Published online Jun 15, 2002. doi: 10.3748/wjg.v8.i3.480
Revised: January 5, 2002
Accepted: January 24, 2002
Published online: June 15, 2002
AIM: In hepatocellular carcinoma (HCC) prevalent areas of China, the point mutation of p53 exon7 is highly correlated with Hepatitis B virus (HBV) infection and aflatoxin B intake. While in non-HCC-prevalent areas of China, these factors are not so important in the etiology of HCC. Therefore, the point mutation of p53 exon7 may also be different than that in HCC-prevalent areas of China. The aim of this study is to investigate the status and carcinogenic role of the point mutation of p53 gene exon7 in hepatocellular carcinoma from Anhui Province, a non-HCC-prevalent area in China.
METHODS: PCR, PCR-SSCP and PCR-RFLP were applied to analyze the homozygous deletion and point mutation of p53 exon7 in HCC samples from Anhui, which were confirmed by DNA sequencing and Genbank comparison.
RESULTS: In the 38 samples of hepatocellular carcinoma, no homozygous deletion of p53 exon7 was detected and point mutations of p53 exon7 were found in 4 cases, which were found to be heterozygous mutation of codon 249 with a mutation rate of 10.53% (4/38). The third base mutation (GiúT) of p53 codon 249 was found by DNA sequencing and Genbank comparison.
CONCLUSION: The incidence of point mutation of p53 codon 249 is lower in hepatocellular carcinoma and the heterozygous mutation of p53 exon7 found in these patients only indicate that they have genetic susceptibility to HCC. p53 codon 249 is a hotspot of p53 exon7 point mutation, suggesting that the point mutation of p53 exon 7 may not play a major role in the carcinogenesis of HCC in Anhui Province, a non-HCC-prevalent area in China.
- Citation: Liu H, Wang Y, Zhou Q, Gui SY, Li X. The point mutation of p53 gene exon7 in hepatocellular carcinoma from Anhui Province, a non HCC prevalent area in China. World J Gastroenterol 2002; 8(3): 480-482
- URL: https://www.wjgnet.com/1007-9327/full/v8/i3/480.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i3.480
Hepatocellular carcinoma is one of the most common cancers in the world. Abnormalities of p53 are the most frequent genetic alterations in human cancers, and the role and mechanism of p53 gene mutations have been well studied in many types of cancer[1-3]. Genetic analysis of 26 HCC samples from North America and Europe revealed a high incidence of an AGG→AGT transversional changes in codon 249 of the p53 gene; and recently exon7 has been proven a hotspot of p53 gene mutation[4-6]. Zhang et al[4] reported the high relationship between HBVx gene and codon 249 mutation of the p53 gene in HCC-prevalence areas in China. Our previous studies have also indicated that hepatitis B virus infection is an important risk factor for HCC[7]. These data indicate that p53 mutations generally occur in the process of HCC carcinogenesis in HCC-prevalent area in China. However, further mutation analyses will be necessary to clarify the status of p53 mutations for HCC in non-HCC-prevalent areas in China.
In this study, we analyzed p53 exon 7 point mutation in HCCs from non-HCC-prevalent areas in China using the polymerase chain reaction (PCR), PCR-single-strand conformational polymorphism (PCR-SSCP), PCR-restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing analysis.
The surgical specimens of HCC were collected from the First Affiliated Hospital of Anhui Medical University, which were confirmed by pathological diagnosis and stored at -80 °C. The patients were born in and permanent residents of different places of the Anhui Province, China.
DNA was extracted from tissues with standard proteinase K-phenol/choloroform methods[9].The primers for p53 gene exon7 amplification were designed according to the sequence of p53 exon7 published[4,5]. 3’primer (GW-XI-1C): 5’CTTGCCACAGGTCTCCCCAA, 5’primer (GW—XI-1D): 5'TGTGCAGGGTGGCAAGTGGC; CDK4 as a control, 3’primer (GW-IV-1K): 5’GGAGGTCGGTACCAGAGTG, 5’primer (GW—IV-1J): 5’CATGTAGACCAGGACAGG. Into 100 ng of DNA template of each sample was added PCR reaction solution (10 mmol/L Tris, 50 mmol/L KCl, 2 mmol/L MgCl2, 0.001% Gelatin, 200 mmol/L dNTPs, 6% DMSO and 0.5 mmol/L primers). Hotstart was performed: 97 °C 5 min; chilled on ice at once. 0.9 U of Taq polymerase was added, which was diluted with 1 × PCR buffer for each sample. Ran PCR: 94 °C 30 s, 60 °C 30 s, 72 °C 30 s, 35 cycles in all and checked with 2% agarose gel electrophoresis stained with ethidium bromide. The result of homozygous deletion should be the one with no specific band of p53 exon7 while its counterpart of CDK4 appeared.
Eight μL of PCR products were aspirated, into which was added equal volumes of deionized formamide and 4 μL of DNA loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol FF, 30% glycerol). They were mixed well, boiled for 5 min, and then chilled on ice for 3 min. The samqles (20 μL in volume) were loaded into separate wells. Samples were run in an 8% non-denaturing polyacrylamide gel at 80 V for 5 hrs. The gel was taken off from the electrophoresis apparatus and readied for silver staining. The gel was submerged in 5% ethanol for 5 min; 3 min in 1% HNO3; 20 min in 0.012 mol·L-1 AgNO3; washed with dd-H2O for about 10 sec; developed with 0.28 mol·L-1 Na2CO3; fixed with 10% acetic acid; and finally washed with dd-H2O. When the bands appeared, photos were taken and the gel was dried with Slab Gel Dryer or wrapped with a membrane and air dried for several days. Na2CO3 was changed 2-4 times when the developing solution turned black.
Into each restrictive endonuclease system was added 2 μL of 10 × Buffer C, 2 μL of DTT (1%), 2 μL of BSA (1%) and 0.25 μL of Hae III (20 U·μL-1). The total volume was brought up to 20 μL with PCR products. They were incubated at 37 °C for 3 hrs and checked with an 8% non-denaturation polyacrylamide gel, electrophoresed at 40 V for 4 hrs and developed with silver staining[8] as described above.
The sample of p53 exon7 mutation was confirmed by PCR-SSCP and RFLP and PCR products of p53 exon7 were sent the to Bioasia Biotechnololy Company, Shanghai, China for DNA sequencing with ABI 377 automatic DNA sequencer.
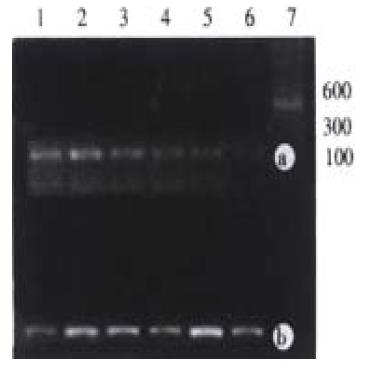
With 100 ng of genomic DNA extracted from surgical HCC tissue as template, p53 exon7 and CDK4 genes were amplified with different specific primers in separate tubes. The products were checked with 2% agarose gel electrophoresis. The results showed that the products amplified with each pair of specific pairs were of the same length with that reported in the literature (Figure 1). No homozygous deletion of p53 exon7 was found in any HCC surgical sample.
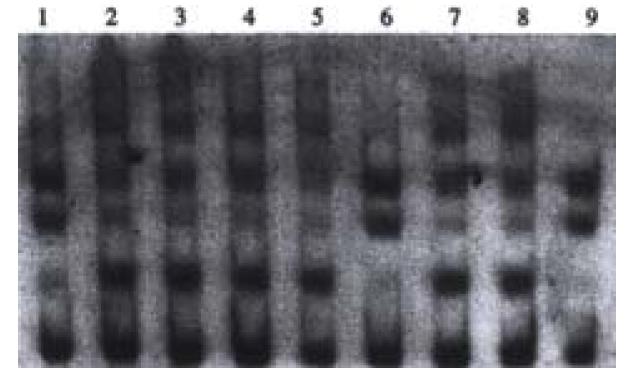
Point mutations of p53 exon7 were found in 4 cases out of the 38 samples of HCC examined. No. 1, 6 and 9 sample had point mutations of p53 exon7 (Figure 2).
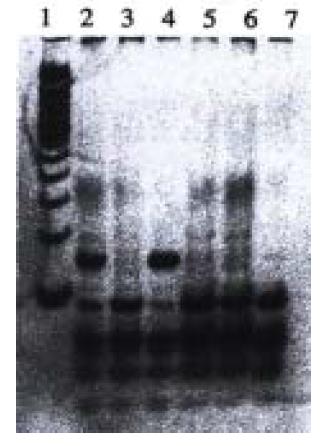
With PCR-RFLP, we found that 4 samples have heterozygous point muation of p53 codon 249, which has a band of 150 bp in addition to wild type bands (40, 60, and 90 bp) as shown by agarose/EB gel electrophoresis (Figure 3). However, no homozygous point mutation was found among these samples, which would have had bands of 40 bp and 150 bp. We found that those samples which have point mutation of p53 codon 249 was the same samples that were found to have point mutation of p53 exon 7 by PCR-SSCP.
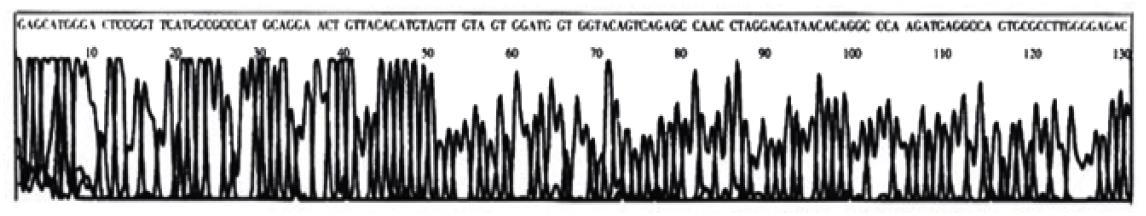
One sample that has been found by PCR-SSCP to have point mutation of p53 exon7 was randomly chosen for DNA sequencing. The DNA sequencing result is shown in the following graph. The sequence was compared with that published by the Genbank (gbAF136270.1 HOMOTSP1), which shows that a point mutation exists in p53 codon 249 with ggAc taken place of ggCc (Figure 4, Figure 5).
The tumor suppressor gene p53 is found to be involved in the carcinogeneses of diverse types of cancer[1-3,10-16]. Some domains of the p53 protein are highly conservated, which also reflect their significance for the function of the p53 protein. The exons that encode the domains are hotspots for point mutations. If point mutations occur in these sites, the peptides that are translated from these templates will affect the correct folding of the p53 protein. Hence, the cell cycle suppressing function of p53 will be affected, which will result loss of cell proliferation control. Recent research has found that p53 exon 7 (codon 249) is a hotspot for point mutation in hepatocellular carcinoma (HCC), and most of the point mutations are G→T changes at the third base of codon 249 (Arginine→Serine), and others are G→C (Arginine→Serine). Point mutations of codon 249 will lead to the loss of the Hae III restricion site in the tumor’s genomic DNA (GGCC→GTCC)[4-6,17,18]. Therefore, PCR-RFLP should be a expedient and convenient method to detect the point mutation of p53 codon 249. In our experiments, the 4 specimens that were found to have point mutation in p53 exon7 by PCR-SSCP were all found to have the point mutation in codon 249 by PCR-RFLP, which confirmed the previous reports discussed above.
It was reported that in Qidong, Jiangsu Province, a HCC prevalent area in China, about half of the HCC patients had point mutations of p53 gene exon 7, and codon 249 was the hotspot of point mutation, while no point mutation was found in other exons[4]. In order to investigate whether it is the same case in non-HCC prevalent areas of China, we collected surgical specimens from Anhui province, a non-HCC prevalent area in China[7], and tried to examine the status of point mutation in p53 exon7, which turned out to be only 4 specimens of point mutation that were all heterozygous p53 codon 249 mutations. No homozygous mutation was found among these specimens. These results showed that p53 codon 249 was the hotspot for the mutation of p53 exon 7 and its mutation was not a frequent event in non-HCC prevalent area. The fact that they are heterozygous mutations only suggests these patients are susceptible to cancer. Therefore, in non-HCC prevalent areas of P.R. China, the point mutation in p53 codon 249 may play some role but not a major role in the carcinogenesis of hepatocellular carcinoma. In HCC prevalent areas in P.R. China such as Qidong, Jiangsu Province, HBV, aflatoxin B and polluted water are the major causes of cancer, therefore the genetic alteration in the HCC patients may have correlations with the point mutations or deletions in some set loci in the genome. In Anhui province, a non HCC prevalent area in China, factors like HBV infection and aflatoxin B intake are not so prominent in the etiology of HCC. Our results suggest that the causes of HCC in non-HCC prevalent areas are multiple and complex and therefore the genetic alterations caused by them are also diverse.
Edited by Pagliarini R1
| 1. | Lee CC, Liu JY, Lin JK, Chu JS, Shew JY. p53 point mutation enhanced by hepatic regeneration in aflatoxin B1-induced rat liver tumors and preneoplastic lesions. Cancer Lett. 1998;125:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 2. | Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1896] [Cited by in RCA: 1960] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 3. | Bressac B, Galvin KM, Liang TJ, Isselbacher KJ, Wands JR, Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:1973-1977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 312] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Zhang F, Zhu Y, Sun Z. [Universal presence of HBVx gene and its close association with hotspot mutation of p53 gene in hepatocellular carcinoma of prevalent area in China]. Zhonghua Zhongliu Zazhi. 1998;20:18-21. [PubMed] |
| 5. | Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 969] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 6. | Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 972] [Cited by in RCA: 872] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 7. | Wang Y, Liu H, Zhou Q, Li X. Analysis of point mutation in site 1896 of HBV precore and its detection in the tissues and serum of HCC patients. World J Gastroenterol. 2000;6:395-397. [PubMed] |
| 8. | LI ZY, Quan XF, Wang YG, Lu H, Chen ZD. The epidemiology sampling survey report of hepatocellular carcinoma in Anhui province in 1990-1992. Gandan Waike Zazhi. 1995;3:166-169. |
| 9. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloninig: A laboratory manual (2nd edition). New York: Cold Spring Harbor Laboratory Press. 1989;9:16-23. |
| 10. | Johnson SM, Shaw JA, Walker RA. Sporadic breast cancer in young women: prevalence of loss of heterozygosity at p53, BRCA1 and BRCA2. Int J Cancer. 2002;98:205-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Feng DY, Chen RX, Peng Y, Zheng H, Yan YH. Effect of HCV NS(3) protein on p53 protein expression in hep atocarcinogenesis. World J Gastroenterol. 1999;5:45-46. [PubMed] |
| 12. | Sullivan A, Yuille M, Repellin C, Reddy A, Reelfs O, Bell A, Dunne B, Gusterson BA, Osin P, Farrell PJ. Concomitant inactivation of p53 and Chk2 in breast cancer. Oncogene. 2002;21:1316-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Reiher F, Ozer O, Pins M, Jovanovic BD, Eggener S, Campbell SC. p53 and microvessel density in primary resection specimens of superficial bladder cancer. J Urol. 2002;167:1469-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Zhang Z, Yuan Y, Gao H, Dong M, Wang L, Gong YH. Apoptosis, proliferation and p53 gene expression of H. pylori associated gastric epithelial lesions. World J Gastroenterol. 2001;7:779-782. [PubMed] |
| 15. | Pollack IF, Finkelstein SD, Woods J, Burnham J, Holmes EJ, Hamilton RL, Yates AJ, Boyett JM, Finlay JL, Sposto R. Expression of p53 and prognosis in children with malignant gliomas. N Engl J Med. 2002;346:420-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 194] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 16. | Garcia CA, Ahmadian A, Gharizadeh B, Lundeberg J, Ronaghi M, Nyrén P. Mutation detection by pyrosequencing: sequencing of exons 5-8 of the p53 tumor suppressor gene. Gene. 2000;253:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Hsu CH, Lee SC, Yang YC, Lian ST, Shin SJ, Lin SR. The p53 codon 249 mutant--derived from human functional adrenal tumors--can modify the cell shape of normal adrenocortical transfected cells. Cancer Lett. 2001;170:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 18. | Hussain SP, Raja K, Amstad PA, Sawyer M, Trudel LJ, Wogan GN, Hofseth LJ, Shields PG, Billiar TR, Trautwein C. Increased p53 mutation load in nontumorous human liver of wilson disease and hemochromatosis: oxyradical overload diseases. Proc Natl Acad Sci USA. 2000;97:12770-12775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 171] [Article Influence: 6.8] [Reference Citation Analysis (0)] |