Published online Oct 15, 2003. doi: 10.3748/wjg.v9.i10.2258
Revised: June 5, 2003
Accepted: June 12, 2003
Published online: October 15, 2003
AIM: To determine the distribution of cagG gene of Helicobacter pylori (H pylori) isolates cultured from patients with various digestive diseases and its relationship with gastroduodenal diseases.
METHODS: cagG was amplified by polymerase chain reaction in 145 H pylori isolates cultured from patients with chronic gastritis (n = 72), duodenal ulcer (n = 48), gastric ulcer (n = 17), or gastric and duodenal ulcer (n = 8), and the relationship between cagG status and the grade of gastric mucosal inflammation was determined.
RESULTS: cagG was present in 91.7% of the 145 H pylori isolates, with the rates were 90.3%, 93.8%, 88.2% and 100.0%, respectively, in those from patients with chronic gastritis, duodenal ulcer, gastric ulcer, and gastric and duodenal ulcer. There was no significant difference among the four groups (P > 0.05). The average grade of gastric mucosal inflammation in the antrum and corpus was 1.819 ± 0.325 and 1.768 ± 0.312, respectively in cagG positive patients, whereas the average inflammation grade was 1.649 ± 0.297, 1.598 ± 0.278 respectively in cagG negative cases (P > 0.05).
CONCLUSION: cagG gene of H pylori was quite conservative, and most H pylori strains in Chinese patients were cagG positive. cagG status was not related to clinical outcome or the degree of gastric mucosal inflammation. Therefore, cagG can not be used as a single marker for discrimination of H pylori strains with respect to a specific digestive disease.
-
Citation: Xu C, Li ZS, Tu ZX, Xu GM, Gong YF, Man XH. Distribution of
cagG gene inHelicobacter pylori isolates from Chinese patients with different gastroduodenal diseases and its clinical and pathological significance. World J Gastroenterol 2003; 9(10): 2258-2260 - URL: https://www.wjgnet.com/1007-9327/full/v9/i10/2258.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i10.2258
Helicobacter pylori is a well-recognized pathogen that chronically infects more than 50% of the world population. H pylori is associated with the development of acute or chronic gastritis, peptic ulcer diseases, gastric adenocarcinoma and gastric mucosa-associated lymphoid tissue (MALT) lymphoma. Most infected subjects will remain asymptomatic throughout life with only about 20% developing peptic ulcer diseases or gastric carcinoma[1-5]. What determines the outcome of an infection remains unclear. The reasons for these different outcomes of H pylori infection may be related to both bacterial factors and host responses. The major H pylori disease-associated genetic factor is the whole cag pathogenicity island (PAI), which contains 25 open reading frames and at least 30 genes. The cag PAI is associated with increased interleukin (IL)-8 production by gastric epithelial cells[6,7].
The cytotoxin-associated gene A (cagA) is located in the most downstream part of the cag PAI. The presence of this gene or its encoded protein, CagA, has been regarded as a marker for the cag PAI. Many clinical studies have demonstrated that cagA gene or CagA protein is associated with a more severe clinical outcome. CagA was reported to increase the risk of development of duodenal ulcer, atrophic gastritis and gastric adenocarcinoma. In contrast to the cagA-negative patients, gastric mucosal inflammation of cagA-positive patients was more severe[8-10]. cagG is located within the cag PAI upstream of cagA. The distribution of this gene in H pylori strains isolated from Chinese digestive patients and its relation with the gastroduodenal diseases remain unclear. In the present study, a set of specific primers were designed to detect the cagG gene in 145 clinical H pylori strains, and the relationship between cagG and different digestive diseases was determined.
H pylori isolates obtained from 145 patients (80 males, 65 females, aged 18-69 years, mean age 42.5 years old) who underwent upper endoscopy in our department were included in this study. These patients were diagnosed endoscopically as chronic gastritis (n = 72), duodenal ulcer (n = 48), gastric ulcer (n = 17), or gastric and duodenal ulcer (n = 8). Informed consents were obtained from all patients. The standard strains CCUG17874 (NCTC11638) and Tx30a were kindly provided by the Italian IRIS Research Center.
Two antral biopsy specimens taken during endoscopy were immediately cultured on the H pylori selective agar plates with 10% defibrillated sheep blood and antibiotics (Merck Company, Germany) at 37 °C under microaerophilic conditions with 5% O2 10% CO2 and 85% N2 for 3-6 d. The colonies were identified as H pylori if Gram stain morphology and biochemical tests were positive for urease, oxidase and catalase. All stock cultures were preserved at -80 °C in Brucella broth with 20% glycerol, and subcultured for genomic DNA extraction. The passage number of H pylori used in this study averaged six.
Two biopsy specimens each taken from the gastric corpus and antrum endoscopically were used for histopathologic examination to grade the severity of gastritis after they were embedded in paraffin and stained with hematoxylin and eosin. The severity of gastritis (i.e. mononuclear cell and polymorphonuclear leukocyte infiltration) was evaluated, and graded on a scale of 0-3 (i.e. 0 = no, 1 = mild, 2 = moderate, and 3 = marked) according to the updated Sydney system[11].
Subcultured H pylori cells were collected from the agar plates, then genomic DNA was extracted and purified from each H pylori isolate using cetyltrimethyl ammonium bromide (CTAB), phenol-chloroform-isoamyl alcohol, and ethanol precipitation.
The primers to amplify cagG gene and give a 497 base pair (bp) product were designed based on the published gene sequence[12]. cagGF: 5’-GCCATGTTAACACCCCCTAG-3’, and cagGR: 5’-TTAATGCGCTAGAATAGTGC-3’. PCR was performed in an Eppendorf thermal cycler using a PCR kit (Takara, Dalian, China) according to the manufacturer’s instructions. Briefly, the reaction was performed in a total volume of 50 μL containing 20 ng genomic DNA as a template and 200 μM each deoxynucleotide, 1.5U Taq polymerase, 0.4 μM each primer and PCR buffer. The PCR amplification program comprised at 95 °C for 5 min, then 35 cycles at 95 °C for 1 min, at 52 °C for 1 min and at 72 °C for 1 min, followed by at 72 °C for 7 min, then cooled at 4 °C. The PCR products were analyzed on 1.5% agarose gels with ethidium bromide. CCUG17874 was taken as a positive control, Tx30a as a negative control, and deionized water as a blank control.
The data were expressed as the mean ± SD. The t test and χ2 test were used for statistical analysis. A P value < 0.05 was considered to be statistically significant.
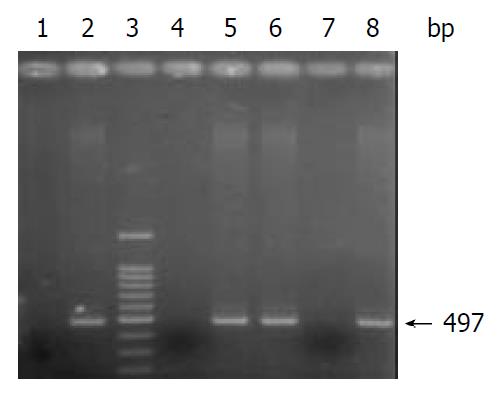
After PCR amplification of the cagG gene, the products were electrophoresed on 1.5% agarose gels, and stained with ethidium bromide. Under ultraviolet light, cagG appeared as a specific band with 497 bp (Figure 1).
H pylori cagG gene was detected in 91.7% of the 145 isolates, the rate was 90.3%, 93.8%, 88.2% and 100.0% in patients with chronic gastritis, duodenal ulcer, gastric ulcer and gastric/duodenal ulcer, respectively (Table 1). There was no significant difference among the groups (P > 0.05).
The average grade of gastric mucosal inflammation in the antrum and corpus was 1.819 ± 0.325 and 1.768 ± 0.312 in cagG positive patients, respectively, whereas the average grade was 1.649 ± 0.297, 1.598 ± 0.278 in cagG negative group, respectively (P > 0.05, both in the antrum and body).
H pylori infects human gastric mucosa which evokes a mucosal inflammatory response by neutrophil recruitment from the microcirculation. Persistent inflammation may lead to the development of digestive diseases such as chronic gastritis, peptic ulcer disease and gastric cancer. Although pathogenicity of H pylori infection is not well understood, there were several putative virulence factors that might contribute to mucosal damage by H pylori infection[13,14].
The cag PAI is an approximately 40-kb cluster of genes in H pylori chromosome, and a quite conservative entity. Many of H pylori strains had an intact cag PAI divided into two regions: cagI in downstream and cagII in upstream, and some with an insert sequence IS605 or IS606[15,16]. There were at least 14 and 16 open reading frames in cagI and cagII, several of which were virulence factors[17]. Several studies suggested that cagA gene could be used as a marker for cag PAI[16]. cagG is another gene in cagI region which is located upstream of cagA gene. The function of this gene is not well known. Recently, Hsu et al[17] reported that an intact cag PAI was identified in 95% and 100% of the strains that possessed cagA and cagG respectively, whereas the cagA and cagG genes were found in 100% and 95% of the strains containing a partial or complete set of cag PAI, indicating that cagA gene is not associated with a complete cag PAI in 5% of the strains, and cannot be considered as an absolute marker for the presence of a complete set of cag PAI, but cagG gene may be a better indicator for the presence of an intact cag PAI.
Extensive studies of cagA gene indicated that CagA protein encoded by cagA gene was associated with severe clinical outcomes, such as peptic ulcer disease and gastric cancer. Therefore, it was considered as a main virulence factor of H pylori[16]. Some reports suggested that the presence of cagE gene within cagI might be related to more severe clinical outcomes. For example, Day et al[18] revealed that H pylori isolates containing cagE were associated with duodenal ulcer in Canadian children. Fallone et al[19] reported that cagE-positive isolates were more prevalent in Canadian adult patients with peptic ulcer or gastric cancer than in those with gastritis only. In the present study, we designed a set of primers to amplify cagG gene of 145 clinical H pylori isolates, and determined the correlation of cagG status with endoscopic presentation, and histological findings. The results showed that cagG was present in 91.7% H pylori isolates examined. 100% of H pylori isolates from patients with gastric and duodenal ulcer were cagG positive, which was higher, but not statistically significant than that in other groups (P > 0.05). Lack of difference in cagG positive rate might be due to patient selection or the relatively small number of patients with gastric and duodenal ulcer. Nevertheless, our study suggested that positive rate of cagG in H pylori was high and cagG was quite conservative in Chinese population, and that there was no difference in the frequencies of cagG-positive isolates among patients with gastritis, duodenal ulcer, gastric ulcer or gastric duodenal ulcer. Our results are supported by the study by Jenks et al[20] who demonstrated that no specific genes within the cag PAI could reliably predict the clinical outcome of H pylori infection in French patients, and also by Hsu et al[17] who concluded that any of the cag PAI genes such as cagE could not predict the clinical presentation in Korean patients.
Hsu et al[17] reported that of the 120 clinical isolates from Korean patients with various gastrointestinal diseases, 86.7% (104/120) were cagG positive. Mizushima et al[21] used PCR and Southern blot to investigate the prevalence of cagG gene in 236 clinical H pylori isolates from Japanese patients, and found that cagG was present in 97% of the isolates. These results were similar with ours. The same Japanese research group[21] further used flow cytometry to assay the ability of H pylori with or without cagG to adhere to KATOIII and ELISA to detect the IL-8 secreted from gastric epithelial cells induced by H pylori. They observed that in comparison with the cagG-positive strains, all cagG-deleted strains decreased adherence to KATOIII cells, and abolished IL-8 induction despite the presence of cagE, which was reported to be essential for IL-8 induction. H pylori genome is known to be diversified and may differ between geographic regions. However, there has no reports so far about cagG gene distribution in the Western countries.
Infection with cagA positive H pylori induces stronger gastric chemokine mRNA expression such as IL-8 in the antral mucosa, which may be relevant to the increased mucosal damage associated with cagA positive H pylori infection. The levels of the chemokines were correlated with cellular infiltration in the antrum and inflammation of the gastric mucosa[22,23]. We compared the severity of gastric mucosal inflammation in the antrum and corpus in cagG-positive and cagG-negative patients, and observed that, the average grade of inflammation was only slightly higher in cagG-positive group than that in cagG negative group both in the antrum and in the corpus (P > 0.05). Therefore, the cagG status has no relation to the severity of gastritis.
In conclusion, cagG gene was quite conservative in clinical H pylori isolates from Chinese patients with different gastroduodenal diseases, since most H pylori isolates were cagG positive. There was no difference in the frequency of cagG -positive isolates among patients with different diseases. The cagG status was not related to gastric mucosal inflammation grade. Therefore, cagG cannot reliably predict the clinical and histological outcomes.
Edited by Xia HHX
| 1. | Kapadia CR. Gastric atrophy, metaplasia, and dysplasia: a clinical perspective. J Clin Gastroenterol. 2003;36:S29-36; discussion S61-62. [PubMed] |
| 2. | Matsuhisa TM, Yamada NY, Kato SK, Matsukura NM. Helicobacter pylori infection, mucosal atrophy and intestinal metaplasia in Asian populations: a comparative study in age-, gender- and endoscopic diagnosis-matched subjects. Helicobacter. 2003;8:29-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 3. | Kelley JR, Duggan JM. Gastric cancer epidemiology and risk factors. J Clin Epidemiol. 2003;56:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 505] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 4. | McColl KE, El-Omar E. How does H. pylori infection cause gastric cancer? Keio J Med. 2002;51 Suppl 2:53-56. [PubMed] |
| 5. | Sepulveda AR, Graham DY. Role of Helicobacter pylori in gastric carcinogenesis. Gastroenterol Clin North Am. 2002;31:517-535, x. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Figura N, Valassina M. Helicobacter pylori determinants of pathogenicity. J Chemother. 1999;11:591-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 7. | Mégraud F. Impact of Helicobacter pylori virulence on the outcome of gastroduodenal diseases: lessons from the microbiologist. Dig Dis. 2001;19:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Audibert C, Burucoa C, Janvier B, Fauchère JL. implication of the structure of the Helicobacter pylori cag pathogenicity island in induction of interleukin-8 secretion. Infect Immun. 2001;69:1625-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Li CQ, Pignatelli B, Ohshima H. Increased oxidative and nitrative stress in human stomach associated with cagA+ Helicobacter pylori infection and inflammation. Dig Dis Sci. 2001;46:836-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 10. | Perez-Perez GI, Peek RM, Legath AJ, Heine PR, Graff LB. The role of CagA status in gastric and extragastric complications of Helicobacter pylori. J Physiol Pharmacol. 1999;50:833-845. [PubMed] |
| 11. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [PubMed] |
| 12. | Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relation between clinical presentation, Helicobacter pylori density, interleukin 1beta and 8 production, and cagA status. Gut. 1999;45:804-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 87] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Suzuki H, Masaoka T, Miyazawa M, Suzuki M, Miura S, Ishii H. Gastric mucosal response to Helicobacter pylori. Keio J Med. 2002;51 Suppl 2:40-44. [PubMed] |
| 14. | McGee DJ, Mobley HL. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr Top Microbiol Immunol. 1999;241:155-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 49] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Zhang M, Zhang J, He L, Guo H, Yin Y, Zhou Z. [Dissemination of insertion sequences IS605, IS606 among clinical isolates of Helicobacter pylori in China]. Zhonghua Liuxingbingxue Zazhi. 2002;23:366-369. [PubMed] |
| 16. | Owen RJ, Peters TM, Varea R, Teare EL, Saverymuttu S. Mo-lecular epidemiology of Helicobacter pylori in England: prevalence of cag pathogenicity island markers and IS605 presence in rela-tion to patient age and severity of gastric disease. FEMS Immunol Med Microbiol. 2001;30:65-71. [RCA] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 17. | Hsu PI, Hwang IR, Cittelly D, Lai KH, El-Zimaity HM, Gutierrez O, Kim JG, Osato MS, Graham DY, Yamaoka Y. Clinical presentation in relation to diversity within the Helicobacter pylori cag pathogenicity island. Am J Gastroenterol. 2002;97:2231-2238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Day AS, Jones NL, Lynett JT, Jennings HA, Fallone CA, Beech R, Sherman PM. cagE is a virulence factor associated with Helicobacter pylori-induced duodenal ulceration in children. J Infect Dis. 2000;181:1370-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Fallone CA, Barkun AN, Göttke MU, Best LM, Loo VG, Veldhuyzen van Zanten S, Nguyen T, Lowe A, Fainsilber T, Kouri K. Association of Helicobacter pylori genotype with gastroesophageal reflux disease and other upper gastrointestinal diseases. Am J Gastroenterol. 2000;95:659-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Jenks PJ, Mégraud F, Labigne A. Clinical outcome after infection with Helicobacter pylori does not appear to be reliably predicted by the presence of any of the genes of the cag pathogenicity island. Gut. 1998;43:752-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 88] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 21. | Mizushima T, Sugiyama T, Kobayashi T, Komatsu Y, Ishizuka J, Kato M, Asaka M. Decreased adherence of cagG-deleted Helicobacter pylori to gastric epithelial cells in Japanese clinical isolates. Helicobacter. 2002;7:22-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 22. | Shimoyama T, Everett SM, Dixon MF, Axon AT, Crabtree JE. Chemokine mRNA expression in gastric mucosa is associated with Helicobacter pylori cagA positivity and severity of gastritis. J Clin Pathol. 1998;51:765-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Yamaoka Y, Kita M, Kodama T, Sawai N, Tanahashi T, Kashima K, Imanishi J. Chemokines in the gastric mucosa in Helicobacter pylori infection. Gut. 1998;42:609-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 162] [Article Influence: 6.0] [Reference Citation Analysis (0)] |