Published online Dec 15, 2000. doi: 10.3748/wjg.v6.i6.800
Revised: May 7, 2000
Accepted: May 12, 2000
Published online: December 15, 2000
AIM: To investigate the hepatitis C virus (HCV) infection in the tissues of carcinoma of extrahepatic bile duct and study their correlation.
METHODS: HCV NS5 protein and HCV RNA were detected by labeled streptavidin biotin (LSAB) method and in situ reverse transcription polymerase chain reaction (IS-RT-PCR) in sections of 51 cases of carcinoma of extrahepatic bile duct and 34 cases of control group (without malignant biliary disease).
RESULTS: In 51 cases of carcinoma of extrahepatic bile duct, HCV NS5 protein was detected in 14 (27.5%), which was clearly stained in the cytoplasm of cancer cell but not in the nucleus or cell membrane. HCV RNA was detected in 18 (35.4%), which was located in the nucleus of cancer cell in 12 cases and in the cytoplasm in 6 cases. HCV NS5 protein and RNA coexistence was found in 2 cases. In 34 cases of control group, HCV RNA was detected in 2 (5.9%). HCV NS5 protein and RNA positive cells were found either scattered or in clusters.
CONCLUSION: The prevalence of hepatitis C viral infection in the tissues of carcinoma of extrahepatic bile duct was significantly higher than in control group (χ² = 9.808, P = 0.002). The findings suggest a correlation between HCV infection and carcinoma of extrahepatic bile duct, which is different from the traditional viewpoint. HCV infection might be involved in the development of carcinoma of extrahepatic bile duct.
- Citation: Chen MY, Huang ZQ, Chen LZ, Gao YB, Peng RY, Wang DW. Detection of hepatitis C virus NS5 protein and genome in Chinese carcinoma of the extrahepatic bile duct and its significance. World J Gastroenterol 2000; 6(6): 800-804
- URL: https://www.wjgnet.com/1007-9327/full/v6/i6/800.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i6.800
Cancer of bile duct arises from malignant transformation of the epithelia of bile duct. It is even less common than gallbladder carcinoma, and seen in 0.01%-0.46% of all autopsies[1] and its pathogenesis has not been fully understood. Cancer of bile duct is different from hepatocellular carcinoma (HCC) in etiologic factors, the former is not correlated to HBV or HCV infection and liver cirrhosis, traditionally[2]. It has been reported that only 10%-20% patients with bile duct cancer accompany liver cirrhosis, but 70%-90% patients with HCC are associated with liver cirrhosis[3-5]. There is a high incidence of bile duct cancer in the southeast Asia, and liver fluke infection due to clonorchis sinensis and opisthorchis viverrini is the most frequently cited cause of bile duct cancer[6,7]. Now the development of bile duct cancer has been linked to hepatolithiasis, clon orchis sinensis, congenital dilatation of bile duct, and chronic inflammatory bowel disease[8-11]. In China, 0.33%-9.7%, patients with hepatolithiasis[12,13], 2.1%-21% with the choledochal cyst[14-17] and 0.22% with clonorchis sinensis infestation[18] are simultaneously complicated with bile duct cancer. Bile duct cancer simultaneously complicated with gallstone, choledochal cyst, and clonorchis sinensis infestation accounts for 6.15%-16.9%[19-22], 7%[15] and 6.4%[23] in total bi le duct cancer of the corresponding period, respectively. But the incidence of bile duct cancer complicated with ulcerative colitis simultaneously is not estimated in literature in China now. These investigations indicated, therefore, that Chinese patients with bile duct cancer suffered from above-mentioned diseases before only account for one third or a half of the total patients with bile duct cancer. In extrahepatic biliary carcinoma in China, carcinoma of extrahepa tic bile duct covered 75.2%[24], the incidence of carcinoma of extrahepatic bile duct tends to increase over the past decade, but its cause is still unclear[24].
Hepatitis C virus (HCV) is a RNA virus with a genomic size of 9.6 kb, and now known to be the chief cause of transfusion-associated non-A, non-B hepatitis, which has been reported to occur in 7%-10% patients who received transfusion[25,26]. More than 50% of individuals exposed to HCV develop chronic infection. Of those chronically infected individuals, about 20%-30% will develop liver cirrhosis and/or HCC when followed for 20 or 30 years[27]. In China, the prevalence of HCV antibodies in blood donors as measured by the second or third generation assays is about 0%-4.6%[28,29], and in a rural population the HCV infection rate is up to 15.3%[30], which indicates that China is a relatively high incidence area of HCV infection. In situ reverse-transcription polymerase chain (IS-RT-PCR) has been successfully applied to the detection and localization of HCV RNA in formalin-fixed paraffin-embedded liver sections[31,32], indicating that it becomes easy to detect the low level of HCV RNA. This study aims to investigate the HCV infection in the tissues of carcinoma of extrahepatic bile duct (51 cases) and control group specimens (34 cases) by detecting HCV NS5 protein and RNA using labeled streptavidin biotin (LSAB) method and IS-RT-PCR, and to determine their correlation.
Fifty-one cases of carcinoma of extrahepatic bile duct, and 34 specimens as control group (including 10 cases of choledochal cyst, 8 cases of hepatolithiasis, 2 cases of congenital dilatation of the intrahepatic bile duct, 2 cases of cystadenoma and 2 cases of adenoma of common bile duct, and 10 cases of wall of extrahepatic bile duct near gallstone) were collected from Department of Hepatobiliary Surgery, General Hospital of People’s Liberation Army (PLA). All specimens were resected from 1995 to 1998, and fixed and embedded routinely. All carcinomas of extrahepatic bile duct were diagnosed as adenocarcinoma by the Department of Pathology, General Hospital of PLA. Five-micrometer thick for malin-fixed paraffin-embedded sections were prepared.
Primers and probe were all located at the highly conserved 5’non-coding region of the HCV genome. The oligonucleotide primers and probe were synthesized, and the probe was labeled with biotin (Sangon Co.Ltd). The sequences of outer primers are: sense, 5’-GGCGACACTCCACCATAGATC-3’ (1-21 nt), antisense, 5’-GGTGCACGGTCTACGAGACCT-3’ (304-324 nt). The sequences of inner primers are: sense, 5’-CTGTGAGGAACTACTGTCTTC-3’ (28-48 nt), antisense, 5’-CCCTATCAGGCAGTACCACAA-3’ (264-284 nt). Probe sequence is: 5’-ACACCGGAATTGCCAGGACGACCGGGTCCTTTCTTG-3’ (142-177 nt).
The sections were dewaxed and rehydrated routinely, and then treated for 5 min with 0.03% hydrogen peroxide in methanol to eliminate endogenous peroxidase activity. Antigen of the tissues was prepared in microwave oven. Sections were incubated with normal goat serum for 5 min, and then incubated with anti-HCV-NS5 IgG (diluted 1∶50) at 4 °C overnight. After washed with phosphate buffered saline (PBS), sections were incubated with biotin-labeled goat anti-mouse IgG (diluted 1∶200) for 45 min at 37 °C. Sections were then incubated with horseradish peroxidase streptavidin (S-A/HRP) for 45 min at 37 °C. After washed with PBS, peroxidase activity was developed using 3,3’-diaminobenzidine (DAB, 0.3 g/L) for 15 min. The DBA detection method yields a yellow precipitate. Sections were subsequently counterstained with hematoxylin.
PBS substituting for anti-HCV-NS5 IgG, biotin-labeled goat anti-mouse IgG, S-A/HRP, and omitting DBA in procedure served as negative controls, respectively.
The sections were deparaffinized with fresh xylene and graded alcohols, followed by PBS for 5 min. The tissues were digested with protinease K (30 mg/L, 37 °C, Sigma) for 15 min and rinsed with DEPC-treated PBS three times, and then treated with Rnase-free Dnase I (700 U/mL, Promega) at room temperature overnight or 37 °C 2 h in a humidified chamber. The sections were then fixed twice in 95% and 100% alcohol each for 3 min. RT was achieved with 30 μL- RT solution for each section (1 × RT buffer, dNTP 250 μM each, antisense of outer primer 1 μm, Rnasin 1 U/μL, AMV reverse transcriptase 0.4 U/μL Promega]) in a humidified chamber at 42 °C for 60 min. The reaction solution was dripped away and was hed with DEPC-treated PBS twice each for 5 min, then fixed twice in 95% and 100% alcohol each for 2 min, and the 50 μL PCR solution for each section was applied which consisted of MgCl2 2.5 mM, 1 × PCR buffer, each primer 1 μM, dNTP 250 μM, Taq-DNA polymerase 4 U/50 μL and BSA 3 g/L.The “hot-start” approach was employed during which Taq DNA ploymerase was added at 80 °C. The in situ amplification of target sequences was performed in a thermal cycler (GeneAmp in situ PCR System 1000 [Perkin Elmer]), using two primer pairs. The cycling conditions used were: the initial denaturation step at 94 °C for 4 min followed by 20 cycles of denaturation at 94 °C for 2 min, annealing at 55 °C for 1.5 min, and final extension of 72 °C for 3 min. The cover slip was removed. The sections were washed with PBS for 5 min and fixed in 100% alcohol for 10 min, then second PCR amplification was made. Except inner primers substituted for outer primers, the remaining steps were the same as the initial PCR amplification. The cover slip was removed. The sections were washed with PBS, and fixed in 100% alcohol for 10 min. Sections in PBS were heated at 80 °C for 10 min, and put on ice. Then hybridization solution (probe 2.5 mg/L, 50% deionized formamide, 5 × SSC, 1 × Denhardt’s solution, sssDNA 100 mg/L) was added on the slides at 37 °C overnight. The section was washed with serial SSC, and covered with 10% normal sheep serum. The specimens covered with S-A/HRP at 37 °C for 45 min. DAB solution was added in slides at 37 °C for 15 min. The sections were incubated in the dark and checked at a 5 min interval. The DBA detection method yields a yellow precipitate. After development, the sections were counterstained with hematoxylin. Positive cells and their histological distribution were examined.
Hepatitis B liver tissues; HCV RNA positive specimens digested by Rnase (10 g/L) at 37 °C for 3 h; HCV RNA positive specimens omitted AMV reverse transcription; HCV RNA positive specimens omitted Taq polymerase; and no probe control.
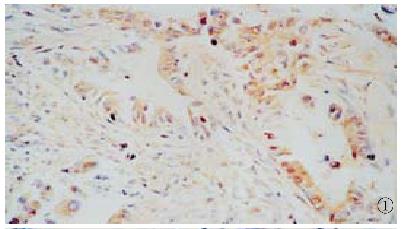
In 51 cases of carcinoma of extrahepatic bile duct, HCV NS5 protein was detected in 14 (27.5%), which was clearly stained in the cytoplasm of cancer cell but not in the nucleus or cell membrane. The positive signals of NS5 protein were distributed diffusely in the cytoplasm (Figure 1). The positive immunochemical reaction was not obtained in the same section when PBS substituted for anti-HCV-NS5 IgG, as did omission of the primary antibody from the staining procedure.
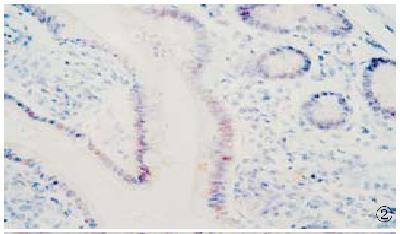
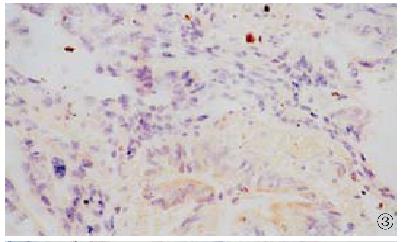
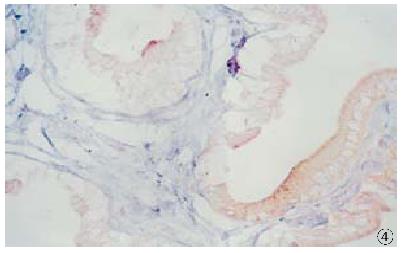
HCV RNA sequence was detected in 18 (35.4%) of 51 cases of carcinoma of extrah epatic bile duct. HCV RNA was located in the nucleus in 12 cases (Figure 2), and in the cytoplasm in 6 cases (Figure 3). In 34 cases of control group, HCV RNA sequences were detected in 2 (5.9%) (Figure 4). The HCV RNA positive signal was located occasionally in the mononuclear cells. After treated by Rnase or omitting AMV, Taq polymerase and probe in the procedure, no sections showed HCV RNA positive signal.
HCV NS5 protein and RNA coexistence were found in 2 cases. HCV NS5 protein and RNA positive cells were found to be either scattered or in clusters. In the cytoplasm, some positive signals of HCV NS5 protein and RNA were so strong that it might be difficult in determining nucleic positive.
Although HCV is considered essentially to be hepatotropic, some studies indicate that it can also be found in the extrahepatic tissue, such as peripheral blood mononuclear cells[33-37], kidney tissue and salivary glands[38]. In the present study, HCV NS5 protein and RNA were found in cells of carcinoma of extrahepatic bile duct, which further demonstrates that HCV has wide host cells, and the main nucleic localization of HCV RNA in cells of carcinoma of extrahepatic bile duct was resembled the localization of HCV RNA in HCC[39]. The incidence of HCV infection in the tissues of carcinoma of extrahepatic bile duct is significantly higher than in control group (χ² = 9.808, P = 0.002). This study indicates a correlation between HCV infection and carcinoma of extrahepatic bile duct, which is different from the traditional viewpoint─HCV infection is not correlated to bile duct cancer[2]. It is inferred that HCV infection, being similar to hepatolithiasis, choledocal cyst, etc., may be one of the risk factors involved in the development of carcinoma of extrahepatic bile duct.
HCV is a plus-strand RNA virus[40]. HCV RNA and proteins can be detected in cells of HCC[41-46], and infection with the HCV is now known to be a major risk factor for the development of HCC[47-51]. But HCV genome does not integrate into the genome of infected cells[52,53]. For hepatitis B virus (HBV), integration of HBV DNA into host chromosomes raises the possibility of a direct carcinogenic effect of HBV through interaction with oncogenes, growth factors, or tumor suppressor genes[54-58]. The mechanism of carcinogenesis of HCV is not fully understood now, which may be involved in proteins HCV gene encoding. It has been noted recently that the HCV core protein demonstrates diverse biological functions, including the regulation of cellular and unrelated viral genes at the transcriptional level, and has some potential direct carcinogenic effects in vitro. HCV core protein could activate human c-myc, early promoter of SV10, Rous sarcoma virus LTR and HIV-1 LTR[59], inhibit cisplatin-mediated apoptosis in human cervical epithelial cells and apoptosis induced by the overexpression of c-myc in Chinese hamster ovarian cells[60], and repress transcriptional activity of p53 promoter[61]. Translocated expression of HCV core protein may also inhibit apoptosis in the tissue of HCC[62]. REF cells cotransfected with HCV core and H-ras genes became transformed and exhibited rapid proliferation, anchor-independent growth, and tumor formation in athymic nude mice[63]. Transformation of NIH3T3 cells to the tumorigenic phenotype by the nonstructural protein NS3 of HCV was demonstrated and the proteinase activity associated with this protein was suggested as the cause of transformation[64]. HCV NS3 protein may exert its hepatocarcinogenic effect in early stage on host cells by endogenous pathway which may bring about mutation of p53 gene and transformation of hepatocytes[65]. NS5 protein from HCV-1b ORF includes NS5A and NS5B. Recently, NS5A protein is reported to be a potent transcriptional activator[66], and can repress the interferon-induced protein kinase through direct interaction with each other[67]. The experimental data suggest that HCV gene products have a function of gene regulation, and can modulate cell growth and differentiation, and may be directly involved in the malignant transformation of HCV-infecting cells. But how HCV infection is involved in the development of carcinoma of extrahepatic bile duct needs further research.
Edited by Ma JY
| 1. | Nahawold DL, Dawes LG. Carcinoma of the bile ducts. surgery: scientific principles and practice. 2nd ed. Philadelphia, New York: Lippincott Raven 1997; 1060-1066. |
| 2. | Anthony PP. Tumours and tumour like lesions of the liver and biliary tract. Pathology of the liver. 2nd ed. Edinburgh: Churchill Livingstone 1987; 574-590. |
| 3. | No authors listed. Primary liver cancer in Japan. The Liver Cancer Study Group of Japan. Cancer. 1984;54:1747-1755. [PubMed] |
| 4. | Moto R, Kawarada Y. Diagnosis and treatment of cholangiocarcinoma and cystic adenocarcinoma of the liver. Neoplasms of the liver. Tokyo: Spring-Verlag 1987; 381-393. |
| 5. | Chearanai O, Plengvanit U, Damrongsak D, Tuchinda S, Damrongsak C, Viranuvatti V. Primary liver cancer, angiographic study of 127 cases. J Med Assoc Thai. 1984;67:482-491. [PubMed] |
| 6. | Schwartz DA. Cholangiocarcinoma associated with liver fluke infection: a preventable source of morbidity in Asian immigrants. Am J Gastroenterol. 1986;81:76-79. [PubMed] |
| 7. | Kurathong S, Lerdverasirikul P, Wongpaitoon V, Pramoolsinsap C, Kanjanapitak A, Varavithya W, Phuapradit P, Bunyaratvej S, Upatham ES, Brockelman WY. Opisthorchis viverrini infection and cholangiocarcinoma. A prospective, case-controlled study. Gastroenterology. 1985;89:151-156. [PubMed] |
| 8. | Colombari R, Tsui WM. Biliary tumors of the liver. Semin Liver Dis. 1995;15:402-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Craig JR, Peters RL, Edmondson HA. Primary malignant epithelial tumors. Atlas of tumor pathology: tumors of the liver and intrahepatic bile ducts. 2nd series. Washington DC: Armed Forces Institute of Pathology 1989; 123-222. |
| 10. | Ishak KG, Anthony PP, Sobin LH. Intrahepatic cholangiocarcinoma. Internationalhistopathological classification of tumors: histological typing of tumors of the liver. 2nd ed. Washington DC: Springer-Verlag 1994; 17-19. |
| 11. | Altaee MY, Johnson PJ, Farrant JM, Williams R. Etiologic and clinical characteristics of peripheral and hilar cholangiocarcinoma. Cancer. 1991;68:2051-2055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Huang ZQ. [The present status of surgical treatment of intrahepatic lithiasis in a nation-wide survey in China of 4197 operative cases 1981-1985]. Zhonghua Wai Ke Za Zhi. 1988;26:513-22, 572. [PubMed] |
| 13. | Wang YS, Yang JZ. Fourty-five cases of cholelithiasis with cholangiocarcinoma. Xin Xiaohuabingxue Zazhi. 1997;5:606. |
| 14. | Wang BS. [Choice of operation in congenital dilatation of the bile duct in adults]. Zhonghua Wai Ke Za Zhi. 1988;26:287-29, 287-29. [PubMed] |
| 15. | Liang LJ, Huang JF, Lu MD, Chao XH. Cancerization of choledochal cyst in adults: report of 12 cases.Zhongguo. Shiyong Waike Zazhi. 1995;15:598-599. |
| 16. | Ji F, Shi WJ. Analysis of five cases of bile duct cysts with bile duct carcinoma.Gandan. Yipi Waike Zazhi. 1997;9:15-16. |
| 17. | Gu HG, Cai JX. Ten cases report on congenital choledochal cyst associated biliary cancer. Gandan Waike Zazhi. 1996;4:49-51. |
| 18. | Zhu SH. [The relationship between clonorchiasis (Clonorchis sinensis) and other hepatobiliary diseases--clinical analysis of 2, 214 cases (author's transl)]. Zhonghua Nei Ke Za Zhi. 1982;21:34-36. [PubMed] |
| 19. | Cao XH, Deng BS, Wan DS. Carcinoma of extrahepatic bile duct. Zhonghua Waike Zazhi. 1982;22:164-165. |
| 20. | Huang ZQ, Huang XQ. Evolution of surgical treatment of intrahepatic lithiasis in China. China Natl J New Gastroenterol. 1997;3:131-133. |
| 21. | Yuan M, Huang ZQ. Cholangiocarcinoma as related to intrahepatic stones. Zhonghua Binglixue Zazhi. 1982;11:95-97. |
| 22. | Huang ZQ. Current progress of biliary tract surgery in China. Huaren Xiaohua Zazhi. 1998;6:1-2. |
| 23. | Li J, Liu XT, Wang KL, Li XL. The relationship between clonorchis sinensis infestion and hepatobiliary tumor. Zhongguo Chuji Weisheng Baojian. 1998;12:39. |
| 24. | Biliary surgery group of surgery association of Chinese medical association. Analysis of 1698 cases of carcinoma ofextrahepatic bile duct in China. Zhonghua Waike Zazhi. 1990;28:516-519. |
| 25. | Esteban JI, González A, Hernández JM, Viladomiu L, Sánchez C, López-Talavera JC, Lucea D, Martin-Vega C, Vidal X, Esteban R. Evaluation of antibodies to hepatitis C virus in a study of transfusion-associated hepatitis. N Engl J Med. 1990;323:1107-1112. [PubMed] |
| 26. | Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo QL, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1225] [Cited by in RCA: 1145] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 27. | Heintges T, Wands JR. Hepatitis C virus: epidemiology and transmission. Hepatology. 1997;26:521-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 139] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Tao QM, Wang Y, Wang H, Chen WR, Sun Y, Meng Q. Investigation of anti-HCV in 391 serum samples in China. Chin Med J (. Engl). 1990;103:616-618. [PubMed] |
| 29. | Tang S. [Seroepidemiological study on hepatitis C virus infection among blood donors from various regions in China]. Zhonghua Liu Xing Bing Xue Za Zhi. 1993;14:271-274. [PubMed] |
| 30. | Ling BH, Zhuang H, Cui YH, An WF, Li ZJ, Wang SP, Zhu WF. A cross-sectional study on HGV infection in a rural population. World J Gastroenterol. 1998;4:489-492. [PubMed] |
| 31. | Nuovo GJ, Lidonnici K, MacConnell P, Lane B. Intracellular localization of polymerase chain reaction (PCR)-amplified hepatitis C cDNA. Am J Surg Pathol. 1993;17:683-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 76] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Lau GK, Fang JW, Wu PC, Davis GL, Lau JY. Detection of hepatitis C virus genome in formalin-fixed paraffin-embedded liver tissue by in situ reverse transcription polymerase chain reaction. J Med Virol. 1994;44:406-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Zignego AL, Macchia D, Monti M, Thiers V, Mazzetti M, Foschi M, Maggi E, Romagnani S, Gentilini P, Bréchot C. Infection of peripheral mononuclear blood cells by hepatitis C virus. J Hepatol. 1992;15:382-386. [PubMed] |
| 34. | Moldvay J, Deny P, Pol S, Brechot C, Lamas E. Detection of hepatitis C virus RNA in peripheral blood mononuclear cells of infected patients by in situ hybridization. Blood. 1994;83:269-273. [PubMed] |
| 35. | Wu HB, Li ZW, Li Y. Clinical significance of detection of positive and negative strands of HCV RNA in peripheral bloodmononuclear cells. Shijie Huaren Xiaohua Zazhi. 1999;7:220-221. |
| 36. | He Y, Liu W, Zeng L. [The effect of interferon in combination with ribavirin on the plus and minus strands of hepatitis C virus RNA in patients with hepatitis]. Zhonghua Nei Ke Za Zhi. 1996;35:32-35. [PubMed] |
| 37. | Zhou P, Cai Q, Chen YC, Zhang MS, Guan J, Li XJ. Hepatitis C virus RNA detection in serum and peripheral bloodmononuclear cells of patients with hepatitis C. China Natl J New Gastroenterol. 1997;3:108-110. |
| 38. | Willson RA. Extrahepatic manifestations of chronic viral hepatitis. Am J Gastroenterol. 1997;92:3-17. [PubMed] |
| 39. | Zeng WZ, Jian MD, Chu GZ. [Detection of hepatitis C virus RNA in cells with hepatocellular carcinoma and its practical significance by in situ polymerase chain reaction]. Zhonghua Nei Ke Za Zhi. 1994;33:747-750. [PubMed] |
| 40. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. |
| 41. | Dai YM, Shou ZP, Ni CR, Wang NJ, Zhang SP. Localization of HCV RNA and capsid protein in human hepatocellular carcinoma. World J Gastroenterol. 2000;6:136-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 42. | Liu YJ, Cong WM, Xie TP, Wang H, Shen F, Guo YJ, Chen H, Wu MC. Detecting the localization of hepatitis B and C virus in hepatocellular carcinoma by double in situ hybridization. China Natl J New Gastroenterol. 1996;2:187-189. |
| 43. | Zhang LF, Peng WW, Yao JL, Tang YH. Immunohistochemical detection of HCV infection in patients with hepatocellular carcinoma and other liver diseases. World J Gastroenterol. 1998;4:64-65. [PubMed] |
| 44. | Yang JM, Wang RQ, Bu BG, Zhou ZC, Fang DC, Luo YH. Effect of HCV infection on expression of several cancer-associated gene products in HCC. World J Gastroenterol. 1999;5:25-27. [PubMed] |
| 45. | Zhao XP, Shen HX, Tian DY, Zhang DS, Peng ZH, Yang DL, Hao LJ. Expression and significance of HCV RNA and HCV NS5 antigen in liver tissues of patients with hepatitis C. Shijie Huaren Xiaohua Zazhi. 1999;7:516-518. |
| 46. | Zhai SH, Liu JB, Liu YM, Zhang LL, Du ZP. Expression of HBsAg, HCV-Ag and AFP in liver cirrhosis and hepatocarcinoma. Shijie Huaren Xiaohua Zazhi. 2000;8:524-527. |
| 47. | Di Bisceglie AM. Hepatitis C and hepatocellular carcinoma. Hepatology. 1997;26:34S-38S. [PubMed] |
| 48. | Liu WW. Etiological studies of hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:93-95. |
| 49. | Du JH, Cha WZ. Interrelation between hepatitis C and primary hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:176. |
| 50. | Wu MC. Clinical research advances in primary liver cancer. World J Gastroenterol. 1998;4:471-474. [PubMed] |
| 51. | Zhang WH, Zhu SN, Lu SL, Cong WM, Wu MC. HBV concomitant infection in HCV associated HCC. Shijie Huaren Xiaohua Zazhi. 2000;8:175-177. |
| 52. | Tsuboi S, Nagamori S, Miyazaki M, Mihara K, Fukaya K, Teruya K, Kosaka T, Tsuji T, Namba M. Persistence of hepatitis C virus RNA in established human hepatocellular carcinoma cell lines. J Med Virol. 1996;48:133-140. [PubMed] |
| 53. | Fong TL, Shindo M, Feinstone SM, Hoofnagle JH, Di Bisceglie AM. Detection of replicative intermediates of hepatitis C viral RNA in liver and serum of patients with chronic hepatitis C. J Clin Invest. 1991;88:1058-1060. [PubMed] |
| 54. | Shafritz DA, Shouval D, Sherman HI, Hadziyannis SJ, Kew MC. Integration of hepatitis B virus DNA into the genome of liver cells in chronic liver disease and hepatocellular carcinoma. Studies in percutaneous liver biopsies and post-mortem tissue specimens. N Engl J Med. 1981;305:1067-1073. [PubMed] |
| 55. | Wang XZ, Tao QM. Hepatitis B X gene and hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:1063-1064. |
| 56. | Gao FG, Sun WS, Cao YL, Zhang LN, Song J, Li HF, Yan SK. HBx-DNA probe preparation and its application in study of hepatocarcinogenesis. World J Gastroenterol. 1998;4:320-322. [PubMed] |
| 57. | Qin LL, Su JJ, Li Y, Yang C, Ban KC, Yian RQ. Expression of IGF- II, p53, p21 and HBxAg in precancerous events of hepatocarcinogenesis induced by AFB1 and/or HBV in tree shrews. World J Gastroenterol. 2000;6:138-139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 58. | Wang Y, Liu H, Zhou Q, Li X. Analysis of point mutation in site 1896 of HBV precore and its detection in the tissues and serum of HCC patients. World J Gastroenterol. 2000;6:395-397. [PubMed] |
| 59. | Ray RB, Lagging LM, Meyer K, Steele R, Ray R. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 1995;37:209-220. [PubMed] |
| 60. | Ray RB, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176-182. [PubMed] |
| 61. | Ray RB, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983-10986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 201] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 62. | Li J, Chen YF, Wang WL, Lin SG. Translocated expression of HCV core protein inhibits apoptosis in the tissue of hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:579-582. |
| 63. | Ray RB, Lagging LM, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438-4443. [PubMed] |
| 64. | Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893-3896. [PubMed] |
| 65. | Feng DY, Chen RX, Peng Y, Zheng H, Yan YH. Effect of HCV NS (3) protein on p53 protein expression in hep atocarcinogenesis. World J Gastroenterol. 1999;5:45-46. [PubMed] |