Published online Oct 15, 2003. doi: 10.3748/wjg.v9.i10.2221
Revised: June 12, 2003
Accepted: June 19, 2003
Published online: October 15, 2003
AIM: To investigate the association between the polymorphism of HLA-DRB1, -DQA1 and -DQB1 alleles and viral hepatitis B.
METHODS: HLA-DRB1, -DQA1 and -DQB1 alleles in 54 patients with chronic hepatitis B, 30 patients with acute hepatitis B and 106 normal control subjects were analyzed by using the polymerase chain reaction/sequence specific primer (PCR/SSP) technique.
RESULTS: The allele frequency of HLA-DRB1*0301 in the chronic hepatitis B group was markedly higher than that in the normal control group (17.31% vs 5.67%), there was a significant correlation between them (χ2 = 12.3068, Pc = 0.0074, RR = 4.15). The allele frequency of HLA-DQA1*0501 in the chronic hepatitis B group was significantly higher than that in the normal control group (25.96% vs 13.68%), there was a significant correlation between them (χ2 = 9.2002, Pc = 0.0157, RR = 2.87). The allele frequency of HLA-DQB1*0301 in the chronic hepatitis B group was notably higher than that in the normal control group (35.58% vs 18.87%), there was a significant correlation between them (χ2 = 15.5938, Pc = 0.0075, RR = 4.07). The allele frequency of HLA-DRB1*1101/1104 in the chronic hepatitis B group was obviously lower than that in the normal control group (0.96% vs 13.33%), there was a significant correlation between them (χ2 = 11.9206, Pc = 0.0145, RR = 18.55). The allele frequency of HLA-DQA1*0301 in the chronic hepatitis B group was remarkably lower than that in the normal control group (14.42% vs 30%), there was a significant correlation between them (χ2 = 8.7396, Pc = 0.0167, RR = 0.35).
CONCLUSION: HLA-DRB1*0301, HLA-DQA1*0501 and HLA-DQB1*0301 are closely related with susceptibility to chronic hepatitis B, and HLA-DRB1*1101/1104 and HLA-DQA1*0301 are closely related with resistance to chronic hepatitis B. These findings suggest that host HLA class II gene is an important factor determining the outcome of HBV infection.
- Citation: Jiang YG, Wang YM, Liu TH, Liu J. Association between HLA class II gene and susceptibility or resistance to chronic hepatitis B. World J Gastroenterol 2003; 9(10): 2221-2225
- URL: https://www.wjgnet.com/1007-9327/full/v9/i10/2221.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i10.2221
The progression of hepatitis B virus (HBV) infection may be influenced by a number of factors including the viral genotype and the level of viremia, but these factors alone do not account for the variability in outcome. There is an increasing awareness that host factors are involved. A great deal of evidences suggest that both cellular and humoral immune responses are required for viral clearance[1-3]. Polymorphisms of human leukocyte antigen (HLA) influence immune responses. Variability in immune response is often associated with HLA polymorphism. HLA genotype of an individual may influence the progression of HBV infection. Patients who have successfully recovered from acute hepatitis B develop strong HLA classes I and II restricted T cell response, whereas these responses are weak or absent in patients with chronic hepatitis B[4,5]. In the present study, we have analyzed the polymorphism of HLA-DRB1, -DQA1 and -DQB1 alleles in patients with chronic and acute hepatitis B and healthy controls using the polymerase chain reaction with sequence specific primers (PCR/SSP). This study aimed at investigating whether these alleles might be associated with susceptibility or resistance to chronic hepatitis B.
Fifty-two patients (43 males, 9 females, mean age: 33.46 years) with chronic hepatitis B and 30 patients (24 males, 6 females, mean age: 33.25 years) with acute hepatitis B, and 106 healthy blood donors (88 males, 18 females, mean age: 31.27 years) were included in this study. All the patients were from the Institute of Infectious Diseases, Southwest Hospital of Third Military Medical University. The diagnosis of all the cases was made according to the criteria established on the Viral Hepatitis Conference held in 2000. All the patients and controls were Chinese Han people without relatives from Chongqing. The subjects were divided into chronic hepatitis B group, acute hepatitis B group and healthy control group.
The polymorphisms of HLA-DRB1, -DQA1 and -DQB1 alleles were assessed by PCR/SSP technique. HLA-DRB1, -DQA1 and -DQB1 loci of specific PCR primers were designed by Olerup et al[6,7], and synthesized by Shanghai Branch, Canadian Sangon Company. The primers amplifying human growth hormone gene (5’-primer: 5’-GCCTTCCCAACCATTCCCTTA-3’, 3’-primer: 5’-TCACGGATTTCTGTTGTGTTTC-3’) were synthesized by Shanghai Branch, Canadian Sangon Company. Taq DNA polyenzyme and dNTP were purchased from Shanghai Branch, Canadian Sangon Company, pBR322/Hand III marker and the ReadyPCRTM whole blood genomic DNA purification system were provided by Sino-American Biotechnology Company.
DNA extraction Genomic DNA was extracted from peripheral blood by using the Ready PCRTM whole blood genomic DNA purification system.
A total amount of 25 μL PCR reaction solution contained 8 pmoles of each sequence specific primer (3.2 μL), 0.8 pmoles of each internal control primer (0.32 μL), 80-100 ng of genomic DNA (2 μL), 2.5 μL of 10 × buffer, 25 mmol/L of MgCl2 (2.5 μL), 10 mmol/L of dNTP (1 μL), 5 unit/μL of Taq polymerase (0.5 μL) and 13 μL of demonized H2O. The PCR cycling parameters of HLA-DRB1 alleles were as follows: predenaturation at 94 °C for 5 min, denaturation at 94 °C for 50 s, annealing at 65 °C for 1 min, extension at 72 °C for 1 min, repetition for 30 cycles and final extension at 72 °C for 5 min. The PCR cycling parameters of HLA-DQA1 and -DQB1 alleles were as follows: predenaturation at 94 °C for 4 min, denaturation at 94 °C for 1 min, annealing at 65 °C for 1 min, extension at 72 °C for 1 min, repetition for 30 cycles and final extension at 72 °C for 2 min. In each PCR reaction a primer pair was included to amplify the human growth hormone gene, which functioned as an internal positive amplification control and gave rise to a 429 base pair fragment.
Detection of PCR products PCR products were loaded in 2% agars gel containing 0.5 μg/mL of ethidium bromide, electrophoresed for 20 min at 15 V/cm, examined under ultraviolet light. The individual alleles were assigned for the specific pattern of appropriately sized bands.
Allele frequencies of HLA-DRB1, -DQA1 and -DQB1 were calculated by direct count. AF for the study group was compared with that for the control group using Chi-square (χ2) test. The Fisher’s exact test was used when χ2 value exceeded 3.84, the P values were corrected for the number of alleles (corrected P = Pc). Relative risk frequencies (RR) were calculated according to Wolf formula.
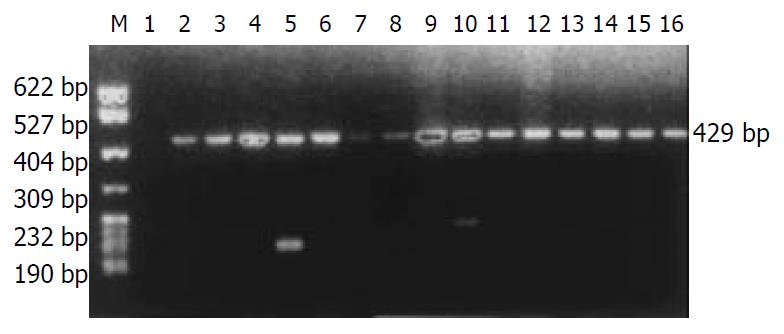
The distribution of HLA-DRB1 alleles is shown in Table 1. The allele frequencies of HLA-DRB1*0301 in the chronic hepatitis B group (17.31%) were markedly higher than those in the normal control group (5.67%), there was a significant correlation between them (χ2 = 12.3068, Pc = 0.0074, RR = 4.15). The allele frequencies of HLA-DRB1*1101/1104 in the chronic hepatitis B group (0.96%) were significantly lower than those in the acute hepatitis B group (13.33%), with significant correlation between them (χ2 = 11.9206, Pc = 0.0145, RR = 18.55). The data of electrophoresis of HLA-DRB1 alleles amplification are shown in Figure 1.
| HLA-DRB1 allele | Normal control (n = 106) | Chronic hepatitis B (n = 52) | Acute hepatitis B (n = 30) | |||
| PN | AF | PN | AF | PN | AF | |
| 0101/0103 | 1 | 0.47 | 1 | 0.96 | 1 | 1.67 |
| 0301* | 12 | 5.66 | 18 | 17.31 | 6 | 10.00 |
| 0401/0411 | 24 | 11.32 | 13 | 12.50 | 7 | 11.67 |
| 0701/0702 | 11 | 5.19 | 8 | 7.69 | 4 | 6.67 |
| 0801/0804 | 9 | 4.25 | 6 | 5.77 | 3 | 5.00 |
| 0901 | 32 | 15.09 | 16 | 15.39 | 8 | 13.33 |
| 1001 | 2 | 0.94 | 2 | 1.92 | 1 | 1.67 |
| 1101/1104** | 13 | 6.13 | 1 | 0.96 | 8 | 13.33 |
| 1201/1202 | 34 | 16.04 | 15 | 14.42 | 8 | 13.33 |
| 1301/1302 | 4 | 1.89 | 1 | 0.96 | 1 | 1.67 |
| 1303/1304 | 1 | 0.47 | 1 | 0.96 | 1 | 1.67 |
| 1401, 1404 | 14 | 6.60 | 6 | 5.77 | 4 | 6.67 |
| 1402, 1403 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| 1501/1502 | 34 | 16.04 | 11 | 10.58 | 5 | 8.33 |
| 1601/1602 | 13 | 6.13 | 2 | 1.92 | 1 | 1.67 |
| Blank | 8 | 3.77 | 3 | 2.89 | 2 | 3.33 |
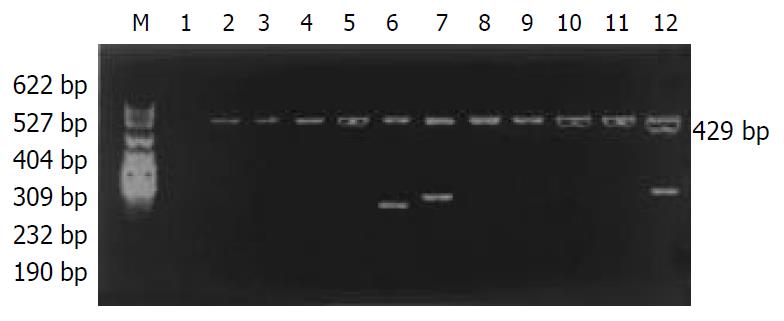
The distribution of HLA-DQA1 alleles is shown in Table 2. The allele frequencies of HLA-DQA1*0501 in the chronic hepatitis B group (25.96%) were markedly higher than those in the normal control group (13.68%), there was a significant correlation between them (χ2 = 9.2002, Pc = 0.0157, RR = 2.87). The allele frequencies of HLA-DQA1*0301 in the chronic hepatitis B group (14.42%) was significantly lower than those in the acute hepatitis B group (30%), there was a significant correlation between them (χ2 = 7.6781, Pc = 0.0388, RR = 3.70). The data of electrophoresis of HLA-DQA1 alleles amplification are shown in Figure 2.
| HLA-DQA1 allele | Normal control (n = 106) | Chronic hepatitis B (n = 52) | Acute hepatitis B (n = 30) | |||
| PN | AF | PN | AF | PN | AF | |
| 0101 | 17 | 8.02 | 9 | 8.65 | 4 | 6.67 |
| 0102 | 45 | 21.23 | 22 | 21.15 | 12 | 20.00 |
| 0103 | 9 | 4.25 | 5 | 4.81 | 2 | 3.33 |
| 0104 | 3 | 1.42 | 1 | 0.96 | 1 | 1.67 |
| 0201 | 7 | 3.30 | 3 | 2.88 | 1 | 1.67 |
| 0301* | 57 | 26.89 | 15 | 14.42 | 18 | 30.00 |
| 0302 | 1 | 0.47 | 0 | 0.00 | 0 | 0.00 |
| 0401 | 2 | 0.49 | 1 | 0.96 | 1 | 1.67 |
| 0501** | 29 | 13.68 | 27 | 25.96 | 10 | 16.67 |
| 0601 | 23 | 10.85 | 12 | 11.54 | 6 | 10.00 |
| Blank | 19 | 8.96 | 9 | 8.65 | 5 | 8.33 |
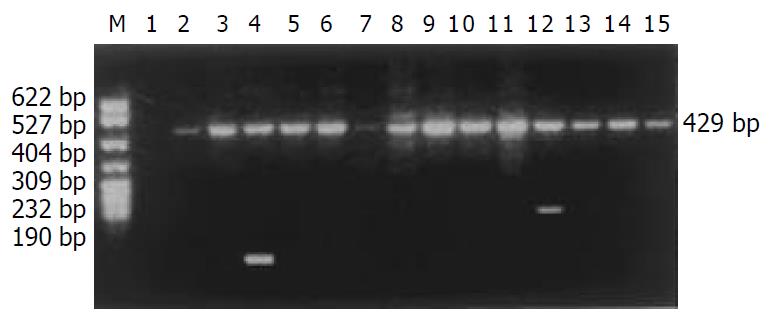
The distribution of HLA-DQB1 alleles is shown in Table 3. The allele frequencies of HLA-DQB1*0301 allele in the chronic hepatitis B group (35.58%) were markedly higher than those in the normal control group (18.87%), there was a significant correlation between them (χ2 = 15.5938, Pc = 0.0075, RR = 4.07). The data of electrophoresis of HLA-DQB1 alleles amplification are shown in Figure 3.
| HLA-DQB1 allele | Normal control (n = 106) | Chronic hepatitis B (n = 52) | Acute hepatitis B (n = 30) | |||
| PN | AF | PN | AF | PN | AF | |
| 0201 | 23 | 10.85 | 10 | 9.62 | 6 | 10.00 |
| 0301* | 40 | 18.87 | 37 | 35.58 | 16 | 26.67 |
| 0302 | 14 | 6.61 | 6 | 5.77 | 3 | 5.00 |
| 0303 | 35 | 16.51 | 15 | 14.42 | 10 | 16.67 |
| 0401 | 11 | 5.19 | 5 | 4.81 | 3 | 5.00 |
| 0402 | 2 | 0.94 | 1 | 0.96 | 1 | 1.67 |
| 0501 | 9 | 4.25 | 3 | 2.88 | 2 | 3.33 |
| 0502 | 20 | 9.43 | 7 | 6.73 | 3 | 5.00 |
| 0503 | 6 | 2.83 | 2 | 1.92 | 1 | 1.67 |
| 0601 | 20 | 9.43 | 7 | 6.73 | 7 | 11.67 |
| 0602 | 12 | 5.66 | 4 | 3.85 | 3 | 5.00 |
| 0603 | 5 | 2.36 | 2 | 1.92 | 1 | 1.67 |
| 0604 | 7 | 3.30 | 2 | 1.92 | 2 | 3.33 |
| Blank | 8 | 3.77 | 3 | 2.89 | 2 | 3.33 |
Host and viral factors undoubtedly influence the clinical expression and behavior of chronic hepatitis B. Attempts to explain the clinical expression and the behavior of chronic hepatitis B by viral factors have shown the importance of viral genotypes and viraemia level for the clinical presentation. However, there remain large inconsistencies, and it is very likely that immune response to hepatitis B virus (HBV) of the host can modify disease outcome[8-10]. HLA is a critical genetic factor that determines individual variations of immune response. The ternary structure of HLA molecules and their roles in the control of immune response have been clearly elucidated. There are many reports about statistical associations between HLA and diseases. HLA gene contributes to the host response against HBV[11-28]. Individuals with different HLA types may differ in susceptibility or resistance to disease[29-35], and associations between HLA polymorphism and susceptibility or resistance to diseases have been identified.
Researches on the correlation between HLA and hepatitis B have been performed for many years. Traditional serological method was used in some investigations, but it has become obsolete and inaccurate. To have a better understanding of the disease, correlation between hepatitis B and HLA should be further studied using nucleotide-typing techniques. Therefore, in the present study, we examined the HLA-DRB1, -DQA1 and -DQB1 alleles by PCR/SSP technique in patients with hepatitis B in an attempt to investigate the association between the polymerase of HLA class II gene and hepatitis B. Fourteen HLA-DRB1 alleles, ten HLA-DQA1 alleles and thirteen HLA-DQB1 alleles were detected. The allele frequencies of HLA-DRB1, -DQA1 and -DQB1 in healthy individuals tallied with genetic characteristics of the Han people in southern region of China.
A previous study showed that the allele frequencies of HLA-B8, DR3, A30, DQA1*0501 in patients with chronic hepatitis B were markedly increased, suggesting that these alleles are associated with chronic hepatitis B[36]. Thio et al[37] found that HBV persistence was significantly associated with two class II alleles, DQA1*0501 (OR = 2.6) and DQB1*0301 (OR = 3.9), the two-locus haplotype consisted of these same two alleles (OR = 3) and the three-locus haplotype consisted of DQA1*0501, DQB1*0301 and DRB1*1102 (OR = 10.7). The study by Shen et al[38] suggested that the susceptibility to chronic hepatitis B was strongly associated with HLA-DRB1*10 allele in northern Chinese patients. In the present study, we found that the allele frequencies of HLA-DRB1*0301, -DQA1*0501 and -DQB1*0301 in the chronic hepatitis B group were markedly higher than those in the normal control group, there was a significant correlation between them (Table 1, Table 2 and Table 3). These findings suggest that HLA-DRB1*0301, -DQA1*0501 and -DQB1*0301 are closely associated with the susceptibility to chronic hepatitis B, and may be the susceptible gene.
Cotrina et al[39] analyzed the HLA-DRB1 genotype in a series of patients with chronic hepatitis B and acute hepatitis B, which further confirmed that HLA-DRB1*1301 and -DRB1*1302 alleles were associated with the clearance of HBV infection and protected people against chronic hepatitis B. Diepolder et al[40] found that a strong virus-specific CD4+ and CD8+ T lymphocyte response to hepatitis B virus was associated with viral clearance, patients with acute hepatitis B carrying HLA-DR13 had a more vigorous CD4+ T cell response to HBV core than patients not carrying HLA-DR13, suggesting that HLA-DR13 is associated with a self-limited course of HBV infection, and the beneficial effect of HLA-DR13 alleles on the outcome of HBV infection could be explained by a more vigorous HBV core-specific CD4+ T cell response, which might be either due to a more proficient antigen presentation by HLA-DR13 molecules themselves or due to a linked polymorphism in a neighboring immunoregulatory gene. In the present study, we found that the allele frequencies of HLA-DRB1*1101/1104 and HLA-DQA1*0301 in the chronic hepatitis B group were markedly lower than those in the acute hepatitis B group, there was a significant correlation between them (Table 1 and Table 2). These findings suggest that HLA-DRB1*1101/1104 and -DQA1*0301 are closely associated with the resistance to chronic hepatitis B, and may be the resistant gene.
The results of the present study suggest that HLA-DRB1*0301, -DQA1*0501 and -DQB1*0301 may be the susceptible gene, and HLA-DRB1*1101/1104 and -DQA1*0301 may be the resistant genes to chronic hepatitis B, and that host HLA class II gene is an important factor determining the outcome of HBV infection, which will give some new clues to the study of pathogenesis of chronic hepatitis B.
Edited by Wang XL
| 1. | Takayama T, Sekine T, Makuuchi M, Yamasaki S, Kosuge T, Yamamoto J, Shimada K, Sakamoto M, Hirohashi S, Ohashi Y. Adoptive immunotherapy to lower postsurgical recurrence rates of hepatocellular carcinoma: a randomised trial. Lancet. 2000;356:802-807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 630] [Cited by in F6Publishing: 626] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 2. | Chiari R, Hames G, Stroobant V, Texier C, Maillère B, Boon T, Coulie PG. Identification of a tumor-specific shared antigen derived from an Eph receptor and presented to CD4 T cells on HLA class II molecules. Cancer Res. 2000;60:4855-4863. [PubMed] [Cited in This Article: ] |
| 3. | Feinmesser M, Sulkes A, Morgenstern S, Sulkes J, Stern S, Okon E. HLA-DR and beta 2 microglobulin expression in medullary and atypical medullary carcinoma of the breast: histopathologically similar but biologically distinct entities. J Clin Pathol. 2000;53:286-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Zhang SL, Liu M, Zhu J, Chai NL. Predominant Th-2 immune response and chronic hepatitis B virus infection. Shijie Huaren Xiaohua Zazhi. 1999;7:513-515. [Cited in This Article: ] |
| 5. | Chen WN, Oon CJ. Mutation "hot spot" in HLA class I-restricted T cell epitope on hepatitis B surface antigen in chronic carriers and hepatocellular carcinoma. Biochem Biophys Res Commun. 1999;262:757-761. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 6. | Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alterna-tive to serological DR typing in clinical practice including do-nor-recipient matching in cadaveric transplantation. Tissue Anti-gens. 1992;39:225-235. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1332] [Cited by in F6Publishing: 1379] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 7. | Olerup O, Aldener A, Fogdell A. HLA-DQB1 and DQA1 typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Tissue Antigens. 1993;41:119-134. [DOI] [Cited in This Article: ] [Cited by in Crossref: 415] [Cited by in F6Publishing: 422] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 8. | Jiang YG, Li QF, Mao Q, WangYM . Primary human fetal hepa-tocytes with HBV infection in vitro. Shijie huaren Xiaohua Zazhi. 2000;8:403-405. [Cited in This Article: ] |
| 9. | Sing G, Butterworth L, Chen X, Bryant A, Cooksley G. Composition of peripheral blood lymphocyte populations during different stages of chronic infection with hepatitis B virus. J Viral Hepat. 1998;5:83-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Cao T, Meuleman P, Desombere I, Sällberg M, Leroux-Roels G. In vivo inhibition of anti-hepatitis B virus core antigen (HBcAg) immunoglobulin G production by HBcAg-specific CD4(+) Th1-type T-cell clones in a hu-PBL-NOD/SCID mouse model. J Virol. 2001;75:11449-11456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Du YP, Deng CS, Lu DY, Huang MF, Guo SF, Hou W. The relation between HLA-DQA1 genes and genetic susceptibility to duodenal ulcer in Wuhan Hans. World J Gastroenterol. 2000;6:107-110. [PubMed] [Cited in This Article: ] |
| 12. | Ding HL, Cheng H, Fu ZZ, Deng QL, YanLAST_NAME& gt; Tang Yan L, Yan T. The relationship of lmp2 and DR3 genes with susceptibility to type I diabetes mellitus in south China Han population. World J Gastroenterol. 2000;6:111-114. [PubMed] [Cited in This Article: ] |
| 13. | Lin J, Deng CS, Sun J, Zheng XG, Huang X, Zhou Y, Xiong P, Wang YP. HLA-DRB1 allele polymorphisms in genetic susceptibility to esophageal carcinoma. World J Gastroenterol. 2003;9:412-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 9] [Cited by in F6Publishing: 11] [Article Influence: 0.5] [Reference Citation Analysis (1)] |
| 14. | Pu J, Yang XS, Zhang YL, Pan LJ, Zhou DY. Expression of HLA-DR in epithelie around lymphofollicle of human gastrointestinal mucosa. Shijie huaren Xiaohua Zazhi. 2000;8:706-707. [Cited in This Article: ] |
| 15. | Zhai SH, Liu JB, Zhu P, Wang YH. CD54, CD80, CD86 and HLA-ABC expressions in liver cirrhosis and hepatocarcinoma. Shijiehuaren Xiaohua Zazhi. 2000;8:292-295. [Cited in This Article: ] |
| 16. | Qu S, Li QF, Deng YZ, Zhang JM, Zhang J. Cloning and expression of HLA-B7 gene. World J Gastroenterol. 1999;5:345-348. [PubMed] [Cited in This Article: ] |
| 17. | Asti M, Martinetti M, Zavaglia C, Cuccia MC, Gusberti L, Tinelli C, Cividini A, Bruno S, Salvaneschi L, Ideo G. Human leukocyte antigen class II and III alleles and severity of hepatitis C virus-related chronic liver disease. Hepatology. 1999;29:1272-1279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Barrett S, Ryan E, Crowe J. Association of the HLA-DRB1*01 allele with spontaneous viral clearance in an Irish cohort infected with hepatitis C virus via contaminated anti-D immunoglobulin. J Hepatol. 1999;30:979-983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Lechmann M, Schneider EM, Giers G, Kaiser R, Dumoulin FL, Sauerbruch T, Spengler U. Increased frequency of the HLA-DR15 (B1*15011) allele in German patients with self-limited hepatitis C virus infection. Eur J Clin Invest. 1999;29:337-343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Mangia A, Gentile R, Cascavilla I, Margaglione M, Villani MR, Stella F, Modola G, Agostiano V, Gaudiano C, Andriulli A. HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J Hepatol. 1999;30:984-989. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 84] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Chang KM, Gruener NH, Southwood S, Sidney J, Pape GR, Chisari FV, Sette A. Identification of HLA-A3 and -B7-restricted CTL response to hepatitis C virus in patients with acute and chronic hepatitis C. J Immunol. 1999;162:1156-1164. [PubMed] [Cited in This Article: ] |
| 22. | Aaltonen LM, Partanen J, Auvinen E, Rihkanen H, Vaheri A. HLA-DQ alleles and human papillomavirus DNA in adult-onset laryngeal papillomatosis. J Infect Dis. 1999;179:682-685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Harcourt G, Hellier S, Bunce M, Satsangi J, Collier J, Chapman R, Phillips R, Klenerman P. Effect of HLA class II genotype on T helper lymphocyte responses and viral control in hepatitis C virus infection. J Viral Hepat. 2001;8:174-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Zhou HC, Xu DZ, Wang XP, Zhang JX, Huang Y, Yan YP, Zhu Y, Jin BQ. Identification of the epitopes on HCV core protein recognized by HLA-A2 restricted cytotoxic T lymphocytes. World J Gastroenterol. 2001;7:583-586. [PubMed] [Cited in This Article: ] |
| 25. | Ma X, Qiu DK. Relationship between autoimmune hepatitis and HLA-DR4 and DRbeta allelic sequences in the third hypervariable region in Chinese. World J Gastroenterol. 2001;7:718-721. [PubMed] [Cited in This Article: ] |
| 26. | Godkin A, Jeanguet N, Thursz M, Openshaw P, Thomas H. Characterization of novel HLA-DR11-restricted HCV epitopes reveals both qualitative and quantitative differences in HCV-specific CD4+ T cell responses in chronically infected and non-viremic patients. Eur J Immunol. 2001;31:1438-1446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 27. | Bosi I, Ancora G, Mantovani W, Miniero R, Verucchi G, Attard L, Venturi V, Papa I, Sandri F, Dallacasa P. HLA DR13 and HCV vertical infection. Pediatr Res. 2002;51:746-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Hüe S, Cacoub P, Renou C, Halfon P, Thibault V, Charlotte F, Picon M, Rifflet H, Piette JC, Pol S. Human leukocyte antigen class II alleles may contribute to the severity of hepatitis C virus-related liver disease. J Infect Dis. 2002;186:106-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | McDermott AB, Cohen SB, Zuckerman JN, Madrigal JA. Human leukocyte antigens influence the immune response to a pre-S/S hepatitis B vaccine. Vaccine. 1999;17:330-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | McDermott AB, Madrigal JA, Sabin CA, Zuckerman JN, Cohen SB. The influence of host factors and immunogenetics on lymphocyte responses to Hepagene vaccination. Vaccine. 1999;17:1329-1337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Wang FS, Xing LH, Liu MX, Zhu CL, Liu HG, Wang HF, Lei ZY. Dysfunction of peripheral blood dendritic cells from patients with chronic hepatitis B virus infection. World J Gastroenterol. 2001;7:537-541. [PubMed] [Cited in This Article: ] |
| 32. | Sobao Y, Sugi K, Tomiyama H, Saito S, Fujiyama S, Morimoto M, Hasuike S, Tsubouchi H, Tanaka K, Takiguch M. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J Hepatol. 2001;34:922-929. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Thimme R, Chang KM, Pemberton J, Sette A, Chisari FV. Degenerate immunogenicity of an HLA-A2-restricted hepatitis B virus nucleocapsid cytotoxic T-lymphocyte epitope that is also presented by HLA-B51. J Virol. 2001;75:3984-3987. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Pellegris G, Ravagnani F, Notti P, Fissi S, Lombardo C. B and C hepatitis viruses, HLA-DQ1 and -DR3 alleles and autoimmunity in patients with hepatocellular carcinoma. J Hepatol. 2002;36:521-526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 35. | Desombere I, Gijbels Y, Verwulgen A, Leroux-Roels G. Characterization of the T cell recognition of hepatitis B surface antigen (HBsAg) by good and poor responders to hepatitis B vaccines. Clin Exp Immunol. 2000;122:390-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 36. | Chen DF, Kliem V, Endres W, Brunkhorst R, Tillmann HL, Koch KM, Manns MP, Stangel W. Relationship between human leuko-cyte antigen determinants and courses of hepatitis B virus infec-tion in Caucasian patients with end-stage renal disease. Scand J Gastroenterol. 1996;31:1211-1215. [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 37. | Thio CL, Carrington M, Marti D, O'Brien SJ, Vlahov D, Nelson KE, Astemborski J, Thomas DL. Class II HLA alleles and hepatitis B virus persistence in African Americans. J Infect Dis. 1999;179:1004-1006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 85] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Shen JJ, Ji Y, Gu XL, Huang RJ, Sun YP. The association of HLA-DRB1 with chronic hepatitis B in Chinese patients. Zhonghua Weishengwuxue He Mianyixue Zazhi. 1999;19:58-59. [Cited in This Article: ] |
| 39. | Cotrina M, Buti M, Jardí R, Rodríguez-Frías F, Campins M, Esteban R, Guardia J. [Study of HLA-II antigens in chronic hepatitis C and B and in acute hepatitis B]. Gastroenterol Hepatol. 1997;20:115-118. [PubMed] [Cited in This Article: ] |
| 40. | Diepolder HM, Jung MC, Keller E, Schraut W, Gerlach JT, Grüner N, Zachoval R, Hoffmann RM, Schirren CA, Scholz S. A vigorous virus-specific CD4+ T cell response may contribute to the association of HLA-DR13 with viral clearance in hepatitis B. Clin Exp Immunol. 1998;113:244-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 66] [Article Influence: 2.5] [Reference Citation Analysis (0)] |