression and promote functional recovery following transplantation in rat model of traumatic brain injury. Neurochem Res. 2012;37:69-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 216. | Chang DJ, Cho HY, Hwang S, Lee N, Choi C, Lee H, Hong KS, Oh SH, Kim HS, Shin DA, Yoon YW, Song J. Therapeutic Effect of BDNF-Overexpressing Human Neural Stem Cells (F3.BDNF) in a Contusion Model of Spinal Cord Injury in Rats. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 217. | Thomsen GM, Avalos P, Ma AA, Alkaslasi M, Cho N, Wyss L, Vit JP, Godoy M, Suezaki P, Shelest O, Bankiewicz KS, Svendsen CN. Transplantation of Neural Progenitor Cells Expressing Glial Cell Line-Derived Neurotrophic Factor into the Motor Cortex as a Strategy to Treat Amyotrophic Lateral Sclerosis. Stem Cells. 2018;36:1122-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 218. | Akhtar AA, Gowing G, Kobritz N, Savinoff SE, Garcia L, Saxon D, Cho N, Kim G, Tom CM, Park H, Lawless G, Shelley BC, Mattis VB, Breunig JJ, Svendsen CN. Inducible Expression of GDNF in Transplanted iPSC-Derived Neural Progenitor Cells. Stem Cell Reports. 2018;10:1696-1704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 219. | Li X, Peng Z, Long L, Tuo Y, Wang L, Zhao X, Le W, Wan Y. Wnt4-modified NSC transplantation promotes functional recovery after spinal cord injury. FASEB J. 2020;34:82-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 220. | Deng M, Xie P, Chen Z, Zhou Y, Liu J, Ming J, Yang J. Mash-1 modified neural stem cells transplantation promotes neural stem cells differentiation into neurons to further improve locomotor functional recovery in spinal cord injury rats. Gene. 2021;781:145528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 221. | Madrazo I, Kopyov O, Ávila-Rodríguez MA, Ostrosky F, Carrasco H, Kopyov A, Avendaño-Estrada A, Jiménez F, Magallón E, Zamorano C, González G, Valenzuela T, Carrillo R, Palma F, Rivera R, Franco-Bourland RE, Guízar-Sahagún G. Transplantation of Human Neural Progenitor Cells (NPC) into Putamina of Parkinsonian Patients: A Case Series Study, Safety and Efficacy Four Years after Surgery. Cell Transplant. 2019;28:269-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 222. | Mazzini L, Gelati M, Profico DC, Sorarù G, Ferrari D, Copetti M, Muzi G, Ricciolini C, Carletti S, Giorgi C, Spera C, Frondizi D, Masiero S, Stecco A, Cisari C, Bersano E, De Marchi F, Sarnelli MF, Querin G, Cantello R, Petruzzelli F, Maglione A, Zalfa C, Binda E, Visioli A, Trombetta D, Torres B, Bernardini L, Gaiani A, Massara M, Paolucci S, Boulis NM, Vescovi AL; ALS-NSCs Trial Study Group. Results from Phase I Clinical Trial with Intraspinal Injection of Neural Stem Cells in Amyotrophic Lateral Sclerosis: A Long-Term Outcome. Stem Cells Transl Med. 2019;8:887-897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 223. | Hunsberger JG, Rao M, Kurtzberg J, Bulte JWM, Atala A, LaFerla FM, Greely HT, Sawa A, Gandy S, Schneider LS, Doraiswamy PM. Accelerating stem cell trials for Alzheimer's disease. Lancet Neurol. 2016;15:219-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 224. | Park HJ, Han A, Kim JY, Choi J, Bae HS, Cho GB, Shin H, Shin EJ, Lee KI, Kim S, Lee JY, Song J. SUPT4H1-edited stem cell therapy rescues neuronal dysfunction in a mouse model for Huntington's disease. NPJ Regen Med. 2022;7:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 225. | Kim SW, Woo HJ, Kim EH, Kim HS, Suh HN, Kim SH, Song JJ, Wulansari N, Kang M, Choi SY, Choi SJ, Jang WH, Lee J, Kim KH, Lee W, Yang J, Kyung J, Lee HS, Park SM, Chang MY, Lee SH. Neural stem cells derived from human midbrain organoids as a stable source for treating Parkinson's disease: Midbrain organoid-NSCs (Og-NSC) as a stable source for PD treatment. Prog Neurobiol. 2021;204:102086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 226. | Duan H, Li X, Wang C, Hao P, Song W, Li M, Zhao W, Gao Y, Yang Z. Functional hyaluronate collagen scaffolds induce NSCs differentiation into functional neurons in repairing the traumatic brain injury. Acta Biomater. 2016;45:182-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 227. | Shi W, Nie D, Jin G, Chen W, Xia L, Wu X, Su X, Xu X, Ni L, Zhang X, Chen J. BDNF blended chitosan scaffolds for human umbilical cord MSC transplants in traumatic brain injury therapy. Biomaterials. 2012;33:3119-3126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 228. | Skop NB, Calderon F, Cho CH, Gandhi CD, Levison SW. Optimizing a multifunctional microsphere scaffold to improve neural precursor cell transplantation for traumatic brain injury repair. J Tissue Eng Regen Med. 2016;10:E419-E432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 229. | Skop NB, Singh S, Antikainen H, Saqcena C, Calderon F, Rothbard DE, Cho CH, Gandhi CD, Levison SW, Dobrowolski R. Subacute Transplantation of Native and Genetically Engineered Neural Progenitors Seeded on Microsphere Scaffolds Promote Repair and Functional Recovery After Traumatic Brain Injury. ASN Neuro. 2019;11:1759091419830186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 230. | Jiang J, Dai C, Liu X, Dai L, Li R, Ma K, Xu H, Zhao F, Zhang Z, He T, Niu X, Chen X, Zhang S. Implantation of regenerative complexes in traumatic brain injury canine models enhances the reconstruction of neural networks and motor function recovery. Theranostics. 2021;11:768-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 231. | Zhang J, Wang RJ, Chen M, Liu XY, Ma K, Xu HY, Deng WS, Ye YC, Li WX, Chen XY, Sun HT. Collagen/heparan sulfate porous scaffolds loaded with neural stem cells improve neurological function in a rat model of traumatic brain injury. Neural Regen Res. 2021;16:1068-1077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 232. | Li X, Fan C, Xiao Z, Zhao Y, Zhang H, Sun J, Zhuang Y, Wu X, Shi J, Chen Y, Dai J. A collagen microchannel scaffold carrying paclitaxel-liposomes induces neuronal differentiation of neural stem cells through Wnt/β-catenin signaling for spinal cord injury repair. Biomaterials. 2018;183:114-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 233. | Li G, Zhang B, Sun JH, Shi LY, Huang MY, Huang LJ, Lin ZJ, Lin QY, Lai BQ, Ma YH, Jiang B, Ding Y, Zhang HB, Li MX, Zhu P, Wang YQ, Zeng X, Zeng YS. An NT-3-releasing bioscaffold supports the formation of TrkC-modified neural stem cell-derived neural network tissue with efficacy in repairing spinal cord injury. Bioact Mater. 2021;6:3766-3781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 234. | Koffler J, Zhu W, Qu X, Platoshyn O, Dulin JN, Brock J, Graham L, Lu P, Sakamoto J, Marsala M, Chen S, Tuszynski MH. Biomimetic 3D-printed scaffolds for spinal cord injury repair. Nat Med. 2019;25:263-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 420] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 235. | Liu X, Hao M, Chen Z, Zhang T, Huang J, Dai J, Zhang Z. 3D bioprinted neural tissue constructs for spinal cord injury repair. Biomaterials. 2021;272:120771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 147] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 236. | Xiao Z, Tang F, Zhao Y, Han G, Yin N, Li X, Chen B, Han S, Jiang X, Yun C, Zhao C, Cheng S, Zhang S, Dai J. Significant Improvement of Acute Complete Spinal Cord Injury Patients Diagnosed by a Combined Criteria Implanted with NeuroRegen Scaffolds and Mesenchymal Stem Cells. Cell Transplant. 2018;27:907-915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 237. | Chen W, Zhang Y, Yang S, Sun J, Qiu H, Hu X, Niu X, Xiao Z, Zhao Y, Zhou Y, Dai J, Chu T. NeuroRegen Scaffolds Combined with Autologous Bone Marrow Mononuclear Cells for the Repair of Acute Complete Spinal Cord Injury: A 3-Year Clinical Study. Cell Transplant. 2020;29:963689720950637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 238. | Kwak SS, Washicosky KJ, Brand E, von Maydell D, Aronson J, Kim S, Capen DE, Cetinbas M, Sadreyev R, Ning S, Bylykbashi E, Xia W, Wagner SL, Choi SH, Tanzi RE, Kim DY. Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer's disease. Nat Commun. 2020;11:1377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 239. | Park J, Wetzel I, Marriott I, Dréau D, D'Avanzo C, Kim DY, Tanzi RE, Cho H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer's disease. Nat Neurosci. 2018;21:941-951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 444] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 240. | Taylor-Whiteley TR, Le Maitre CL, Duce JA, Dalton CF, Smith DP. Recapitulating Parkinson's disease pathology in a three-dimensional human neural cell culture model. Dis Model Mech. 2019;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 241. | Kim H, Park HJ, Choi H, Chang Y, Park H, Shin J, Kim J, Lengner CJ, Lee YK. Modeling G2019S-LRRK2 Sporadic Parkinson's Disease in 3D Midbrain Organoids. Stem Cell Reports. 2019;12:518-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 244] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 242. | Chen X, Sun G, Tian E, Zhang M, Davtyan H, Beach TG, Reiman EM, Blurton-Jones M, Holtzman DM, Shi Y. Modeling Sporadic Alzheimer's Disease in Human Brain Organoids under Serum Exposure. Adv Sci (Weinh). 2021;8:e2101462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 243. | Haremaki T, Metzger JJ, Rito T, Ozair MZ, Etoc F, Brivanlou AH. Self-organizing neuruloids model developmental aspects of Huntington's disease in the ectodermal compartment. Nat Biotechnol. 2019;37:1198-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 244. | Pașca AM, Park JY, Shin HW, Qi Q, Revah O, Krasnoff R, O'Hara R, Willsey AJ, Palmer TD, Pașca SP. Human 3D cellular model of hypoxic brain injury of prematurity. Nat Med. 2019;25:784-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 245. | Sood D, Tang-Schomer M, Pouli D, Mizzoni C, Raia N, Tai A, Arkun K, Wu J, Black LD 3rd, Scheffler B, Georgakoudi I, Steindler DA, Kaplan DL. 3D extracellular matrix microenvironment in bioengineered tissue models of primary pediatric and adult brain tumors. Nat Commun. 2019;10:4529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 246. | Meinhardt A, Eberle D, Tazaki A, Ranga A, Niesche M, Wilsch-Bräuninger M, Stec A, Schackert G, Lutolf M, Tanaka EM. 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Reports. 2014;3:987-999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 156] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 247. | Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, Vaccarino FM. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770-12775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 388] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 248. | Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Pääbo S, Huttner WB, Treutlein B. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112:15672-15677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 763] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 249. | Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Rep. 2016;17:3369-3384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 263] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 250. | Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14:743-751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 538] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 251. | Simão D, Terrasso AP, Teixeira AP, Brito C, Sonnewald U, Alves PM. Functional metabolic interactions of human neuron-astrocyte 3D in vitro networks. Sci Rep. 2016;6:33285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 252. | Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1082] [Cited by in RCA: 998] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 253. | Liu D, Pavathuparambil Abdul Manaph N, Al-Hawwas M, Zhou XF, Liao H. Small Molecules for Neural Stem Cell Induction. Stem Cells Dev. 2018;27:297-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 254. | Weltner J, Balboa D, Katayama S, Bespalov M, Krjutškov K, Jouhilahti EM, Trokovic R, Kere J, Otonkoski T. Human pluripotent reprogramming with CRISPR activators. Nat Commun. 2018;9:2643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 255. | de Luzy IR, Law KCL, Moriarty N, Hunt CPJ, Durnall JC, Thompson LH, Nagy A, Parish CL. Human stem cells harboring a suicide gene improve the safety and standardisation of neural transplants in Parkinsonian rats. Nat Commun. 2021;12:3275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 256. | Chen S. Screening-Based Chemical Approaches to Unravel Stem Cell Biology. Stem Cell Reports. 2018;11:1312-1323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 257. | Jorfi M, D'Avanzo C, Tanzi RE, Kim DY, Irimia D. Human Neurospheroid Arrays for In Vitro Studies of Alzheimer's Disease. Sci Rep. 2018;8:2450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 258. | Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, Elliott DA, Porrello ER, Hudson JE. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci USA. 2017;114:E8372-E8381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 348] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 259. | Mills RJ, Parker BL, Quaife-Ryan GA, Voges HK, Needham EJ, Bornot A, Ding M, Andersson H, Polla M, Elliott DA, Drowley L, Clausen M, Plowright AT, Barrett IP, Wang QD, James DE, Porrello ER, Hudson JE. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell. 2019;24:895-907.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 205] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 260. | Czerniecki SM, Cruz NM, Harder JL, Menon R, Annis J, Otto EA, Gulieva RE, Islas LV, Kim YK, Tran LM, Martins TJ, Pippin JW, Fu H, Kretzler M, Shankland SJ, Himmelfarb J, Moon RT, Paragas N, Freedman BS. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell. 2018;22:929-940.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 331] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 261. | Renner H, Grabos M, Becker KJ, Kagermeier TE, Wu J, Otto M, Peischard S, Zeuschner D, TsyTsyura Y, Disse P, Klingauf J, Leidel SA, Seebohm G, Schöler HR, Bruder JM. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 262. | Shin H, Jeong S, Lee JH, Sun W, Choi N, Cho IJ. 3D high-density microelectrode array with optical stimulation and drug delivery for investigating neural circuit dynamics. Nat Commun. 2021;12:492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 263. | Lam D, Fischer NO, Enright HA. Probing function in 3D neuronal cultures: A survey of 3D multielectrode array advances. Curr Opin Pharmacol. 2021;60:255-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 264. | Renner H, Schöler HR, Bruder JM. Combining Automated Organoid Workflows with Artificial Intelligence-Based Analyses: Opportunities to Build a New Generation of Interdisciplinary High-Throughput Screens for Parkinson's Disease and Beyond. Mov Disord. 2021;36:2745-2762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 265. | Schudel BR, Harmon B, Abhyankar VV, Pruitt BW, Negrete OA, Singh AK. Microfluidic platforms for RNA interference screening of virus-host interactions. Lab Chip. 2013;13:811-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 266. | Wang G, Björk SM, Huang M, Liu Q, Campbell K, Nielsen J, Joensson HN, Petranovic D. RNAi expression tuning, microfluidic screening, and genome recombineering for improved protein production in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2019;116:9324-9332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 267. | Han X, Liu Z, Zhao L, Wang F, Yu Y, Yang J, Chen R, Qin L. Microfluidic Cell Deformability Assay for Rapid and Efficient Kinase Screening with the CRISPR-Cas9 System. Angew Chem Int Ed Engl. 2016;55:8561-8565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 268. | Titmarsh DM, Glass NR, Mills RJ, Hidalgo A, Wolvetang EJ, Porrello ER, Hudson JE, Cooper-White JJ. Induction of Human iPSC-Derived Cardiomyocyte Proliferation Revealed by Combinatorial Screening in High Density Microbioreactor Arrays. Sci Rep. 2016;6:24637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 269. | Schuster B, Junkin M, Kashaf SS, Romero-Calvo I, Kirby K, Matthews J, Weber CR, Rzhetsky A, White KP, Tay S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun. 2020;11:5271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 229] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 270. | Tumarkin E, Tzadu L, Csaszar E, Seo M, Zhang H, Lee A, Peerani R, Purpura K, Zandstra PW, Kumacheva E. High-throughput combinatorial cell co-culture using microfluidics. Integr Biol (Camb). 2011;3:653-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 271. | Wevers NR, van Vught R, Wilschut KJ, Nicolas A, Chiang C, Lanz HL, Trietsch SJ, Joore J, Vulto P. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci Rep. 2016;6:38856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 272. | Lee SR, Hyung S, Bang S, Lee Y, Ko J, Lee S, Kim HJ, Jeon NL. Modeling neural circuit, blood-brain barrier, and myelination on a microfluidic 96 well plate. Biofabrication. 2019;11:035013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 273. | Xu Z, Fang P, Xu B, Lu Y, Xiong J, Gao F, Wang X, Fan J, Shi P. High-throughput three-dimensional chemotactic assays reveal steepness-dependent complexity in neuronal sensation to molecular gradients. Nat Commun. 2018;9:4745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 274. | Rifes P, Isaksson M, Rathore GS, Aldrin-Kirk P, Møller OK, Barzaghi G, Lee J, Egerod KL, Rausch DM, Parmar M, Pers TH, Laurell T, Kirkeby A. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat Biotechnol. 2020;38:1265-1273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 275. | Mullard A. Microfluidics platform lowers barrier to drug combination screening. Nat Rev Drug Discov. 2018;17:691-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 276. | Azari H, Reynolds BA. In Vitro Models for Neurogenesis. Cold Spring Harb Perspect Biol. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |