Published online Mar 15, 2003. doi: 10.3748/wjg.v9.i3.474
Revised: July 3, 2002
Accepted: July 11, 2002
Published online: March 15, 2003
AIM: To study the effect of hepatitis C virus nonstructural protein 3 c-terminal deleted protein (HCV NS3-5’) on hepatocyte transformation and tumor development.
METHODS: QSG7701 cells were transfected with plasmid pRcHCNS3-5’ (expressing HCV NS3 c-terminal deleted protein) by lipofectamine and selected in G418. The expression of HCV NS3 gene and protein was determined by PCR and immunohistochemistry respectively. Biological behavior of transfected cells was observed through cell proliferation assay, anchorage-independent growth and tumor development in nude mice. The expression of HCV NS3 and c-myc proteins in the induced tumor was evaluated by immunohistochemistry.
RESULTS: HCV NS3 was strongly expressed in QSG7701 cells transfected with plasmid pRcHCNS3-5’ and the positive signal was located in cytoplasm. Cell proliferation assay showed that the population doubling time in pRcHCNS3-5’ transfected cells was much shorter than that in pRcCMV and non-transfected cells (24 h, 26 h, 28 h respectively). The cloning ratio of cells transfected with pRcHCNS3-5’, pRcCMV and non-transfected cells was 33%, 1.46%, 1.11%, respectively, the former one was higher than that in the rest two groups (P < 0.01). Tumor development was seen in nude mice inoculated with pRcHCNS3-5’ transfected cells after 15 days. HE staining showed its feature of hepatocarcinoma, and immunohistochemistry confirmed the expressions of HCV NS3 and c-myc proteins in tumor tissue. The positive control group inoculated with HepG2 also showed tumor development, while no tumor developed in the nude mice injected with pRcCMV and non-transfected cells after 40 days.
CONCLUSION: 1.HCV NS3 c-terminal deleted protein has transforming and oncogenic potential. 2. Human liver cell line QSG7701 may be used as a good model to study HCV NS3 pathogenesis.
- Citation: He QQ, Cheng RX, Sun Y, Feng DY, Chen ZC, Zheng H. Hepatocyte transformation and tumor development induced by hepatitis C virus NS3 C-terminal deleted protein. World J Gastroenterol 2003; 9(3): 474-478
- URL: https://www.wjgnet.com/1007-9327/full/v9/i3/474.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i3.474
HCV infection is a major worldwide health problem. Persistent infection with HCV is a critical risk for the development of hepatocellular carcinoma (HCC)[1-3]. It has been reported that the Core protein, NS3, NS4B, and NS5A have oncogenic potential[2,4-6]. HCV NS3 protein (nucleotide 3420 to 5312 with 631 amino acid residues) is a multi-functional viral protein. In addition to serine proteinase activity, which is located in the one-third of the NS3 protein at the N-terminal end, helicase and nucleotide triphosphatase activities are identified in the c-terminal half of the NS3 protein[7]. HCV NS3 that plays a key role in the life cycle of virus and interacts with host cellular protein has been one of hot spots in recent research. We found that the NIH3T3 cell has stronger telomerase activity after transfected with HCV NS3 plasmid, indicating that HCV NS3 may be an important part in the hepatocarcinogenesis[8]. Most studies on NS proteins have been carried out by expression of single or multiple NS proteins in cultured none hepatocyte, despite the fact that HCV is a hepacivirus. In order to reflect the relation between HCV NS3 and host cell transformation more clearly, here we tried to transfect human liver cell line QSG7701 with eukaryotic cell plasmid pRcHCNS3-5’ (expressing NS3 c-terminal deleted protein), and then inoculated nude mice with transfected cells to investigate it’s biological behaviors and carcinogenesis.
Hepatocyte cell line QSG7701 was got from Shanghai Institute of Cell Biology, Chinese Academe of Sciences (CAS) and HepG2 cell line was got from Cell Center of Central South University. Nude mice (BALB/cA-nude) were got from Shanghai Laboratorial Animal Center, CAS. The plasmid pRcHCNS3-5’ (expressing HCV NS3 c-terminal deleted protein) was the kind gift from professor Takegami[9]. Non-expressive plasmid pRcCMV was purchased from Sigma Com USA. LipofectaminTM reagent, G418 and Dulbecco’s modified Eagle medium (DMEM) were products of GIBCO BRL (Germany). Xbal, buffer and PCR kit, marker were bought from Sino-American Biotechnique INC (Shanghai, P.R.China). Anti-HCV NS3 protein MAb, anti-c-myc protein MAb and S-P detection kit were purchased from Boshide Com (Wuhan, P. R.China) and Maxim Biotech INC (Fuzhou, P.R.China), respectively. New-born calf serum was got from Sijiqing Bioengineering Ltd. (Hangzhou, P.R.China). PCR primers for amplifying HCVNS3-5’ gene were synthesized at Shanghai Sangon Com (Shanghai, P.R.China).
Experimental groups Group 1: QSG7701 parental cells; Group 2: QSG7701 transfected with blank plasmid pRcCMV; Group 3: QSG7701 transfected with plasmid pRcHCNS3-5’; Group 4: HepG2 cell line used as positive control.
Cell culture Cell lines were maintained in DMEM with 10% heat-inactivated new-born calf serum, 100 unit/mL penicillin and 100 unit/mL streptomycin at 37 °C in a humidified atmosphere of 5% CO2.
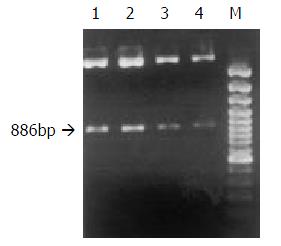
Preparation, purification and identification of plasmids pRcHCNS3-5’ and pRcCMV Plasmids pRcCMV and pRcHCNS3-5’ were transferred into competent E coli JM109 respectively. JM109 was cultured to amplify the two plasmids. Little plasmids were prepared from JM109 to identify the specificity of the plasmids. The plasmid pRcHCNS3-5’ was digested with Xbal, resolved with agarose gel electrophoresis, and stained with ethidium bromide (EB). As shown in Figure 1, the electrophoresis analysis revealed the major 886bp fragment of the plasmid pRcHCNS3-5’ as being expected. Then the plasmids were massively extracted and purified for transfecting QSG7701 cells.
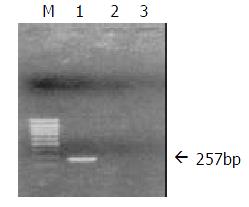
Transfection of QSG7701 cells with plasmids pRcHCNS3-5’ and pRcCMV QSG7701 cells were transfected with the plasmids pRcHCNS3-5’ and pRcCMV, respectively according to the instruction of Lipofectamine reagent. Cells were seeded into selection medium containing 400 μg/mL G418 until G418-resistant clones were obtained, and then cells were maintained in G418 with concentration of 200 μg/mL (Figure 2). QSG7701 parental cells were used for the parallel control. To detect cDNA in stable transfectants, total genomic DNA was extracted according to standard methods and subjected to PCR and agarose gel electrophoresis analysis. The primers for amplifying HCVNS3-5’ gene were based on published sequences[9]. PCR conditions were 35 cycles of three steps (94 °C 30 sec, 57 °C 30 sec, 72 °C 40 sec) and then 72 °C 5 min in a 50 μL reaction mixture containing 5 μL 10×buffer, 5 μL 2 mM dNTPs, 0.5 μL of each primer (25 pmol/μL), 1 μL (100 ng)DNA, 0.5 μL 5U/μL Taq DNA polymerase, and 37.5 μL distilled water. PCR products were subjected to electrophoresis on a 0.8% agarose gel, visualized by EB staining. As shown in Figure 3, 257bp fragment was specifically amplified from DNA of the QSG7701 cells transfected with plasmid pRcHCNS3-5’, but no fragment was amplified from DNA of the pRcCMV transfected cells and parental cells.
Immunohistochemistry S-P method was used to detect the expression of HCV NS3 protein in parental QSG7701 cells and QSG7701 cells transfected with plasmid pRcHCNS3-5’, pRcCMV. PBS substituting Mabs was used as blank control. HCC expressing HCV NS3 protein was used as positive control.
Cell proliferation assay QSG parental cells and cells transfected with pRcHCNS3-5’, pRcCMV (6 × 104 cells per well) were seeded in 24-well plates and counted daily with a haemocytometer respectively. Experiment was lasted for 7 days and was repeated three times.
Anchorage-independent growth Cells were suspended at a density of 2 × 103/mL containing 0.3% granulated agar. The mixture was added over a layer of 0.7% agar in medium. After 4 weeks, the morphology of transfected cells was analysed microscopically. Colonies including more than 50 cells were scored. Experiment was repeated three times.
Tumor development in athymic nude mice 12 nude mice (BALB/cA-nude, females, 4-6 weeks, 18-22 g) were divided into 4 groups of 3 nude mice each and inoculated subcutaneously in the right flank with pRcHCNS3-5’, pRcCMV and parental cells (1 × 107cells in 0.2 ml of phosphate-buffered saline) and monitored for tumor development. HepG2 cells were used as the positive control. Tumor size and animal weight were measured weekly. The nude mice were sacrificed and the tumors were removed 40 days after inoculating. Tumor tissue was fixed by 10% formalin, embedded with paraffin wax, sliced successively 5 μm per section and stained by HE. Immunohistochemistry S-P method was used to detect the expressions of HCV NS3 protein and c-myc protein in tumor tissue.
F test was used. P value of less than 0.05 was considered as statistically significant.
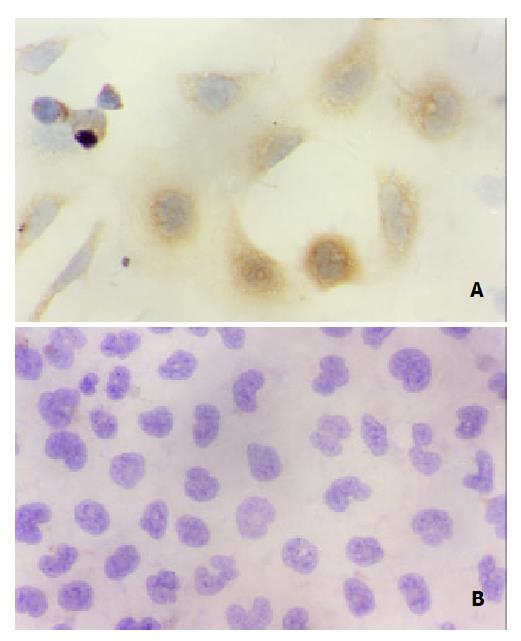
Immunohistochemical staining showed that QSG7701 cells transfected with pRcHCNS3-5’ strongly expressed HCV NS3 protein. The positive signal was in cytoplasm (Figure 4). The positive signal was also found in the positive control group, but not in the blank and negative control groups.
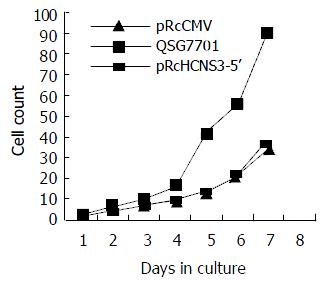
The population doubling time in cells transfected with pRcHCNS3-5’ (12 h) was much shorter than that in those transfected with pRcCMV and parental QSG7701 cells (26 h, 28 h respectively). (Figure 5).
The cloning efficiencies in pRcHCNS3-5’, pRcCMV transfected cells and parental cells were 33%, 1.46%, 1.11%, respectively (P < 0.01, the former vs the rest two).
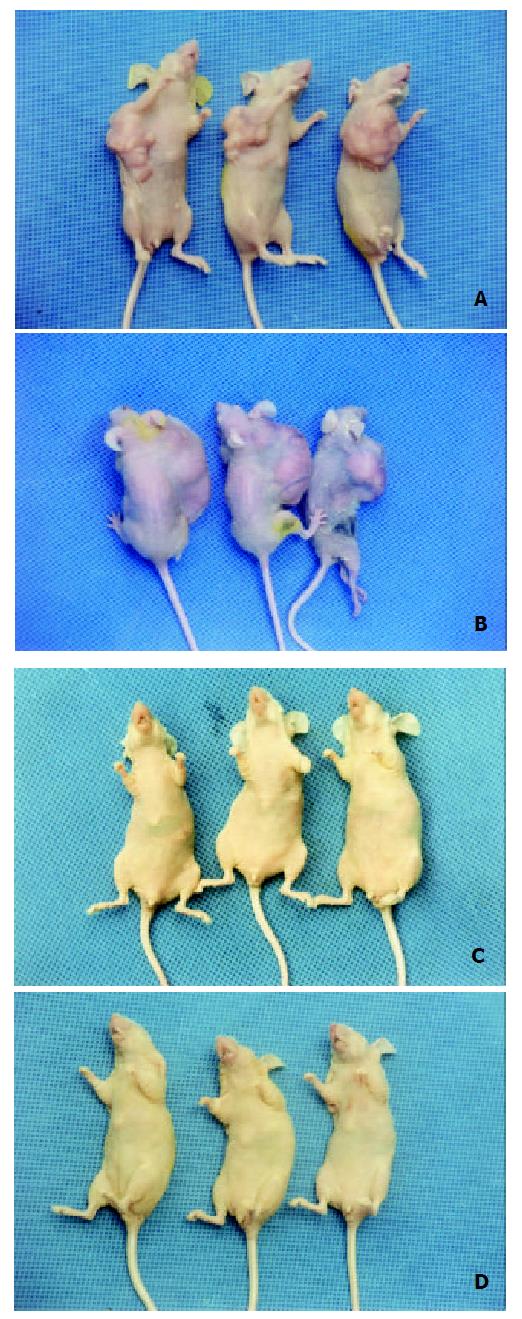
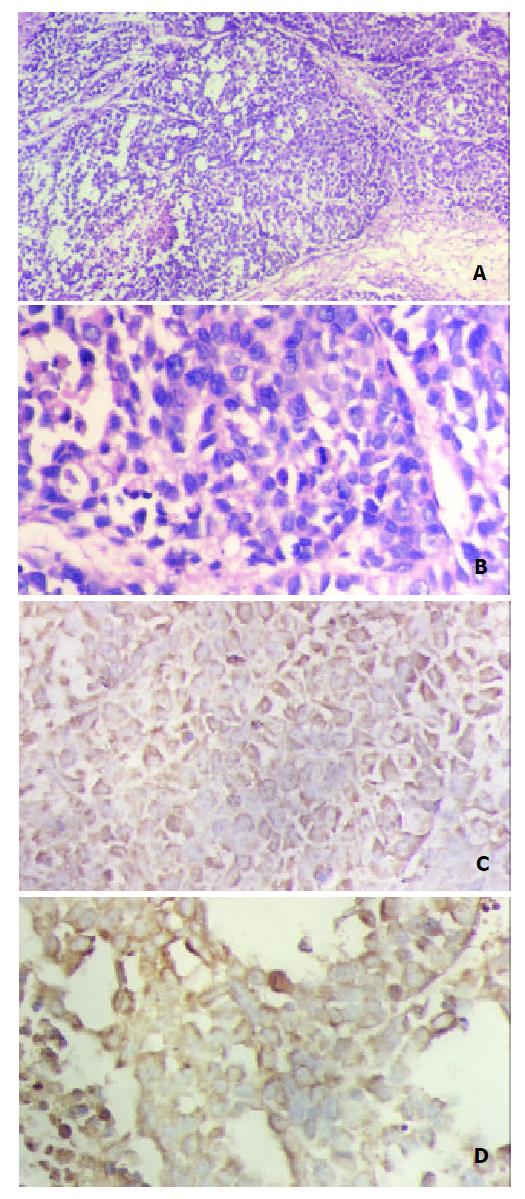
In the nude mice inoculated with pRcHCNS3-5’transfected cells, the first evidence of well-defined and distinct subcutaneous tumors appeared on day 15. The mean tumor size and weight on day 40 were 3.08 cm3 and 3.13 g, respectively. The HepG2 cells, as positive control, also induced tumor in the nude mice. But no tumor developed in the mice inoculated with pRcCMV and none-transfected QSG7701 cells until 40 days after inoculating (Figure 6). Macroscopically, the tumors induced by pRcHCNS3-5’ showed irregular yellow-brown node and appeared to be encapsulated. Central necrosis could be seen on the cut surface. Microscopically, the tumor cells exhibited similarity to hepatocyte except for heterogenesis and they arranged as trabecular with intervening sinusoids. Cancer cell nests with abundance granular necrosis were separated by fibrous tissue (Figure 7(a,b)). They showed the histologically feature of hepatocellular carcinoma. Immunohistochemical staining showed that tumor tissue expressed HCV NS3 protein and c-myc protein. The positive signal was located in cytoplasm (Figure 7(c,d)).
It has been reported that persistent infection with HCV is associated with HCC, and HCV replication and protein expression can be observed in HCC tissue[1,10-11], although the pathogenesis of HCV infection-associated HCC is still unknown. HCV, as a RNA virus without any RT activity, is replicated in the cytoplasm and does not integrate with host genome like HBV. Furthermore, unlike EBV or HPV, the HCV genome itself, does not contain any known oncogene. Most studies indicate that the interaction between the HCV protein an d liver cell gene/p ro tein h as been inv olved in hepatocarcinogenesis. Zemel et al reported significant mutations of amino acids that occur at the catalytic domain of the NS3 serine protease gene isolated from HCC tissue will affect the activity and substrate specificity of serine protease[12]. Sakamuro[9] and Zemel[2] et al founded that HCV NS3 could transform NIH3T3 cell and non-tumorigenic rat fibroblast (RF) cell respectively. HCV NS3 can suppress actinomycin D induced apoptosis[13], inhibit cAMP-dependent protein kinase[14] and repress P53 function[15-17]. But the cell lines used by most researchers were not based on HCV natural host cell-hepatocyte. Here non-tumorigenic human liver cell line QSG7701 was used in our study. QSG7701 cells that were immortally normal hepatocytes came from liver tissue 6 cm far from HCC and their karyotype number was 57[18]. QSG7701 cells with normal hepatocyte phenotype were stably transfected with an expression vector containing cDNA for NS3 proteinase activity. The transfected cells grew rapidly, showed anchorage -independent growth in soft agar and induced significant tumor formation in nude mice. Our study firstly showed that the protease-coding gene of HCV induced malignant transformation of nontumorigenic human liver cells. Moreover injecting pRcHCNS3-5’ transfected cells into nude mice led to significant tumor formation. The tumor obtained from the nude mice showed the histologically feature of hepatocellular carcinoma. The results suggest the involvement of proteinase activity of HCV NS3 N terminal peptide in cellular transforming and oncogenic potential.
Current studies show that HCV NS3 may be directly involved in hepatocarcinogenesis by disturbing the regulation of cell proliferation. But the mechanism of carcinogenesis is still perplexing. Carcinogenesis, including HCC, is the result of the abnormal proliferation of cancer cells. It is reported that various oncogenic products are related to functional abnormalities of intracellular signal transduction pathway, which have been proved to be one of the proliferative mechanism of cancer cells. Great progress has been made that HCV proteins regulate signal transduction in the recent studies. Ras/Raf/MAPK pathway is regarded to have close relation to HCV associated HCC.
Constitutive activation of the Ras/Raf/MAPK pathway is important for the transformation of mammalian cells[19,20]. Indeed, studies have demonstrated that HCC development and progression is associated with the activation of Ras/Raf/MAPK pathway in humans and rodents[21,22] Our lab also found that MAPK activity is higher in HCC than that in the adjacent noncancerous lesions, suggesting a progression of HCC through Ras/Raf/MAPK activation[23]. Recently, the development of anticancer drugs that target signaling proteins of MAPK pathway has been a major goal in the cancer treatment[24,25]. There are several reports concerning the relationship between HCV protein and Ras/Raf/MAPK signal transduction pathway. For example, some evidences suggest that HCV C protein directly or indirectly activates Ras/Raf/MAPK signal transduction pathway elements (including Raf-1, ERK, MEK, Elk, SRE, AP-1) in different cells such as HepG2, CCL13, COS7, MCF-7, BALB/3T3 and NIH3T3[26-31]. Borowski et al reported an arginine-rich domain located in the NS3 region (residues1487 to 1500 of HCV polyprotein) which strongly resembles the autoinhibitory sequence of the PKA R subunit. It binds to the C subunit of PKA and inhibits the translocation of C subunit into nucleus. Consequently, PKA functions are arrested[32]. Marshall identified that specific sites between Ras and Raf-1 are essential to plasma membrane localization and Raf activation. The formation of Ras/ Raf-1 complex is negatively regulated by PKA through phosphorylation of Raf-1 N terminal serine 43, which is believed to cause an N-terminal cap structure to cover the Ras docking site which ultimately leads to inhibit Raf-1 activation[33,34]. HCV NS3 may activate Raf-1 by interfering the function of PKA. We previously demonstrated that HCV NS3 N-terminal peptide (residues 1020 -1295) positively regulates ERK1/2 phosphorylation. Because the N terminal peptide of HCV NS3 used in our study does not include the arginine-rich domain. We preclude that either HCV NS3 interacts with PKA through other domain or HCV NS3 involves in Ras/Raf/MAPK signal transduction pathway through other targets. Nuclear transcriptional factors c-myc as a target factor regulated by downstream of the MAPK casade may increased when MAPK is activated. Overexpression of c-myc in tumor tissue of nude mice was detected which was consistent with other’s report[35]. It is supposed that HCV NS3 may be involved in hepatocarcinogenesis through Ras/ Raf/MAPK/c-myc signal transduction.
We thank professor Takegami for his generous gift of plasmid pRcHCNS3-5’.
Edited by Xu XQ
| 1. | Li L, Wang W, Yu X. [Detection of hepatitis C virus RNA in the tissue of hepatocellular carcinoma by multiple detection system]. Zhonghua Shi Yan He Linchuangbing Duxue Zazhi. 2000;14:47-51, 101. [PubMed] |
| 2. | Zemel R, Gerechet S, Greif H, Bachmatove L, Birk Y, Golan-Goldhirsh A, Kunin M, Berdichevsky Y, Benhar I, Tur-Kaspa R. Cell transformation induced by hepatitis C virus NS3 serine protease. J Viral Hepat. 2001;8:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 3. | Koike K, Tsutsumi T, Fujie H, Shintani Y, Kyoji M. Molecular mechanism of viral hepatocarcinogenesis. Oncology. 2002;62 Suppl 1:29-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 93] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Park JS, Yang JM, Min MK. Hepatitis C virus nonstructural protein NS4B transforms NIH3T3 cells in cooperation with the Ha-ras oncogene. Biochem Biophys Res Commun. 2000;267:581-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 5. | Ghosh AK, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol. 1999;80:1179-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 146] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 912] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 7. | Kato N. Molecular virology of hepatitis C virus. Acta Med Okayama. 2001;55:133-159. [PubMed] |
| 8. | Feng D, Cheng R, Ouyang X, Zheng H, Tsutomu T. Hepatitis C virus nonstructural protein NS(3) and telomerase activity. Chin Med J (Engl). 2002;115:597-602. [PubMed] |
| 9. | Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893-3896. [PubMed] |
| 10. | Yang JM, Wang RQ, Bu BG, Zhou ZC, Fang DC, Luo YH. Effect of HCV infection on expression of several cancer-associated gene products in HCC. World J Gastroenterol. 1999;5:25-27. [PubMed] |
| 11. | Ohishi M, Sakisaka S, Harada M, Koga H, Taniguchi E, Kawaguchi T, Sasatomi K, Sata M, Kurohiji T, Tanikawa K. Detection of hepatitis-C virus and hepatitis-C virus replication in hepatocellular carcinoma by in situ hybridization. Scand J Gastroenterol. 1999;34:432-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 12. | Zemel R, Kazatsker A, Greif F, Ben-Ari Z, Greif H, Almog O, Tur-Kaspa R. Mutations at vicinity of catalytic sites of hepatitis C virus NS3 serine protease gene isolated from hepatocellular carcinoma tissue. Dig Dis Sci. 2000;45:2199-2202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 13. | Fujita T, Ishido S, Muramatsu S, Itoh M, Hotta H. Suppression of actinomycin D-induced apoptosis by the NS3 protein of hepatitis C virus. Biochem Biophys Res Commun. 1996;229:825-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 65] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Borowski P, Heiland M, Oehlmann K, Becker B, Kornetzky L, Feucht H, Laufs R. Non-structural protein 3 of hepatitis C virus inhibits phosphorylation mediated by cAMP-dependent protein kinase. Eur J Biochem. 1996;237:611-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Okada F, Shiraki T, Maekawa M, Sato S. A p53 polymorphism associated with increased risk of hepatitis C virus infection. Cancer Lett. 2001;172:137-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Kwun HJ, Jung EY, Ahn JY, Lee MN, Jang KL. p53-dependent transcriptional repression of p21(waf1) by hepatitis C virus NS3. J Gen Virol. 2001;82:2235-2241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Feng DY, Chen RX, Peng Y, Zheng H, Yan YH. Effect of HCV NS(3) protein on p53 protein expression in hep atocarcinogenesis. World J Gastroenterol. 1999;5:45-46. [PubMed] |
| 18. | Zhu DH, Wang JB. The culture of HCC host hepatocyte QSG7701 cell line and as compared with hepatocarcinoma cell. Zhongliu Fangzhi Yanjiu. 1979;5:7-9. |
| 19. | Pinkas J, Leder P. MEK1 signaling mediates transformation and metastasis of EpH4 mammary epithelial cells independent of an epithelial to mesenchymal transition. Cancer Res. 2002;62:4781-4790. [PubMed] |
| 20. | Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, Der CJ, Counter CM. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045-2057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 321] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 21. | Zhu J, Leng X, Dong N, Liu Y, Li G, Du R. [Expression of mitogen-activated protein kinase and its upstream regulated signal in human hepatocellular carcinoma]. Zhonghua Waike Zazhi. 2002;40:1-16. [PubMed] |
| 22. | Ostrowski J, Woszczynski M, Kowalczyk P, Wocial T, Hennig E, Trzeciak L, Janik P, Bomsztyk K. Increased activity of MAP, p70S6 and p90rs kinases is associated with AP-1 activation in spontaneous liver tumours, but not in adjacent tissue in mice. Br J Cancer. 2000;82:1041-1050. [PubMed] |
| 23. | Feng DY, Zheng H, Tan Y, Cheng RX. Effect of phosphorylation of MAPK and Stat3 and expression of c-fos and c-jun proteins on hepatocarcinogenesis and their clinical significance. World J Gastroenterol. 2001;7:33-36. [PubMed] |
| 24. | Shapiro P. Ras-MAP kinase signaling pathways and control of cell proliferation: relevance to cancer therapy. Crit Rev Clin Lab Sci. 2002;39:285-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 25. | Sebolt-Leopold JS. Development of anticancer drugs targeting the MAP kinase pathway. Oncogene. 2000;19:6594-6599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 238] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 26. | Aoki H, Hayashi J, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein interacts with 14-3-3 protein and activates the kinase Raf-1. J Virol. 2000;74:1736-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 120] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Giambartolomei S, Covone F, Levrero M, Balsano C. Sustained activation of the Raf/MEK/Erk pathway in response to EGF in stable cell lines expressing the Hepatitis C Virus (HCV) core protein. Oncogene. 2001;20:2606-2610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 112] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 28. | Tsuchihara K, Hijikata M, Fukuda K, Kuroki T, Yamamoto N, Shimotohno K. Hepatitis C virus core protein regulates cell growth and signal transduction pathway transmitting growth stimuli. Virology. 1999;258:100-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Fukuda K, Tsuchihara K, Hijikata M, Nishiguchi S, Kuroki T, Shimotohno K. Hepatitis C virus core protein enhances the activation of the transcription factor, Elk1, in response to mitogenic stimuli. Hepatology. 2001;33:159-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Kato N, Yoshida H, Ono-Nita SK, Kato J, Goto T, Otsuka M, Lan K, Matsushima K, Shiratori Y, Omata M. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology. 2000;32:405-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 173] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 31. | Shrivastava A, Manna SK, Ray R, Aggarwal BB. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J Virol. 1998;72:9722-9728. [PubMed] |
| 32. | Borowski P, Heiland M, Feucht H, Laufs R. Characterisation of non-structural protein 3 of hepatitis C virus as modulator of protein phosphorylation mediated by PKA and PKC: evidences for action on the level of substrate and enzyme. Arch Virol. 1999;144:687-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Marshall M. Interactions between Ras and Raf: key regulatory proteins in cellular transformation. Mol Reprod Dev. 1995;42:493-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Häfner S, Adler HS, Mischak H, Janosch P, Heidecker G, Wolfman A, Pippig S, Lohse M, Ueffing M, Kolch W. Mechanism of inhibition of Raf-1 by protein kinase A. Mol Cell Biol. 1994;14:6696-6703. [PubMed] |
| 35. | Fang Y, Huang B, Liang Q, Li H, Huang C. [Clinical significance of c-myc oncogene amplification in primary hepatocellular carcinoma by interphase fluorescence in situ hybridization]. Zhonghua Binglixue Zazhi. 2001;30:180-182. [PubMed] |