INTRODUCTION
With the practice of diagnosis of primary liver cancer at early stage, surgical resection of small hepatocellular carcinoma (HCC) and other improvements in medical diagnosis, imaging, nonsurgical therapies, etc, important progresses have been made toward the control of liver cancer. For example, surgical resection of small HCC has resulted in a marked increase in 5-year survival rate from 20%-30% to 40%-60%. In our institute, the 5-year survival rate of 963 patients with small HCC ( ≤ 5 cm) resection was 65.1%. However, recurrence and metastasis still remain to be major challenges for clinical workers[1-3]. The main obstacle to control of metastasis for liver cancer is the lack of sensitive predictive biomarkers and novel molecular targets for biotherapies[4,5]. The discovery of metastasis suppressor genes for liver cancer will undoubtedly be of vital importance to the prognostic diagnosis and intervention therapy for overcoming the metastasis of liver cancer.
Our previous study suggested that there may harbor metastasis suppressor genes on human chromosome 8p[6]. To seek functional evidence for this possibility, we tried to introduce the normal human chromosome 8 into the highly metastatic rat liver cancer cell line C5F[3] through microcell mediated chromosome transfer (MMCT). The positive microcell hybrids were screened through drug selection and confirmed by sequence tagged sites- polymerase chain reaction (STS-PCR) and whole chromosome painting-fluorescence in situ hybridization (WCP-FISH).
MATERIALS AND METHODS
Cell lines
The highly metastatic rat liver cancer cell line C5F[7] was generously provided by Dr.Kumiko Ogawa (the First Department of Pathology, Nagoya City University, Nagoya, Japan). The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The human chromosome 8 donor cell line A9 (neo8) was purchased from Japanese Collection of Research Bioresources (JCRB) Cell Bank, Osaka, Japan. A9 (neo8) is a mouse fribroblast cell line that contains a human chromosome 8. The human chromsome 8 was randomly marked with neo resistant gene. The A9 (neo8) cells were cultrured in DMEM containing 10% FBS and 800 μg·mL-1 geneticin (G418).
Reageants and instruments
Colcemid, cytochalasin B, phytohemagglutinin A (PHA), polyethylene glycol (PEG, MW1300~1600, Hybrid-Max®) and Dimethylsulfoxide (DMSO, Hybrid-Max®) were purchased from Sigma, USA. DMEM high glucose, 100 × HAT (10 mM hyproxanthin, 40 μM aminopterin, 1.6 mM thymidine) and G418 were ordered from Life Technologies, GIBCO BRL, USA. IsoporeTM polycarbonate membranes were purchased from Millipore, USA. WCP paint DNA probes for chromosome 8 (SpectrumGreenTM) were purchased from Vysis Inc, IL, USA. Primers were ordered from Sangon Biotechnology Company, Shanghai, China. Taq polymerase, dNTP and 100bp DNA ladder marker were the products of Sino- America Biotechnology Company, China. Fetal bovine serum (FBS, define) and Bovine Calf Serum (BCS) were purchased from Hyclone, USA. PE-2400 thermocycler was the product of Perkin-Elmar Company, USA. The high speed Hitachi himac CR21G centrifuge was the product of Hitachi, Japan. Cyto VisionTM Chromosome Analysis System (Cytovision TM Image Analysis Workstations, USA) was the product of Applied Imagining, USA.
Micronuleation of A9(neo8) A9(neo8) cells were seeded in six straight-neck T25 flasks respectively. Colcemid was added to the culture to the final concentration of 0.05 μg·mL-1 when the cells reached confluence of 80%. The cells were subsequently incubated at 37 °C in 50 mL·L-1 CO2 incubator for 40-48 h.
Enucleation and filtration Media was removed from culture and cells were washed twice with PBS. The flasks were filled to the neck with DMEM containing 20 μg·mL-1 cytochalasin B which had been prewarmed at 37 °C. Flasks were sealed with parafilm, and then put into a R12A3 rotor (fixed-angle 28°) to centrifuge at 7500 × g at 36 °C for 75 min. Pellets were collected and resuspended with DMEM. Suspension was serially filtered through 8 μm, 5 μm and 5 μm polycarbonate membranes. Microcells were observed and counted on hemacytometer under microscope. Microcells were collected again by centrifuging at 2000 × g at 4 °C for 20 min and thereafter resuspended with 2 mL of DMEM with 100 μg·mL-1 PHA-P.
Microcell fusion & drug selection of microcell hybrids The recipient cell line C5F was trypsinized to make single cell suspension and counted on a hematocytometer for its cell density. C5F cells equivalent to 1/10 of microcells were taken and washed twice with PBS to get rid of remnant serum and thereby collected by centrifugation and resuspended with the PHA-P solution containing the microcells. The mixture was incubated at 37 °C for 15 min and subsequently centrifuged at 2000 × g for 15 min. Supernatant was removed as much as possible. Microcells were fused with the recipient cells for 30-60 s with 1 mL of 50% PEG, 10% DMSO in DMEM. Fusion reaction were terminated immediately by adding 10 mL of DMEM containing 5% DMSO to remove the PEG medium. Pellets were resuspended with DMEM supplemented with 10% FBS. The culture was recovered at 37 °C for 48 h and replaced with selective media of 1 × HAT and 800 μg·mL-1 G418. The selective medium was refreshed twice per week.
Single cell Isolation cloning[
11]
Viable cells in selective media of G418 and HAT were trypsinized to make single cell suspension. Cell density was examined on a hematocytometer. Fifty to one hundred cells were added into a P100 culture plate with serum free DMEM. Single cells were picked up with a P20 pipette under microscope in laminar flow. It was assured that there was only one cell in view under a low-power objective to exclude the possibility of aspirating more than one cell each time. The opening of tip was pointed by the side of the selected cell and 5 μL media was aspirated each time. The single cell could be seen disappear into the tip under the low magnification microscope. Single cells were added to 96-well cell culture plate containing 0.1 mL of selective media (DMEM supplemented with 20% FBS containing 1 × HAT and 800 μg·mL-1 G418) in each well. After about 10 days, clones that are actively proliferating were picked up and transferred to a 24-well plate. After about 7 days, cells were transferred again to T25 flask. When cells reached large quantity, they were freezed down in liquid nitrogen to keep in stock.
Genomic DNA extraction from cell lines[
12]
About 1 × 106 cultured cells were harvested by trypsinization and centrifugation, into which 0.5 mL of cell lysis buffer (100 mmol·L-1 NaCl , 1 mmol·L-1 EDTA, 0.5% SDS, 50 mmol·L-1 Tris-HCl, pH 8.0) and 10 µL of Proteinase K (2 g × L-1) were added. The mixture was incubated at 37 °C in water bath for 6 h. Genomic DNA was extracted with phenol and chloroform, precipitated with ethanol, dissolved with 20 µL of sterile dd-H2O, quantitated by spectrophotometer, diluted to 10 µg·µL-1 with sterile dd-H2O, and finally aliquoted and stored at -20 °C.
A human STS, D8S277, which located at 8p23.3-p22, was randomly chosen to confirm the result of MMCT. Primers were designed according to the published sequences on NCBI UniSTS database (http://www.ncbi.nlm.nih.gov/genome/sts/). Forward primer: CCAGGTGAGTTTATCAATTCCTGAG; reverse primer: TGAGAGGT CTGAGT GAC ATCCG. PCR product size: 148-180 (bp). PCR reaction solution (10 mmol·L-1 Tris, 50 mmol·L-1 KCl, 2 mmol·L-1 MgCl2, 0.001% Gelatin, 200 mmol·L-1 dNTPs, 0.5 mmol·L-1 primers and 2U of Taq polymerase) was added into 50 ng of DNA template for each sample respectively. PCR program was run as: 94 °C 2 min, 1 cycle; 95 °C 40 s 60 °C 40 s 72 °C 40 s, 35 cycles in all; 72 °C 4 min, 1 cycle; keep at 4 °C. Results were checked on 2% agarose gel electrophoresis stained with ethidium bromide.
Chromosome slide preparation[
15]
Microcell hybrids at logarithmic phase were treated with colcemid at a final concentration of 0.02 μg·mL-1 for 45 to 60 min at 37 °C and trypsinized subsequently to make single cell suspension. Cells were pelleted and then exposed to 5 mL of hypotonic solution (0.075 M KCl) by incubating for 10 to 15 min at 37 °C in a water bath. Chromosomes were repetitively fixed with methanol: glacial acetic acid (3:1, volume ratio) for 3 times, and finally resuspended with 0.5-1 mL of fixative. Three to four drops were dropped evenly on a cold wet slide, which was allowed to dry later. It was examined under low magnification phase objective (10 × or 16 ×) to check the cell density and spread of metaphase chromosomes.
WCP-FISH was performed according to the protocol provided by Vysis Inc (IL, USA). Briefly chromosome slides were immersed in denaturation solution (700 mL·L-1 formamide/ 2 × SSC) at 73 °C ± 1 °C in water bath for 5 min and subsequently dehydrated serially in 700 mL·L-1, 850 mL·L-1 and 1000 mL·L-1 ethanol and dried at 45 °C- 50 °C. The WCP probe mix (7 μL of WCP hybridization buffer, 2 μL of purified H2O and 1 μL of WCP paint DNA probe for human chromosome 8 Spectrum GreenTM) was incubated at 73 °C ± 1 °C in water bath for 5 min and placed at 45 °C~50 °C ready for use. The probe mix was applied to the target area and coverslip was immediately applied which was sealed with rubber cement. Slides were placed in a prewarmed humidified box, which were put into a 37 °C incubator for 16 h. Coverslips were removed from slides, which were immediately immersed in 0.4 × SSC/ 3 mL·L-1 NP-40 prewarmed to 73 °C ± 1 °C agitating for 1-3 s. Slides were removed after 2 min and immersed into 2 × SSC/ 1 mL·L-1 NP-40 at ambient temperature agitating for 1-3 s and removed after 5 s to 1 min. Slides were air dried in darkness. 10 μL of DAPI counterstain was applied to the target area of slide and coverslip was applied. Slides were viewed using DAPI/Green Vysis filter set on an optimally performing fluorescence microscope and images were captured using CytoVisionTM Chromosome Analysis System (Applied Imaging).
RESULTS
Selective screening of microcell hybrids and single cell isolation cloning
Three to four weeks after MMCT, viable floating cells were observed among dead cells in selective media containing G418 and HAT. Figure 1. The viable cells had good refraction and round shape while the dead cells had poor refraction and morphologic shrinkage.
Figure 1 Double selection of microcell hybrids in HAT and G418 (10 × 25 magnification).
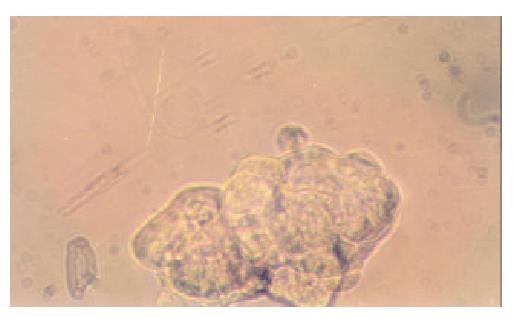
Fifteen microcell hybrid clones were obtained by single cell isolation cloning, which were named neo8/C5F-1~15 respectively. It took about 4 weeks for the progress of enlargement culture serially from 96- to 24-well cell culture plate and finally to T25 flask. Feeder cells are proven unnecessary for the single cell isolation cloning of C5F microcell hybrids. Figure 2 showed the microcell hyrid clone obtained in 96-well plate containing selective media of HAT (1 × ) and G418 (800 μg·mL-1) one week after single cell isolation.
Figure 2 Single-cell isolation cloning in 96-well cell culture plate (10 × 25 magnification).
STS-PCR
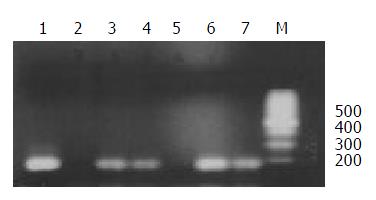
The chromosome donor cell line A9 (neo8) was taken as positive control and the recipient cell line C5F as negative control. DNA extracted from A9 (neo8), C5F and microcell hybrids was used as template. PCR was performed with the primers for D8S277, which is a unique STS located on human chromosome 8p23.3-p22. PCR products, when checked on 2% agarose gel elctrophoresis, were found of the same length with those reported in UniSTS database of NCBI. Figure 3 showed that neo8/C5F-3 had the deletion of D8S277.
Figure 3 STS-PCR Amplification of D8S277.
1: chromosome donor cell line A9(neo8); 2: recipient cell line C5F; 3-7: microcell hybrids neo8/C5F-1-5; M: 100bp ladders.
WCP-FISH
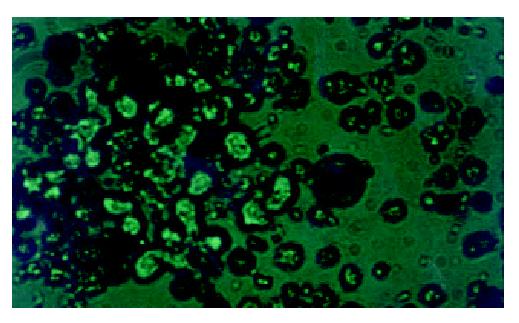
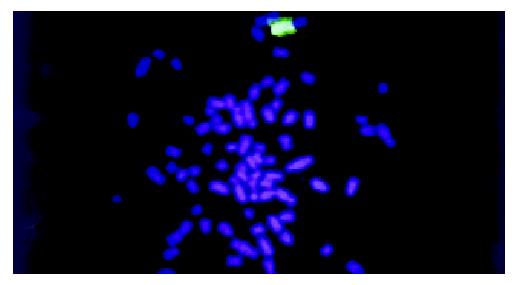
WCP-FISH was performed using whole chromosome painting DNA probe for human chromosome 8 (WCP 8 probe SpectrumGreenTM, Vysis) to confirm its presence. Figure 4 and Figure 5 showed that the human chromosome 8 had been successfully introduced into rat liver cancer cell line. Meanwhile, the recombination of human chromosome 8 with rat chromosome could be observed clearly by comparing the probe with its DAPI counterstain image. This recombination was found to be consistent in different metaphase cells.
Figure 4 WCP-FISH analysis of neo8/C5F microcell hybrids.
Figure 5 WCP-FISH of neo8/C5F microcell hybrids DAPI counterstain.
DISCUSSION
Metastasis is a major problem puzzling both specialists of cancer biology and of clinical oncology. Thousands of cancer patients die of it each year while clinicians can do little to deal with it. The main reason is that the molecular mechanisms of metastasis have not been totally clarified yet. Researchers fascinated by metastasis hope that the discovery of metastasis suppressor genes could shed light on the solution of this problem, while hitherto few of them have been discovered[17,18].
Metastasis suppressor genes suppress the formation of spontaneous, macroscopic metastases without affecting the growth rate of the primary cancer. Until presently, only five genes, nm23, KAI1, KISS1, MKK4 and BrMS1, have been proved to meet this criteria[17,18]. Despite the first metastasis suppressor gene nm23 was discovered by subtractive hybridization, most of encoding regions of metastasis suppressor genes have been discovered with the methodology of MMCT[18], which is thought to be the methodology particularly suitable for the functional localization of metastasis suppressor genes[19]. MMCT is established on somatic cell hybrid. Before MMCT was introduced, researchers had found that somatic cell fusion of metastatic cell line with normal cells could change its metastatic phenotype[20]. Microcells that contain only one single human chromosome are obtained by the micronucleation and enucleation o f human monochromosome donor cells, which include only one intact human chromosome labeled with selective markers such as the neo gene. The target monochromosomes are thereby introduced into recipient cells through the fusion of microcells with recipient cells and the cloning screening of microcell hybrids based on drug resistance conferred. Due to unknown reasons, the introduced chromosome is unstable and random loss or recombination can occur, which result in a series of microcell hybrid clones with different regions of the target chromosome. In this study, the human chromosome 8 introduced into C5F was found to not only have random loss by STS-PCR but also have recombined with rat chromosome by FISH. Those clones in all are called human monochromosome somatic cell panel[21]. In order to judge which region on the target chromosome harbors the gene of deserved function, spontaneous metastasis assay will be performed with these microcell hybrids. Based on the comparison of changes of metastatic phenotype and different chromosome regions each microcell hybrid contains, the target gene can be narrowed down to specific chromosome region.
Most of current researches for the discovery of metastasis suppressor genes focused on the functional localization of metastasis suppressor genes for prostate cancer, melanoma and breast cancer[19,22-31]. However, there hasn’t been any report of such work on liver cancer. Our previous study on the surgical samples of primary liver cancer through comparative genomic hybridization (CGH) showed that the loss of the short arm of human chromosome 8 had correlation with metastasis, which suggested that there could be metastasis suppressor gene(s) on chromosome 8p[6]. This discovery made our present research work imperative. In this study, the highly metastatic rat liver cancer cell line C5F, whose incidence of subcutaneous tumorigenecity is 100% and that of lung metastasis is 89%, was chosen as the recipient. The genetic background of rat facilitated STS-PCR screening of the regions remained on the chromosome introduced, as STS is a unique genetic marker different between species. One STS on human chromosome 8p23.3-22, D8S277, was chosen to confirm the introduction of human chromosome with the genomic DNA of microcell hybrids as templates. The results indicated that the microcell hybrids contained the STS specific for human chromosome 8, which could be taken as a proof for the successful chromosome transfer. Meanwhile some clones had deletions of STS, which showed that random losses of regions on human chromosome had occurred. This phenomenon built the basis for construction of human monochromosome panel. The target chromosome was randomly labeled with the G418 resistance gene, neo, which facilitated the selection of positive microcell hybrid clones. In the mean time, the chromosome donor cell line A9 is hypoxanthine-guanine phosphoribosyl transferase (HGPRT) deficient. Therefore A9 can’t survive in the media with HAT. Double selections of G418 and HAT were applied in this study to exclude the fake clones from either chromosome donor cell line or recipient cell line. Based on the acquisition of drug resistance, we can reasonably infer that the target chromosome has been transferred into C5F. The results of FISH provide the most direct evidence for the successful chromosome transfer. Recombination between human chromosome and rat chromosome was also observed in different metaphase cells, which showed that the recombination had been stabilized. The introduction of human chromosome can cause transient genetic instability, but with the selection the genetic change will become stable, which is the foundation for further analysis. All in all, the human chromosome 8 has been successfully transferred into the highly metastatic rat liver cancer cell line C5F, which builds solid basis for future researches on the discovery of metastasis suppressor genes for liver cancer.
ACKNOWLEDGEMENTS
We are much grateful to Dr. Kumiko Ogawa (First Department of Pathology, Nagoaya City University, Japan) for his generous offer of C5F cell line, Prof. Hong-Xuan Lin (Plant Physiology and Ecology Insitute, Chinese Academy of Science, Shanghai, China) for his warmhearted provision and assistance in the usage of Hitachi high speed centrifuge and Dr. M.Z.Zdzienicka and Wouter Wiegant (Leiden Univerity Medical College, Netherland) for kindly providing us the detailed protocol of MMCT.