Published online Jul 15, 2003. doi: 10.3748/wjg.v9.i7.1521
Revised: January 4, 2003
Accepted: February 18, 2003
Published online: July 15, 2003
AIM: To investigate whether the formation of aggregated HBx has a potential linking with its cellular responses.
METHODS: Recombinant HBx was expressed in Escherichia coli and purified by Ni-NTA metal-affinity chromatography. Anti-HBx monoclonal antibody was developed for immunocytochemical detection. Bicistronic expression vector harboring full-length DNA of HBx was employed for transfection of human HepG2 cells. Immunocytochemical staining was used to examine the intracellular HBx aggregates in cells. The effects of HBx aggregation on cell cycle and apoptosis were assessed by flow cytometry.
RESULTS: Immunocytochemical staining revealed most of the HBx was formed intracellular aggregate in cytoplasm and frequently accumulated in large granules. Flow cytometry analysis showed that HepG2 cells transfected with vector harboring HBx significantly increased apoptosis and largely accumulated in the G0-G1 phase by maintenance in serum medium for 36 h. Control cells without HBx aggregates in the presence of serum entered S phase and proliferated more rapidly at the same time. EGFP fluorescence in HBx expression cells was significantly decreased.
CONCLUSION: Our observations show that cells with HBx aggregate undergo growth arrest and apoptosis, whereas control cells without HBx remain in growth and progression into S phase. Our data may provide helpful information to understand the biological effects of HBx aggregates on cells.
- Citation: Song CZ, Bai ZL, Song CC, Wang QW. Aggregate formation of hepatitis B virus X protein affects cell cycle and apoptosis. World J Gastroenterol 2003; 9(7): 1521-1524
- URL: https://www.wjgnet.com/1007-9327/full/v9/i7/1521.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i7.1521
Hepatitis B virus (HBV) causes transient and chronic infections of the liver. Transient infections may produce serious illness, and approximately 0.5% terminates with fatal, fulminant hepatitis. Chronic infections may also have serious consequences: nearly 25% terminate in untreatable liver cancer. Worldwide deaths from liver cancer caused by HBV infection probably exceed one million per year. X gene is a unique fourth open reading frame of HBV. X gene codes for a 16.5-kDa protein (X protein, HBx) and is well conserved among the mammalian hepadnaviruses[1]. HBx is a multifunctional viral regulator that modulates transcription, cell responses to genotoxic stress, protein degradation, and signaling pathways[2]. The ability of HBx to modulate cell survival is potentially relevant to viral pathogenicity in acute and chronic HBV infection as well as to the late development of hepatocellular carcinoma[3]. HBx activates signal-transduction cascades such as the Ras/Raf mitogen-activated protein kinase, Src kinase, c-Jun NH2-terminal kinase and Janus family tyrosine kinases/signal transducer and activators of transcription[4,5]. HBx targets mitochondrial calcium and activates cytosolic calcium-dependent proline-rich tyrosine kinase-2[6,7]. HBx may directly interact with transcription factors[8]. HBx is also known to play an important role in alterating gene expression, sensitizing cells to apoptosis and affect cell cycle checkpoints[2]. The fate of infected cells expressing HBx is likely to be determined by the balance between apoptotic and anti-apoptotic signals of viral, cellular, and environmental origin.
HBx expression in different cells results in distinct and opposing cellular function responses of cell cycle and apoptosis[9,10]. Most of the investigations described the effects of HBx through cellular signal-transduction pathways. Some reports suggested that HBx could induce cell death when it was expressed at high levels[11-13]. The nine residues of cysteine among 154 amino acids of HBX might involve in disulfide bridge formation and be in favor of aggregate formation[14]. Protein aggregation leads to cell cycle arrest and initiates cell death[15]. We propose that intracellular deposition of aggregated HBx may have a potential linking to its cellular responses. In this study, we reported the cytoplasmic aggregates of HBx and its effect on cell cycle and apoptosis.
Restriction enzymes and T4 DNA ligase were obtained from TaKaRa Biotech (Japan). QIA express Kit including pQE-60 Vector, E. coli strain M15 [pREP4] and Ni-NTA Superflow was purchased from QIAGEN (USA). GeneJammer transfection reagent was from Stratagene (USA). The plasmid pSPX46, a gift of Dr. Curtis C. Harris (National Institutes of Health, USA), encodes full-length HBx of the adr subtype. The bicistronic expression vector pIRES-EGFP-HBx harboring X gene (subtype ayw) was kindly provided by Drs. Jingyu Diao and Christopher D. Richardson (University of Toronto, Canada). SABC immunocytochemical detection kit was from Wuhan Boster Biological Technology Co. (China). Other chemicals of analytical grade were from Sigma.
The coding DNA fragment was amplified by polymerase chain reaction (PCR) using the pSPX46 as a template and the 5'-PCR primer (5'-TTTCCATGGCTGCTCGGGTGTGC-3') carrying the Nco I site before and the 3'-PCR primer (5'-GCGAGATCTGGCAGAGGTGAAAAAGTTG-3') carrying the Bgl II site after the X reading frame. The PCR product was digested with Nco I and Bgl II and ligated into pQE-60. According to the cloning strategy, recombinant construct based on the pQE-60 vector was produced by placing the 6xHis tag at the carboxy-terminus of HBx with the protein beginning with its natural ATG start codon. pQE-60X was obtained as an expression system for biosynthesis of HBx.
Recombinant pQE-60X was transformed into E. coli strain M15 [pREP4]. The culture was induced with 1 mM isopropyl β-D-thiogalactopyranoside for 4.5 h at 37 °C. The bacteria were harvested and lysed in a buffer containing 6 mol/L guanidine hydrochloride. The purification procedure of QIAexpress Kit was optimized. Elution with 250 mmol/L imidazole could effectively separate HBx from nickel-nitrilotriacetic acid (Ni-NTA) resins. Recombinant HBx was analysed by sodium dodecyl sulfate -polyacrylamide gel electrophoresis (SDS-PAGE). Electrophoresis was performed on 15% polyacrylamide gel. After electrophoresis, gels were fixed in 30% ethanol, 10% acetic acid, and stained with Coomassie brilliant blue.
Balb/c mice were immunized repeatedly. Spleen cells from the most responding mouse were fused with myeloma cells (Sp-2/0) according to the routine method described by Yang et al[16]. Positive hybrids were selected on hypoxanthine, aminopterin and thymidine containing medium.
Human hepatoma cell line HepG2 was grown on coverslips or 60-mm dish and maintained in Dulbecco's modified Eagle minimal medium containing penicillin (100 IU/ml) and streptomycin (100 mg/ml) and supplemented with 10% fetal calf serum. Bicistronic expression vector pIRES-EGFP-HBx derived from pIRES-EGFP by adding the DNA fragments of HBx[17]. Cells at 70% confluence were respectively transfected with pIRES-EGFP-HBx and pIRES-EGFP using GeneJammer transfection reagent. DNA transfections were performed according to the protocols supplied with the reagents.
After transfection for 36 h, HepG2 cells were washed in phosphate-buffered saline (PBS) and fixed with 90% ethanol. SABC detection kit was used for immunocytochemical analysis. Cells were incubated with HBx monoclonal antibody for 1 hour at 37 °C. After washed three times with PBS, cells were incubated with anti-mouse biotinylated secondary antibody. Following extensive washes with PBS, color development was demonstrated with diaminobenzidine tetrahy-drochloride chromagen. Finally, the slides were counterstained with hematoxylin. Stained cells were examined using light microscopy and photographed.
To assess the effect of HBx aggregation on cell cycle, apoptosis and fluorescence of enhanced green fluorescent protein (EGFP), we analyzed the transfected cells by flow cytometry for GFP fluorescence and DNA content[18]. The efficiency of transfection was verified by fluorescent signal. Cells were released by trypsinization, resuspended in PBS at a density of 2 × 106 cells/ml, and analyzed on a Becton Dickinson flow cytometer using CellQuest software.
In order to develop an HBx: anti-HBx antibody system for immunocytochemical purposes, an efficient E. coli expression system to produce HBx in large quantity was established. HBx was expressed in E. coli cells harboring pQE-60X as HBx carboxy-terminally fused to six histidine residues. pQE-60X was confirmed by PCR, restriction enzymes analysis and DNA sequencing.
Recombinant HBx was expressed in Escherichia coli. Because of the high stability of HBx aggregates, the purification procedure was optimized. E. coli cells were lysed in a buffer containing 6 mol/L guanidine hydrochloride. Six consecutive histidine residues that placed at the carboxy-terminus of HBx facilitated Ni-NTA metal-affinity chromatography. Elution with 250 mmol/L imidazole could effectively separate HBx from Ni-NTA resins. HBx was eluted as a pure protein (Figure 1). Starting with 1L of culture, only one milligram of recombinant HBx was attained under our experimental conditions. An anti-HBx monoclonal antibody was established following the conventional approach. The anti-HBx monoclonal antibody recognized the recombinant antigen as proved by high optical density measured in a simple binding enzyme-linked immunosorbent assay. Monoclonal antibody secreted by hybridoma clone was alike optimal for immunocytochemistry.
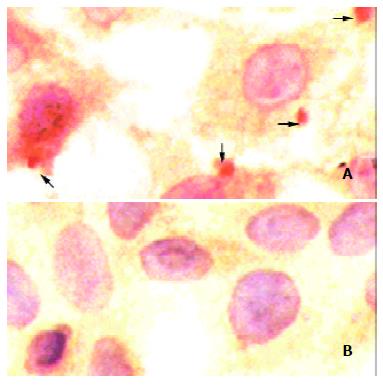
HBx aggregate and its distribution were verified by immunocytochemistry. HBx-bearing cells exhibited positively staining by anti-HBx monoclonal antibody. Cytoplasmic HBx characteristically accumulated in large granules or strongly stained aggregates (Figure 2A, arrows). No immunoreactivity could be detected in a negative control stained by the same antibody under the same conditions (Figure 2B).
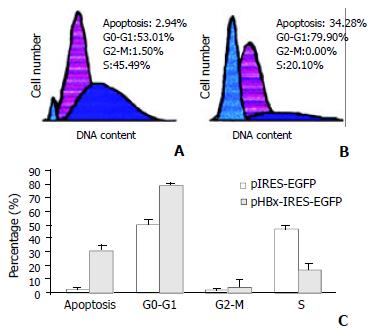
A transient expression plasmid, pHBx-IRES-EGFP was used in this experiment. The vector contained HBx and EGFP reporter genes under control of a cytomegalovirus promoter and the element of internal ribosome entry site (IRES), respectively[17]. More than 80% of human HepG2 cells could be transfected with pIRES-EGFP-HBx and pIRES-EGFP respectively as shown by fluorescence flow cytometry. A representative flow cytometric profile at the 36th is shown in Figure 3A and 3B. Results were expressed as percentage of HepG2 cells in G0-G1, G2-M and S phases of the cell cycle (Figure 3C). Apoptosis was also monitored by flow cytometry (Figure 3C). Each value corresponded to the mean ± standard deviation of two independent experiments. Using Student's t-test, statistical analysis was performed. HepG2 cells transfected with vector harboring HBx were accumulated largely in the G0-G1 phase after maintenance in serum medium for 36 h. Control cells that could not synthesize HBx protein in the presence of serum proliferated more rapidly and entered S phase at the same time.
The mean fluorescence of 104 EGFP positive cells was measured by fluorescence cytometer. The EGFP fluorescence value was 4.43 ± 0.58 and 3.45 ± 0.15 of experiments in triplicate for HepG2 cells transfected with pIRES-EGFP and pIRES-EGFP-HBx, respectively. EGFP fluorescence in HBx expression cells was significantly decreased as compared with control pIRES-EGFP (P < 0.05).
The X open reading frame of HBV is highly conserved among all mammalian hepadnaviruses. HBx might regulate the expression of certain viral and cellular genes which are important for the creation of an environment suitable for viral propagation. HBx inhibits clonal outgrowth of cells and induces apoptosis by a p53-independent pathway. HBx expression can induce a late G1 cell cycle block prior to their counterselection by apoptosis. Furthermore, mutations in the HBx-gene evolving in hepatocellular carcinoma can abolish both HBx-induced growth arrest and apoptosis. Abrogation of the anti-proliferative and apoptotic effects of HBx by naturally occurring mutations might render the hepatocytes susceptible to uncontrolled growth and contribute to multistep hepatocarcinogenesis associated with HBV infection[18]. HBx has been shown to be able to regulate cell cycle. HBx can activate the cyclin A promoter, induce cyclin A-cyclin-dependent kinase 2 complexes, and promote cycling of growth-arrested cells into G1 through a pathway involving activation of Src tyrosine kinases[5]. HBx stimulation of Src kinases and cyclin gene expression was found to force growth-arrested cells to transit through G1 but to stall at the junction with S phase, which may be important for viral replication.
The structure feature of HBx may affect its biophysical properties including solubility, tendency toward aggregation with itself and with other proteins. Gupta et al[19] purified HBx from E. coli, subjected the protein to air oxidation, and analyzed intramolecular disulfide bonds. They found that eight of the nine cysteines were disulphide linked in a sequential manner. The disulphide linkages were between Cys7 and Cys78, Cys17 and Cys115, Cys61 and Cys137, Cys69 and Cys143 while Cys148 was free. The disulphide arrangement in HBx followed a pattern where each cysteine was joined to the fourth cysteine. HBx is able to form intramolecular disulfide bonds even under the reducing conditions of the cytosol. With analysis of purified HBx from eukaryotic sources by using electrospray ionization mass spectrometry, Urban et al[14] uncovered some components: the unmodified, monomeric, fully oxidized form with five intramolecular disulfide bridges, and its N-acetylated modification. It is not very clear whether the disulfide bridge formation is a prerequisite of HBx function, but a point mutation of Cys-69 to Ala abolishes its transactivation activity[20]. Intracellular deposition of aggregated proteins is a prominent cytopathological feature of most neurodegenerative diseases[21]. Protein aggregation is a central event in the initiation of cell death[6]. Protein aggregation directly impaires the function of the ubiquitin-proteasome system and leads to cell cycle arrest and cell death.
In view of the investigations, we proposed apoptotic effects of HBx be related to its aggregate formation. Immunohistochemical examination revealed that only a minor immunological signal of HBx was detectable in a soluble form, whereas most of the protein formed intracellular aggregates in cytoplasm. HBx frequently accumulated in large granules. Our observation could suggest that HBx transfected cells underwent growth arrest and apoptosis, whereas control cells without HBx remained in growth and progression into S phase. In addition, we used the reporter molecule EGFP to reflect the situation of HBx expression indirectly. By counting equal number of EGFP positive cells, pIRES-EGFP-HBx transfected cells showed a weak fluorescence as compared with pIRES-EGFP transfected cells. Diao et al[17] also reported that human primary hepatocytes transfected with pIRES-EGFP-HBx showed less intense fluorescent signal. HBx aggregates may affect the fluorescence intensity of EGFP in the same cell.
HBx were detected in aggregated structures and it was further showed that HBx aggregates mediated growth arrest and cell death. Our data may provide helpful information on the biological effects of HBx aggregates. The precise mechanism by which HBx aggregates could affect cell cycle and apoptosis remains to be established.
Edited by Xu XQ
| 1. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1094] [Cited by in RCA: 1115] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 2. | Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 260] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 3. | Rui E, de Moura PR, Kobarg J. Expression of deletion mutants of the hepatitis B virus protein HBx in E. coli and characterization of their RNA binding activities. Virus Res. 2001;74:59-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Arbuthnot P, Capovilla A, Kew M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J Gastroenterol Hepatol. 2000;15:357-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 151] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Bouchard M, Giannakopoulos S, Wang EH, Tanese N, Schneider RJ. Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J Virol. 2001;75:4247-4257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 78] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 327] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 7. | Diao J, Garces R, Richardson CD. X protein of hepatitis B virus modulates cytokine and growth factor related signal transduction pathways during the course of viral infections and hepatocarcinogenesis. Cytokine Growth Factor Rev. 2001;12:189-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 125] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 8. | Wei W, Dorjsuren D, Lin Y, Qin W, Nomura T, Hayashi N, Murakami S. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J Biol Chem. 2001;276:12266-12273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Su F, Schneider RJ. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc Natl Acad Sci U S A. 1997;94:8744-8749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 251] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 10. | Lee S, Tarn C, Wang WH, Chen S, Hullinger RL, Andrisani OM. Hepatitis B virus X protein differentially regulates cell cycle progression in X-transforming versus nontransforming hepatocyte (AML12) cell lines. J Biol Chem. 2002;277:8730-8740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Chirillo P, Pagano S, Natoli G, Puri PL, Burgio VL, Balsano C, Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci U S A. 1997;94:8162-8167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 157] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 12. | Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon ML, Tiollais P, Buendia MA. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115-2123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 136] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Shintani Y, Yotsuyanagi H, Moriya K, Fujie H, Tsutsumi T, Kanegae Y, Kimura S, Saito I, Koike K. Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J Gen Virol. 1999;80:3257-3265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 61] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Urban S, Hildt E, Eckerskorn C, Sirma H, Kekulé A, Hofschneider PH. Isolation and molecular characterization of hepatitis B virus X-protein from a baculovirus expression system. Hepatology. 1997;26:1045-1053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1622] [Cited by in RCA: 1617] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 16. | Yang LJ, Sui YF, Chen ZN. Preparation and activity of conjugate of monoclonal antibody HAb18 against hepatoma F(ab')(2) fragment and staphylococcal enterotoxin A. World J Gastroenterol. 2001;7:216-221. [PubMed] |
| 17. | Diao J, Khine AA, Sarangi F, Hsu E, Iorio C, Tibbles LA, Woodgett JR, Penninger J, Richardson CD. X protein of hepatitis B virus inhibits Fas-mediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J Biol Chem. 2001;276:8328-8340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 18. | Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Bréchot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848-4859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 161] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 19. | Gupta A, Mal TK, Jayasuryan N, Chauhan VS. Assignment of disulphide bonds in the X protein (HBx) of hepatitis B virus. Biochem Biophys Res Commun. 1995;212:919-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Arii M, Takada S, Koike K. Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene. 1992;7:397-403. [PubMed] |
| 21. | Schulz JB, Dichgans J. Molecular pathogenesis of movement disorders: are protein aggregates a common link in neuronal degeneration. Curr Opin Neurol. 1999;12:433-439. [PubMed] |