INTRODUCTION
Ornithine decarboxylase (ODC) is the first key enzyme of the biosynthesis of polyamine which catalyzes the decarboxylation of the amino acid ornithine to the diamine putrescine. Its activation regulates the metabolism of spermidine, spermine and their precursor putrescine. Activity of polyamine biosynthesis is closely associated with the proceeding of physiological cell growth, proliferation and regeneration[1] and pathological proliferation[2]. It is necessary for cell to progress into S phase, or polyamine depletion arrest cells in G1[3]. ODC activity and polyamine concentration in colorectal cancers are significantly elevated compared with that in normal adjacent and healthy control tissues on rodents[4-7] and human beings. ODC repressor (e.g. Difluoromethylornithine) has been considered to be one of the molecular targeted interventions of colon cancer[8]. The changing of ODC activity is an early event during the expression of malignancy. In this study, an ODC expression vector expressing a 6His-tag fusion protein was successfully constructed. The 6His-tag enabled us to purify the fusion protein with high purity.
MATERIALS AND METHODS
Materials
Trizol, RNA extract reagent, were purchased from Life Technologies Inc. RT-PCR kit, T-A clone kit, DNA marker and all restrictive enzymes were purchased from TaKaRa Shuzo Co.Ltd. Primers were synthesized by Sangon. The QIAquick Gel Extraction Kit, and expression system were got from QIAGEN. Protein marker was purchased from Shanghai Lizhudongfeng biotechnologies Co.Ltd. Standard ODC was purchased from Sigma.
Tissues
Colorectal carcinoma and respective adjacent normal colorectal mucus were obtained during surgery. Once the specimens were removed during operation, the necrotic and ulcerated tumors were removed and the normal mucosa was dissociated from the muscle and connective tissue. All specimens were then kept in liquid nitrogen until further use.
Extraction of total RNA
The total cellular RNA was extracted from normal and cancer tissues, respectively. The method of RNA extraction was similar to the Trizol RNA extraction protocol (Life Technologies Inc.). The concentration of RNA extracted was determined at wavelength of 260 nm using U-2000 spectrophotometer (HITACH Ltd, Tokyo, Japan).
Reverse transcription polymerase chain reaction (RT-PCR)
The sequence of ODC primers was as follows, up-stream primer: 5’-gcaggatccaccatgaacaactttggtaa; down-stream primer: 5’-gaagtcgacctacacattaatactagccg. The 5’primer recognized the start codon of ODC in exon 3, and the 3’primer recognized the end-codon in exon 12. Restriction sites were BamH I and Sal I. The first strand of cDNA was synthesized at 55 °C for 30 min in the presence of AMV reverse transcriptase (0.5 unit/µl), RNase inhibitor 1 unit/mL, dNTP 1.0 mM, Mg2+ 2.5 mM. The PCR was processed through 35 cycles of denature (1 min at 95 °C), annealing (1.5 min at 58 °C), and extension (1 min at 72 °C) (Perkin-Elmer2400 PCR apparatus).
Purification of PCR product and T-A cloning
The PCR products were separated in 1% agarose gel, and the band containing ODC cDNA was cut off and placed into the QIAquick spin column, the ODC cDNA was purified and linked to plasmid pMD-18 with a polyA linker. The recombinant was transformed into E. coli. DH5α and selected by selective culture medium containing ampicillin.
Construction of pQE30-ODC
The pMD-ODC and pQE30 were digested by restrictive enzymes BamH I and Sal I. The inserted fragment of pMD-ODC was collected from electrophoretic gel, then it was ligated with the linearized pQE30 by T4 Ligase at 18 °C overnight. The recombinant was transformed into E. coli DH5α by CaCl2 method and selected by agar plate containing ampicillin and confirmed by restriction enzyme mapping. The positive recombinant was transformed into E. coli. M15.The sequence of inserted fragment was confirmed by DNA sequencing (Shanghai Sangon Bioengineering Co.Ltd.).
The expression of ODC fusion protein
The ampicillin-resistant colony of E. coli cells transformed with plasmid were cultured in LB cultural medium containing 100 mg/L ampicillin and 25 mg/L Kanamycin, and induced by 1 mM IPTG. The cultured cells were harvested at 1, 2, 3, 4 hr after culture, respectively. The optimum time of maximum expression of proteins was analyzed through SDS-PAGE. The expressed ODC protein was tested through Western blot with specific antiserum.
RESULTS
RT-PCR amplification of ODC encoding sequence
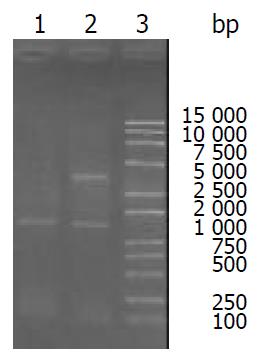
RT-PCR was done with total RNA template extracted from human colon cancer. The designed primers include encoding sequence of ODC. Electrophoresis of RT-PCR products confirmed the length of RT-PCR fragment (1480 bp) (Figure 1).
Figure 1 RT-PCR product on 1% agarose gel.
Lane 1: ODC RT-PCR product, 1480 bp in length; Lane 2: pQE30-ODC plas-mid digested by restriction enzyme BamH I and Sal I.
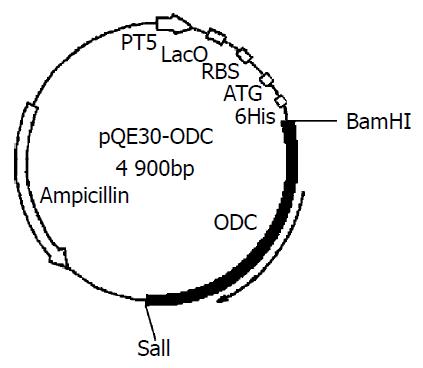
The purified ODC cDNA was ligated to pMD-18 by T-A complimentary pairing. ODC cDNA was inserted into pQE30 at BamH I and Sal I sites (Figure 2).
Figure 2 The pQE30-ODC expression vector including early promoter T5, Lac operator and 6His-Tag.
Sequence analysis
Sequence of inserted DNA was analyzed with automatic sequence analyzer and found to be about 1500 bp in size and showed 99% affinity in comparison with DNA sequence published on line [gi: 4505488].
SDS-PAGE and western blotting
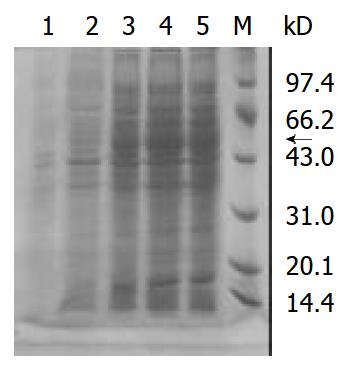
Inserted ODC gene was expressed significantly in the prokaryotic expression system, and specific strip at 50 kDa was demonstrated in Western blot. The optimum induction period is 4 hr after administration of IPTG (Figure 3, Figure 4).
Figure 3 Expressed ODC protein on SDS-PAGE.
Lane 1: M15 negative control; Lane 2-5: Expressed ODC proteins in M15 1 to 4 hour(s) after induction of IPTG; M: Protein marker; The arrow showed the expressed ODC protein.
Figure 4 Western blot shows a strip of 50 kD indicating the expression of ODC protein.
DISCUSSION
The polyamines are naturally occurring aliphatic polycations found in almost all living cells[9]. They are positively charged at neutral pH and the charge is distributed along the length of the molecule. This facilitates their interaction with anionic molecules such as DNA and RNA[9,10]. Polyamines have been shown to be essential for optimal rates of cell growth and differentiation, with high concentrations being found in rapidly growing cells and tissues[11].
Cancer cells always have a higher intracellular polyamine content than the equivalent normal tissue[12-15]. In addition to changes in polyamine content, ODC activity has also been found to be increased significantly in colon adenocarcinoma tissue compared to microscopically normal tissue from the same patients[13,16]. Similar findings were observed in human colonic surgical specimens with both ODC activity and polyamine content of the malignant tissue being increased[16,17]. Intratumour content of spermidine and spermine was increased in line with the increase in ODC activity.
Ornithine decarboxylase (ODC) is a rate-limiting enzyme in the biosynthesis of polyamines, and is induced in response to many growth stimuli such as hormones, growth factors, and tumor promoters, it has a rapid turnover rate with half-life at 15 minutes[18]. Numerous studies have demonstrated that regulation of ODC can occur at multiple levels, including the transcription of protooncogene c-myc[19-22], mRNA translation[10,23,24], protein turnover[10,25,26], and post-transcriptional interactions and modifications[27-30]. Recent studies focus intensely on ODC and polyamine regulation as therapeutic targets. Inhibitors of ODC were found to suppress tunor formation in experimental models of bladder, breast, colon, and skin carcinogenesis[31-35].
Colorectal cancer is a major health problem in the western world and is associated with significant morbidity and mortality. First-line therapy is radical surgery with adjuvant chemotherapy commonly being treatment with antimetabolites such as 5-fluorouracil[30]. Although great clinical efforts have been made for the therapy of colorectal cancer with new technologies, such as CT or MRI colography[4,14], there is still much to be done on the early diagnosis and treatment. It is suggest that ODC activity and polyamine content may have interest in the diagnosis of malignancy and prognosis[15]. As the change of ODC activity is an early event in the development of the disease, it may be helpful for early screening and diagnosis of colon cancer.
We designed two primers from human ODC gene including the start codon (ATG) in exon 3 and the termination codon (TAG) so as to amplify the whole encoding sequence about 1480 bp of ODC. The exons encode a protein is identical to the 461-amino acid sequence derived from human ODC.
The restriction fragment mapping of the recombinant, pQE 30-ODC, indicated that the inserted fragment was about 1.5 kb, which was consistent with the encoding sequence of ODC. ODC activity in the lysate of transformed M15 was demonstrated through Western blot. The expressed fusion protein is about 50 kDa, which is similar to the known ODC protein. The construction and expression of recombinant ODC provide a tool for ODC related further study.