Published online Dec 15, 2002. doi: 10.3748/wjg.v8.i6.1117
Revised: February 26, 2002
Accepted: March 5, 2002
Published online: December 15, 2002
AIM: To study the effects of hydrogen peroxide on mitochondrial gene expression of intestinal epithelial cells in in vitro model of hydrogen peroxide-stimulated SW-480 cells.
METHODS: RNA of hydrogen peroxide-induced SW-480 cells was isolated, and reverse-transcriptional polymerase chain reaction was performed to study gene expression of ATPase subunit 6, ATPase subunit 8, cytochrome c oxidase subunit I (COI), cytochrome coxidase subuit II (COII) and cytochrome c oxidase subunit III (COIII). Mitochondria were isolated and activities of mitochondrial cytochrome c oxidase and ATPase were also measured simultaneously.
RESULTS: Hydrogen peroxide led to differential expression of mitochondrial genes with some genes up-regulated or down-regulated in a dose dependent manner. Differences were very obvious in expressions of mitochondrial genes of cells treated with hydrogen peroxide in a concentration of 400 μmol/L or 4 mmol/L. In general, differential expression of mitochondrial genes was characterized by up-regulation of mitochondrial genes in the concentration of 400 μmol/L and down-regulation in the concentration of 4 mmol/L. In consistence with changes in mitochondrial gene expressions, hydrogen peroxide resulted in decreased activities of cytochrome c oxidase and ATPase.
CONCLUSION: The differential expression of mitochondrial genes encoding cytochrome c oxidase and ATPase is involved in apoptosis of intestinal epithelial cells by affecting activities of cytochorme c oxidase and ATPase.
- Citation: Li JM, Cai Q, Zhou H, Xiao GX. Effects of hydrogen peroxide on mitochondrial gene expression of intestinal epithelial cells. World J Gastroenterol 2002; 8(6): 1117-1122
- URL: https://www.wjgnet.com/1007-9327/full/v8/i6/1117.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i6.1117
Hidden injuries of gut during early stage of severe burn may occur, and lead to endogenous translocation of intestinal endotoxin or bacteria. But factors contributing to gut barrier dysfunction are multiple and its mechanism is unclear[1-10]. Apoptosis of intestinal epithelial cells induced by excessive reactive oxygen species released by activated polymorphonuclear cells and vascular endothelial cells plays a role in the pathogenesis of intestinal mucosal dysfunction. The role of mitochondria in the development of apoptosis has been well clarified recently[11-15]. As we know, there are some proteins related to mitochondrial electron transport chain such as cytochrome coxidase subunits and ATP synthase subunits encoded by mitochondrial genome, which may be involved in mitochondrial injuries of intestinal epithelial cells. Our study focused on the effects of hydrogen peroxide on mitochondrial gene expression of intestinal epithelial cells.
Human intestinal epithelial cell line SW-480 stored routinely in our laboratory was cultured in RPMI1640 supplemented with 10% (v/v) heat inactivated newborn calf serum (Hyclone), 100 units/mL of penicillin, 0.1 mg/mL streptomycin and 2 mM L-glutamine at 37 °C in a humidified 5% CO2, 95% air incubator. Confluent cells were prepared for further studies.
Cells were treated with 4 mmol/L or 400 μmol/L hydrogen peroxide. Cells without any hydrogen peroxide were prepared as control.
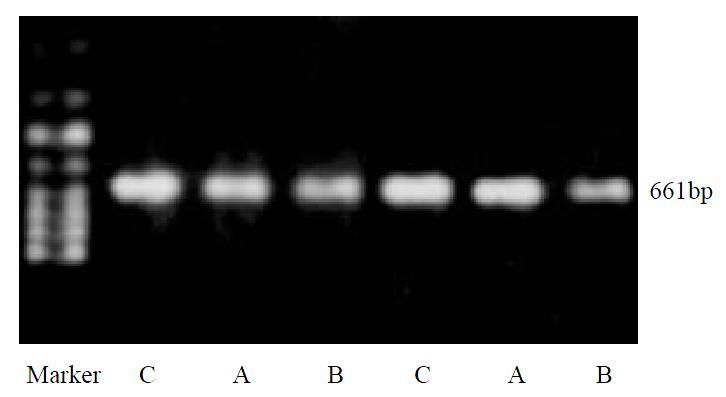
WWW primer picking software (primer3) from Whitehead Institute for Biomedical Research was used to design the primers of ATPase6, ATPase8, COI, COII and COIII genes encoded by mitochondrial genome in accordance with the latest Human Mitochondrial DNA “Cambridge” Sequence (Table 1). Housekeeping gene β-actin was regarded as internal control.
| Target genes | Sequence of base pairs | Size of PCR products(bp) |
| COI | 5’-GTTGTAGCCCACTTCCAC-3’ | 222 |
| 5’-CATCGGGGTAGTCCGAGTAA-3’ | ||
| COII | 5’-TTCATGATCACGCCCTCATA-3’ | 187 |
| 5’-TAAAGGATGCGTAGGGATGG-3’ | ||
| COIII | 5’-AAAGCACATACCAAGGCCAC-3’ | 195 |
| 5’-CTTCTAGGGGATTTAGCGGG-3’ | ||
| ATPase 6 | 5’-GCCCTAGCCCACTTCTTACC-3’ | 256 |
| 5’-TTAAGGCGACAGCGATTTCT-3’ | ||
| ATPase 8 | 5’-CCCACCATAATTACCCCCAT-3’ | 102 |
| 5’-TTTTATGGGCTTTGGTGAGG-3’ | ||
| β-actin | 5’-TGACGGGGTCACCCACACTGTGCCCATCTA-3’ | 661 |
| 5’-CTAGAAGCATTGCGGTGGACGATGGAGGG-3’ |
Total RNA was isolated by Tripure isolation reagent (Roche) according to manual provided by the kit. Any subject used in RNA isolation was treated with DEPC (Sigma) followed by vapor sterilization. A260/A280 of RNA samples were between 1.6 and 2.0.
Titan One Tube RT-PCR Kit (Roche) was applied according to manual for studying expression of mRNA of ATPase6, ATPase8, COI, COII and COIII genes. The PCR profile was as follows: a reverse-transcription reaction at 50 °C for 30 min and initial denaturation at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, primer extension at 68 °C for 45 s and a final extension at 68 °C for 10 min. PCR products were separated by electrophoresis on a 1% vertical agarose electrophoresis and observed under ultraviolet and stored in computer by gel imaging system. Relative density of genes was calculated by dividing density of respective gene with density of β-actin.
Mitochondria were isolated as described previously[16]. Cells about 1 × 107 were typsined followed by washing for one time, and then cells suspended in ice-cold isolation buffer (0.25 M sucrose, 10 mM Tris-HCl, pH 7.4) were homogenized with a glass homogenizer for 10 strokes in ice. The supernatants was further centrifuged at 1500 × g for 10 min at 4 °C, the pellet was suspended again in ice-cold isolation buffer and homogenized in ice. Two supernatants were mixed and centrifuged at 17000 × g for 10 min at 4 °C. Fresh isolated mitochondria was suspended in isolation buffer, protein concentration were determined by Folin-Phen assay and adjusted to 4 mg/mL. Activities of cytochrome c oxidase and ATPase were measured within 6 h after isolation of mitochondria.
In 1 mL reaction volume, 70 μL reduced ferrocytochrome c, 10 μL isolation mitochondria (4 mg/mL) and 920 μL 10 mM K2HPO4 (pH 7.4) were mixed at room temperature, and the absorbance was measured every 30 seconds. The decreased absorbance represents activity of cytochrome c oxidase[17].
Ten micro-liter of isolated mitochondria (4 mg/mL) and 1 mL of 10% TCA were mixed in Eppendorf tubes at 37 °C for 60 min, and the reaction was terminated by incubation in ice. Then 250 μL of 0.02 mol/L ATP-Na2 and 100 μL of 0.05 mol/L MgCl2 were added respectively, and the reaction was ended by adding 1 mL of 10% TCA after 5 min at 37 °C, followed by centrifugation at 3000 rpm for 15 min at 4 °C. The contents of inorganic phosphorus in supernatants representing ATPase activity were determined[18].
Data were expressed as mean ± SD. To analyze the data statistically, student’s t test was used to determine statistical differences, and P < 0.05 was considered significant.
After treating with hydrogen peroxide, no significant changes of house-keeping gene β-actin mRNA could be observed, indicating its value as an internal control (Figure 1).
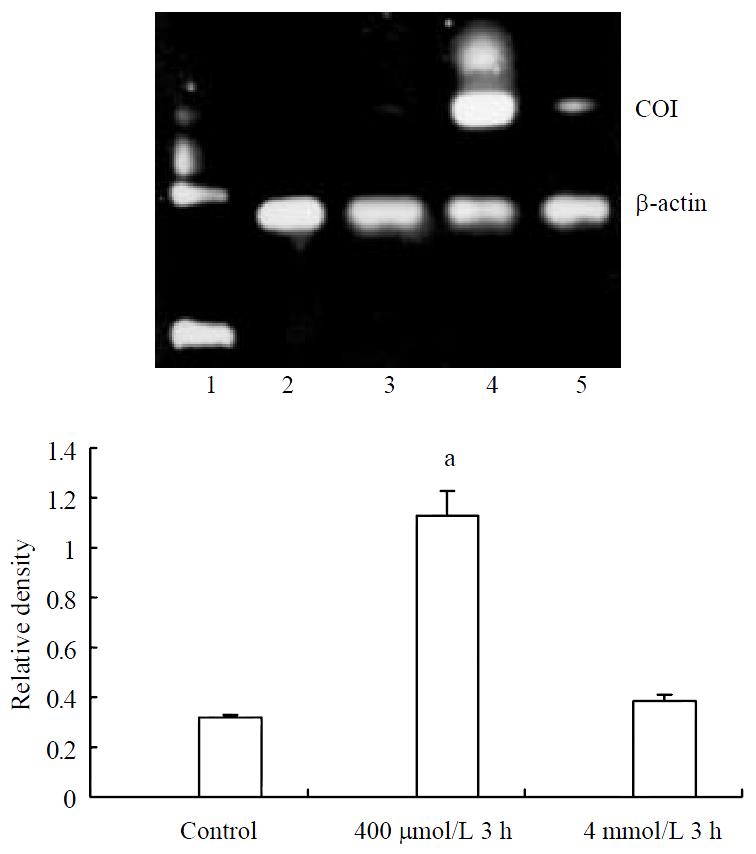
Cells from normal control expressed certain amount of COI mRNA. Increasing expression of COI mRNA was found after treating with hydrogen peroxide in the concentration of 400 μmol/L, but no significant changes could be observed in the concentration of 4 mmol/L (Figure 2).
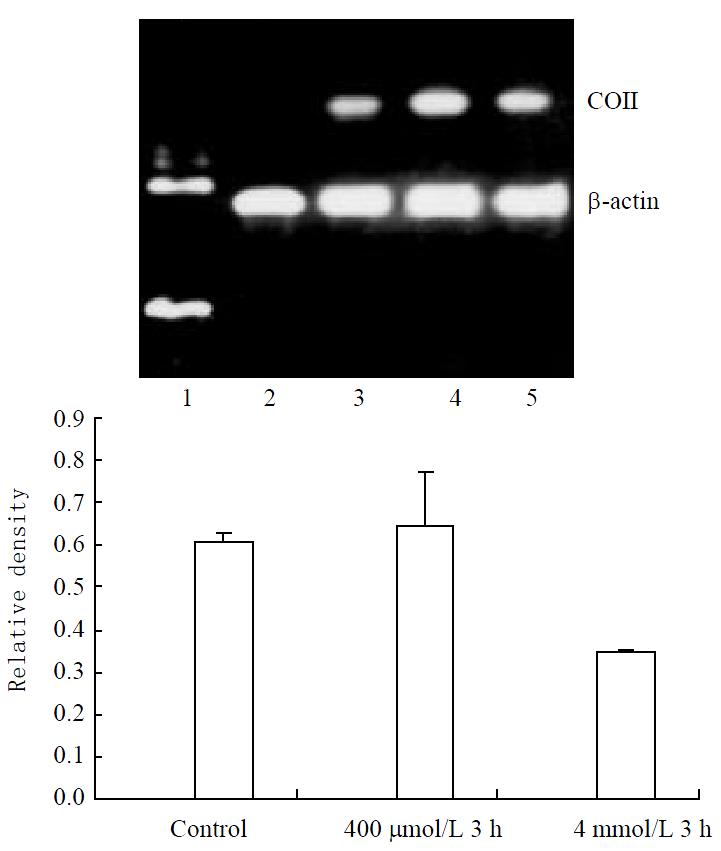
Weak expression of COII mRNA was found in control cells. Significant increase could be found in both 400 μmol/L and 4 mmol/L hydrogen peroxide-stimulated cells (Figure 3).
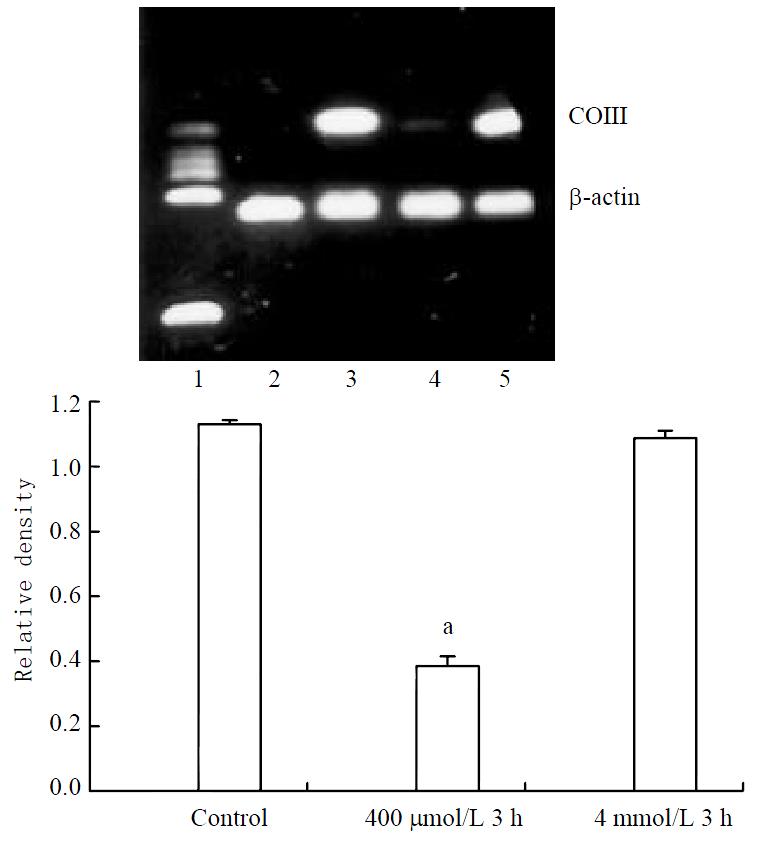
Stable expression of COIII mRNA was found in control cells. Significant decrease could be found in 400 μmol/L hydrogen peroxide-stimulated cells (Figure 4).
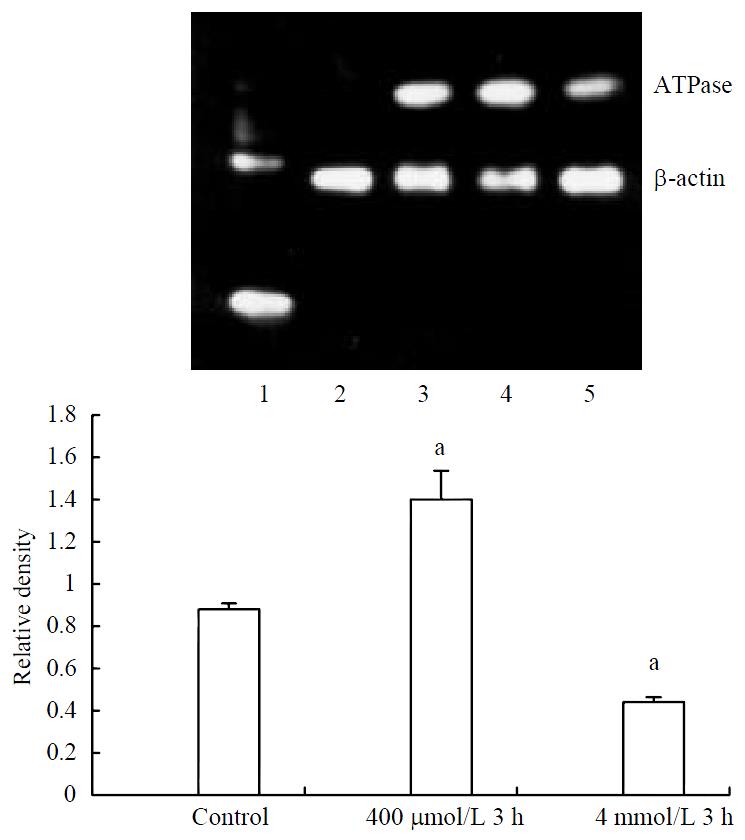
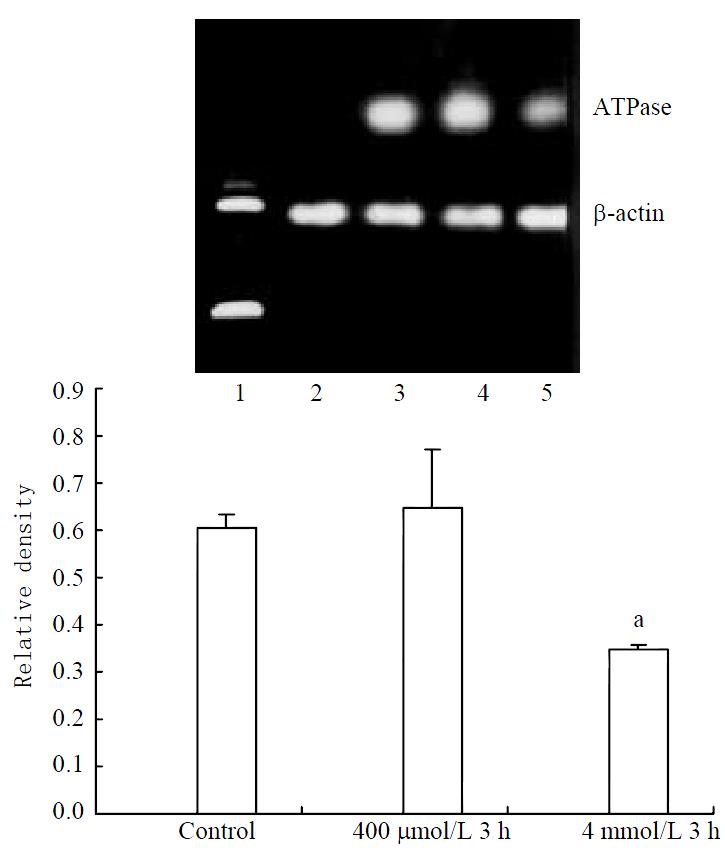
Expression of ATPase6 mRNA increased significantly in 400 μmol/L hydrogen peroxide stimulated-cells, meanwhile decreased expression could also be found in 4 mmol/L hydrogen peroxide-stimulated cells (Figure 5).
Stable expression of ATPase8 mRNA was found in control cells. No significant increase could be found in 400 μmol/L hydrogen peroxide-stimulated cells. Decreased expression of ATPase8 mRNA in 4 mmol/L hydrogen peroxide-stimulated cells was found (Figure 6).
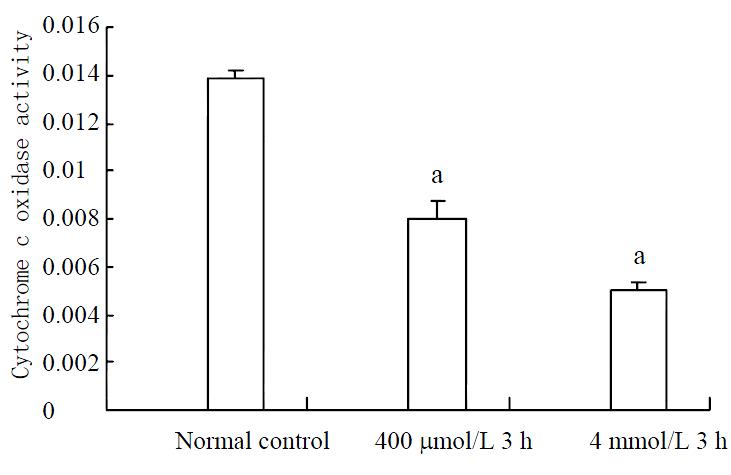
Activities of cytochrome c oxidase decreased significantly in both 400 μmol/L and 4 mmol/L hydrogen peroxide-stimulated cells (Figure 7).
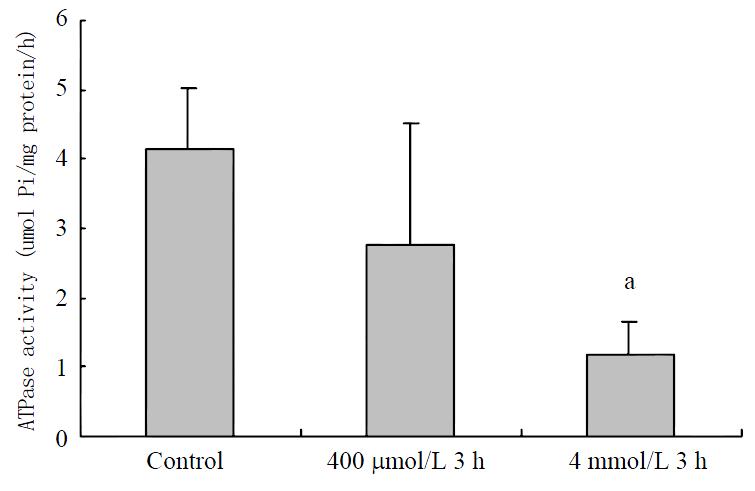
Although only ATPase activity in 4 mM hydrogen peroxide-stimulated cells decreased significantly on the view of statistics, the trend is very obvious (Figure 8).
The role of mitochondrion in the pathogenesis of apoptosis has been well defined. Mitochondrion, a kind of organelle controlling growing, breeding and dying of eukaryocyte, embodies its functions through: (1) production of ATP; (2) production of ROS, which regulates nuclear gene expression; (3) link between cytoplasm and nucleus; (4) sensitive response to stimulator or signals outside cytoplasm; (5) triggering of cell death[19,20]. Evidences show the central role of mitochondria in the development of apoptosis. Many stimulators like ROS, Ca2+ and cytokines lead to activation of caspases by inducing release of cytochrome C and thereby trigger a cascade in apoptosis[11].
Release of mitochondrial apoptosis-inducing substance caused by collapse of mitochondrial membrane potential and open of mitochondrial permeability transition pore controlled by Bcl 2 family is the key step to initiate apoptosis. Mitochondrial respiration depends on maintenance of mitochondrial membrane potential.
Now the relationship between DNA injuries and apoptosis has been recognized[21]. Although mitochondrial genome was sequenced in 1981[22], there is little study on function of mitochondrial genes. Thirteen proteins encoded by mitochondrial genome make up parts of enzymes involving mitochondrial oxidative phosphorylation. Without protection of histone, mitochondrial genome is much easier to be injured compared with nuclear genome. Our research also confirmed this (results to be published). Many researches indicated that oxidative stress led to mutation of mitochondrial genes[23-32]. The relationship between mutation of DNA and expression of genes needs to be well investigated.
Hydrogen peroxide led to differential expression of mitochondrial genes with some genes up-regulated or down-regulated in a dose-dependent manner. Differences were very obvious in expression of mitochondrial genes of cells treated with hydrogen peroxide in the concentration of 400 μmol/L or 4 mmol/L. In general, differential expression of mitochondrial genes was characterized by up-regulation of mitochondrial genes in the concentration of 400 μmol/L and down-regulation in the concentration of 4 mmol/L. In consistence with changes of mitochondrial gene expression, hydrogen peroxide led to similar decreased activities of cytochrome C oxidase and ATPase. Differential expression reflected the difference in the sensitivity of gene responding to oxidative stress, which may relate greatly to cellular physiological process next. In rat lung epithelial cells treated with hydrogen peroxide for for 8 or 24 h, the differential display of genes showed that mutation of genes from mitochondrial genome encoding NADH dehydrognase subunit 5, subunit 6 and 16 S ribosome could be detected; the results also suggested that mitochondrial genes played a role in the control of apoptosis[33]. Researches showed that there were some links between mitochondrial dysfunction and injuries of mitochondrial DNA or expression of mitochondrial genes in hydrogen peroxide-stimulated vascular endothelial cells and smooth muscle cells[34].
Meanwhile we also found a large scale of point mutation of mitocondrial gene such as ATPase8 subunit in 4 mmol/L hydrogen peroxide treated SW-480 cells, suggesting its correlation with down-regulation of ATPase expression.
It is considered now that the mitochondrial permeability transition (MPT) plays a key role in the pathogenesis of apoptosis and necrosis[35-38]. If MPT initiates rapidly with depletion of ATP, cells undergo necrosis, and if MPT occurs without depletion of ATP, cells evolved into apoptosis. So we may conclude that complete inhibition of ATPase would lead to necrosis and partially inhibition of ATPase would lead to apoptosis. Consistent with this conclusion, our study proved a partially inhibition of ATPase in both 4 mmol/L and 400 μmol/ L hydrogen peroxide-treated SW-480 cells, suggesting its potential role in apoptosis. But there are still many problems to be clarified, including the relationship between different expressions of mitochondrial genes, activities of cytochrome C oxdiase and ATPase, oxidative phosphorylation and regulation of apoptosis.
Studies including ours showed that mitochondrial genes played the role in apoptosis. To establish mtDNA deletion cells (ρ0 cells) provided in our studies more direct evidences[39-42].
It is unclear about the pathway leading to increasing expression of mitochondrial genes. Defects of mitochondrial energy supply may be one of the compensatory mechanisms leading to increasing expression of mitochondrial genes[43].
Our results showed that differential expression seemed to be incompatible with decreasing activities of cytochorome C oxidase or ATP synthase. To answer this question, we should pay attention to mitochondrial proteins encoded by the nuclear genome, which form main parts of mitochondrial proteins. For example, only subunits 6 and 8 of ATP synthase were encoded by mitochondrial genome, which may also be controlled by some nuclear transcriptional factors[44-46]. Meanwhile most parts of ATPase were encoded by the nuclear genome. Cross-talk between mitochondrial genes and nuclear genes will be the next key or difficult question to be answered[47-50]. It will become available to study cross-talk between mitochondrial genes and nuclear genes from the view of genome and proteome by the developing techniques of genome and proteome.
Edited by Lu HM
| 1. | Zuo GQ, Gong JP, Liu CA, Li SW, Wu XC, Yang K, Li Y. Expression of lipopolysaccharide binding protein and its receptor CD14 in experimental alcoholic liver disease. World J Gastroenterol. 2001;7:836-840. [PubMed] |
| 2. | Meng AH, Ling YL, Zhang XP, Zhao XY, Zhang JL. CCK-8 inhibits expression of TNF-alpha in the spleen of endotoxic shock rats and signal transduction mechanism of p38 MAPK. World J Gastroenterol. 2002;8:139-143. [PubMed] |
| 3. | Wu RQ, Xu YX, Song XH, Chen LJ, Meng XJ. Relationship between cytokine mRNA expression and organ damage following cecal ligation and puncture. World J Gastroenterol. 2002;8:131-134. [PubMed] |
| 4. | Li SW, Gong JP, Wu CX, Shi YJ, Liu CA. Lipopolysaccharide induced synthesis of CD14 proteins and its gene expression in hepatocytes during endotoxemia. World J Gastroenterol. 2002;8:124-127. [PubMed] |
| 5. | Yu PW, Xiao GX, Qin Xj LX, Wang ZQ. The effects of PAF antagonist on intestinal mucosal microcirculation after burn in rats. World J Gastroenterol. 2000;6:906-908. [PubMed] |
| 6. | Qin RY, Zou SQ, Wu ZD, Qiu FZ. Influence of splanchnic vascular infusion on the content of endotoxins in plasma and the translocation of intestinal bacteria in rats with acute hemorrhage necrosis pancreatitis. World J Gastroenterol. 2000;6:577-580. [PubMed] |
| 7. | Wang QG, He LY, Chen YW, Hu SL. Enzymohistochemical study on burn effect on rat intestinal NOS. World J Gastroenterol. 2000;6:421-423. [PubMed] |
| 8. | Fu XB, Yang YH, Sun TZ, Gu XM, Jiang LX, Sun XQ, Sheng ZY. Effect of intestinal ischemia-reperfusion on expressions of endogenous basic fibroblast growth factor and transforming growth factor betain lung and its relation with lung repair. World J Gastroenterol. 2000;6:353-355. [PubMed] |
| 9. | Fu WL, Xiao GX, Yue XL, Hua C, Lei MP. Tracing method study of bacterial translocation in vivo. World J Gastroenterol. 2000;6:153-155. [PubMed] |
| 10. | Yang YH, Fu XB, Sun TZ, Jiang LX, Gu XM. bFGF and TGFbeta expression in rat kidneys after ischemic/ reperfusional gut injury and its relationship with tissue repair. World J Gastroenterol. 2000;6:147-149. [PubMed] |
| 11. | Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6842] [Cited by in RCA: 6815] [Article Influence: 252.4] [Reference Citation Analysis (0)] |
| 12. | Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5259] [Cited by in RCA: 5204] [Article Influence: 208.2] [Reference Citation Analysis (0)] |
| 13. | Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1428] [Cited by in RCA: 1466] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 14. | Petit PX, Susin SA, Zamzami N, Mignotte B, Kroemer G. Mitochondria and programmed cell death: back to the future. FEBS Lett. 1996;396:7-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 368] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 15. | Finkel E. The mitochondrion: is it central to apoptosis. Science. 2001;292:624-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 149] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 16. | Koya RC, Fujita H, Shimizu S, Ohtsu M, Takimoto M, Tsujimoto Y, Kuzumaki N. Gelsolin inhibits apoptosis by blocking mitochondrial membrane potential loss and cytochrome c release. J Biol Chem. 2000;275:15343-15349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 189] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 17. | Barazzoni R, Short KR, Nair KS. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J Biol Chem. 2000;275:3343-3347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 275] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 18. | Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J Biol Chem. 1998;273:22983-22989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 188] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3574] [Cited by in RCA: 3547] [Article Influence: 126.7] [Reference Citation Analysis (0)] |
| 20. | Susin SA, Zamzami N, Castedo M, Hirsch T, Marchetti P, Macho A, Daugas E, Geuskens M, Kroemer G. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J Exp Med. 1996;184:1331-1341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 842] [Cited by in RCA: 818] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 21. | Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 448] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 22. | Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6507] [Cited by in RCA: 6451] [Article Influence: 146.6] [Reference Citation Analysis (0)] |
| 23. | Simon DK, Lin MT, Ahn CH, Liu GJ, Gibson GE, Beal MF, Johns DR. Low mutational burden of individual acquired mitochondrial DNA mutations in brain. Genomics. 2001;73:113-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found Symp. 2001;235:247-263; discussion 263-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 110] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 25. | Chang SW, Zhang D, Chung HD, Zassenhaus HP. The frequency of point mutations in mitochondrial DNA is elevated in the Alzheimer's brain. Biochem Biophys Res Commun. 2000;273:203-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci USA. 1999;96:4820-4825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 465] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 27. | Lu CY, Lee HC, Fahn HJ, Wei YH. Oxidative damage elicited by imbalance of free radical scavenging enzymes is associated with large-scale mtDNA deletions in aging human skin. Mutat Res. 1999;423:11-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 117] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 28. | Bhat HK, Hiatt WR, Hoppel CL, Brass EP. Skeletal muscle mitochondrial DNA injury in patients with unilateral peripheral arterial disease. Circulation. 1999;99:807-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Swerdlow RH, Parks JK, Cassarino DS, Shilling AT, Bennett JP, Harrison MB, Parker WD. Characterization of cybrid cell lines containing mtDNA from Huntington's disease patients. Biochem Biophys Res Commun. 1999;261:701-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Wei YH. Oxidative stress and mitochondrial DNA mutations in human aging. Proc Soc Exp Biol Med. 1998;217:53-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 221] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 31. | Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res. 2001;88:529-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 517] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 32. | Swerdlow RH, Parks JK, Davis JN, Cassarino DS, Trimmer PA, Currie LJ, Dougherty J, Bridges WS, Bennett JP, Wooten GF. Matrilineal inheritance of complex I dysfunction in a multigenerational Parkinson's disease family. Ann Neurol. 1998;44:873-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 102] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 33. | Janssen YM, Driscoll KE, Timblin CR, Hassenbein D, Mossman BT. Modulation of mitochondrial gene expression in pulmonary epithelial cells exposed to oxidants. Environ Health Perspect. 1998;106 Suppl 5:1191-1195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 34. | Ballinger SW, Patterson C, Yan CN, Doan R, Burow DL, Young CG, Yakes FM, Van Houten B, Ballinger CA, Freeman BA. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res. 2000;86:960-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 311] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 35. | Hatano E, Bradham CA, Stark A, Iimuro Y, Lemasters JJ, Brenner DA. The mitochondrial permeability transition augments Fas-induced apoptosis in mouse hepatocytes. J Biol Chem. 2000;275:11814-11823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 110] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 36. | Lemasters JJ. V. Necrapoptosis and the mitochondrial permeability transition: shared pathways to necrosis and apoptosis. Am J Physiol. 1999;276:G1-G6. [PubMed] |
| 37. | Lemasters JJ, Qian T, Bradham CA, Brenner DA, Cascio WE, Trost LC, Nishimura Y, Nieminen AL, Herman B. Mitochondrial dysfunction in the pathogenesis of necrotic and apoptotic cell death. J Bioenerg Biomembr. 1999;31:305-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 38. | Pastorino JG, Tafani M, Rothman RJ, Marcinkeviciute A, Hoek JB, Farber JL. Functional consequences of the sustained or transient activation by Bax of the mitochondrial permeability transition pore. J Biol Chem. 1999;274:31734-31739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 225] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 39. | Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci U S A. 2001;98:4038-4043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 193] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 40. | Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei YH, Murphy MP. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur J Biochem. 1999;262:108-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 140] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 41. | Tang JT, Yamazaki H, Inoue T, Koizumi M, Yoshida K, Ozeki S, Inoue T. Mitochondrial DNA influences radiation sensitivity and induction of apoptosis in human fibroblasts. Anticancer Res. 1999;19:4959-4964. [PubMed] |
| 42. | Suzuki A, Tsutomi Y, Yamamoto N, Shibutani T, Akahane K. Mitochondrial regulation of cell death: mitochondria are essential for procaspase 3-p21 complex formation to resist Fas-mediated cell death. Mol Cell Biol. 1999;19:3842-3847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 93] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 43. | Wiesner RJ, Hornung TV, Garman JD, Clayton DA, O'Gorman E, Wallimann T. Stimulation of mitochondrial gene expression and proliferation of mitochondria following impairment of cellular energy transfer by inhibition of the phosphocreatine circuit in rat hearts. J Bioenerg Biomembr. 1999;31:559-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 44. | Herzig RP, Scacco S, Scarpulla RC. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem. 2000;275:13134-13141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 142] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 45. | Miranda S, Foncea R, Guerrero J, Leighton F. Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted HeLa cells. Biochem Biophys Res Commun. 1999;258:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 124] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 46. | Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3385] [Cited by in RCA: 3239] [Article Influence: 124.6] [Reference Citation Analysis (0)] |
| 47. | Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J Biol Chem. 2001;276:4020-4027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 165] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 48. | Kaasik A, Veksler V, Boehm E, Novotova M, Minajeva A, Ventura-Clapier R. Energetic crosstalk between organelles: architectural integration of energy production and utilization. Circ Res. 2001;89:153-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 196] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 49. | Poyton RO, Dagsgaard CJ. Mitochondrial-nuclear crosstalk is involved in oxygen-regulated gene expression in yeast. Adv Exp Med Biol. 2000;475:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 50. | Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563-607. [PubMed] |