INTRODUCTION
Calmodulin (CaM), widely distributed in almost all eukaryotic cells, is a major intracellular calcium receptor responsible for mediating the Ca2+ signal to a multitude of different enzyme systems and is thought to play a vital role in the regulation of cell proliferative cycle[1,2]. Recently, many studies showed that CaM is also present in extracellular fluid such as cell culture media and normal body fluid and has been reported to stimulate proliferation in a range of normal and neoplastic cells, apparently acting as an autocrine growth factor[3-11]. In 1988, Crocker et al[7] reported for the first time that addition of extracellular pure pig brain CaM could promote DNA synthesis and cell proliferation in K562 human leukaemic lymphocytes. After th at, more and more research was done on extracellular CaM and evidences demonstra ted that extracellular CaM could also stimulate cell proliferation in normal human umbilical vein endothelial cells[5], keratinocytes[4], suspen sion-cultured cells of Angelica Dahurica, etc[6].CaM is a monome ric protein of 148 amino acids that contains four homologous Ca2+ binding domains. CaM has been highly conserved throughout the evolution. Only 1 out of 148 amino acids of human CaM is different from that of fish CaM. Complementary DNAs encoding rat, eel, chicken, human, and trypanosome CaM have been cloned.
In this paper, we describe the expression and purification of recombinant human CaM (rhCaM) and the role of extracellular rhCaM in SP2/0 mouse myeloma cell proliferation.
MATERIALS AND METHODS
Construction of expression plasmid
PCR amplification was used for the insertion of the coding sequence of human CaM III (hCaM III) between BamH I and EcoR I sites of the expre ssion vector pBV220 (constructed by Dr. Zhang[12,13]). The primers for P CR based on the reported sequence of hCaM III were synthesized by DNA synthesizer (Sangon). Upper primer:5’-CGGAATTCATATGGCTGACCAGCTGAC-3’, containing EcoRI restriction site. Down primers: 5’-CGGGATCCTTACTTTGCAGTCATCATC-3’, containing BamHI restriction site. The vector pUC/hCaM III (a generous gift from Dr.Strehler EE[14]) was used as template. PCR amplif ications were performed on a thermocycler (PE 9600). The PCR conditions used were 95 °C for 1 min, 58 °C for 1 min, 72 °C for 1 min, for 30 cycles.The product was a single band of about 400 bases detected by 1.2% agarose gel electrophoresis. After purifying the PCR product by High Pure PCR Product Purification Kit (Boehr inger Mannheim) and isolating the vector pBV220 by the Wizard Plus Minipreps DNA Purification Kit (Promega), the amplified gene fragments of hCaM III digested with EcoRI and BamHI were ligated into the pBV220 vector that had been previously digested with the same enzymes. The recombinant construct (pBV220/hCaM III) was then used to transform E.coli DH5α (a ge nerous gift from Beijing Institute of Basic Medical Sciences) competent cells by the CaCl2 method.
Expression and purification of rhCaM
A single colony from E.coli DH5α cell harboring the pBV220/hCaM III construct was used to inoculate a 5-mL LB culture. After overnight growth a t 30 °C, the 5 mL culture was used to inoculate a 1-L culture in LB medium and grown at 30 °C for 4-6 h until the absorbance reached 0.5 at 600 nm. The bact erial culture was then induced at 42 °C for additional 5 h. Stable rhCaM-express ing recombinant E.coli was harvested by centrifugation at 5000 r/min for 5 m in. The pellets were washed twice with PBS, resuspended in 0.05 mol/L Tris-HCl buffer (pH7.5, containing 0.25 mmol/L PMSF, 2 mmol/L EGTA and 2 mmol/L β-merca ptoethanol) and disrupted by sonication (Cyclone/Tempest IQ2) at 100 W for 30 min with 10 s of pulse and 10 s of pulse off time. Supernatant fractions were o btained by centrifugation at 12000 r/min for 30 min. CaCl2 solution 0.1 mol/L was added to the supernatant to adjust to 5 mmol/L CaCl2 final concentration. The supernatant was applied to a Phenyl-sepharose CL-4B affinity column (Pharm acia) equilibrated with buffer I (0.05 mol/L Tris-HCl buffer, pH7.5, 0.1 mmol/L CaCl2), then the column was washed with buffer I followed by another wash with buffer I containing 0.5 mol/L NaCl. CaM then was eluted from the column with b uffer I containing 1 mmol/L EGTA. The solution containing purified rhCaM was dialysed against ddH2O for 48 h. The protein concentration was determined by Brad ford assay. RhCaM was filtered through micropore filter membrane (0.22 µm) to remove bacteria and stored at -20 °C.
SDS-PAGE and immunoblot assay
Expression and purified protein were analyzed by 15% SDS-PAGE. Gels were stained with Coomassie brilliant blue G. For immunoblot analysis, protein were separated by 15% SDS-PAGE and electroblotted on to nitrocellulose (NC) membrane (Bio R ad). After blotting, nonspecific protein binding sites were blocked with 3 g/ L B SA in 50 mmol/L Tris-HCl, pH 8, and 150 mmol/L NaCl, 0.5% Tween 20. The NC memb rane were incubated with a 1:200 dilution of anti-CaM McAb (Sigma). After wash ing, the membrane were incubated for 1 h with HRP conjugated goat anti-mouse immunoglobulinantibody(1:200,BoehringerMannheim). Color development was obtai ned by adding DAB substrate.
Amino acid composition
The amino acid composition of purified expression product was analyzed using an automatic amino acid analyzer, Hitachi 835-5Q.
DNA sequence analysis
The DNA sequence was determined with ABI PRISMTM 377 DNA sequenser.
NAD kinase assay
The activity of CaM-dependent NAD kinase(NADK) was detected as described by Harmon[15].
Cell culture
SP2/0 cells were cultured in RPMI 1640 medium (GIBCO), supplemented with 20% neon atal calf serum (NCS) under 37 °C, 5% CO2 conditions.
Measurement of cell proliferative rate
Cell proliferative rate was determined by MTT colorimetric assay[16,17]. SP2/0 cells were collected at the logarithmic growth phase by centrifugation. The cells were washed twice and seeded in 96-well plate and cultured for 48 h with rhCaM and CaM- antagonist trifluoperazine (TFP). MTT 10 µL (Sigma, 0.5 mg M TT in 1 mL PBS) was added to each well of 96-well plate and cultured for another 4 h. After the formazan was dissolved with 100 µL DMSO, absorbance value of each well at 490 nm (A490nm) was read on a BioRad Model 550 microplate reader.
Proliferative rate = A490 nm value of experimental group/A490 nm value of control group × 100%. Improved proliferative rate = Proliferative rate - 100%.
RESULTS
Expression of rhCaM in E.coli
The results of both restriction enzyme digestion of pBV220/hCaM III a nd PCR identification in which pBV220/hCaM III was used as template s howed a specific single band with the same molecular weight as reported hCaM gen e on agarose gel (Figure 1). DNA sequence analysis also indicated that the recom binant vector had been constructed successively. After heat induction, the total extract of the E.coli DH5α cell harboring pBV220/hCaM III was a nalyzed by 15% SDS-PAGE under reducing conditions. As shown in Figure 2, a unique protein band with an apparent molecular weight (17000) was similar to that o f standard human brain CaM. This protein accounted for over 20% of the total cell ular protein. The study on solubility of expression protein indicated that CaM protein was expressed predominantly in the soluble form. Western blot analysis sh owed that anti-CaM McAb specifically bound to the 17000 band of expression product. The expression product purified by phenyl-sepharose CL-4B affinity chrom a tography was shown as a single band on argarose gel by SDS-PAGE. The protein concentration was determined by Bradford method and approximately 3-4 mg of the purified protein were obtained from 1 L of bacterial culture.
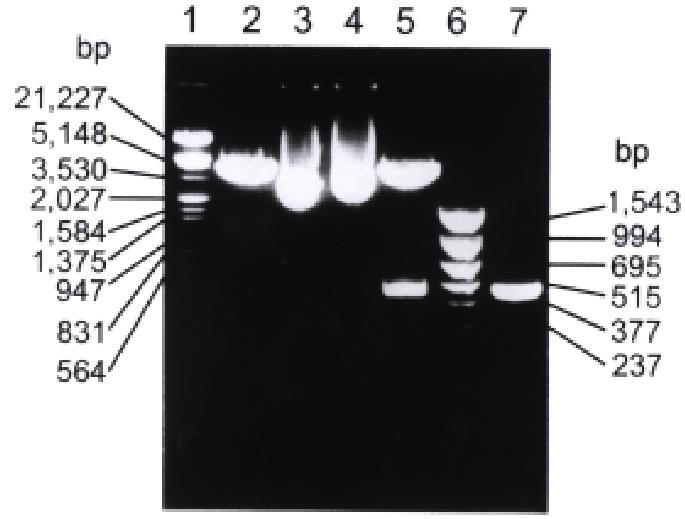
Figure 1 Restriction enzyme digestion and PCR analysis of recombinant plasmid pBV220/hCaM III.
1. λ DNA/Hind III+EcoR I marker; 2. pBV220 digested with EcoR I and BamH I; 3. pBV220; 4. pBV220/hCaM III; 5. pBV220/hCaM III digested with EcoR I and BamH I; 6. pBR322/Hinf I marker; 7. PCR product of hCaM III.
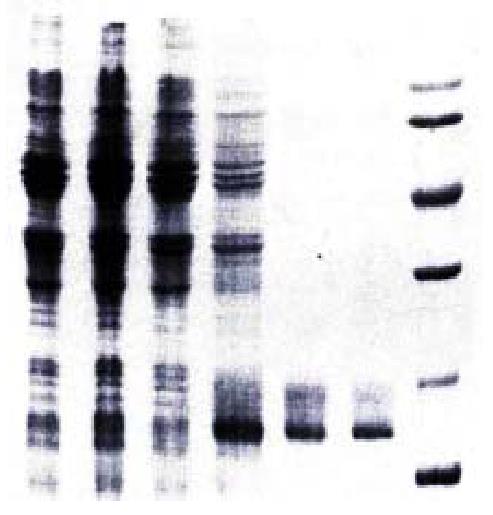
Figure 2 SDS-PAGE analysis of hCaM expression and purification.
1. Uninduced DH5α/pBV220; 2. Induced DH5α/pBV220; 3. Uninduced DH5α/pBV220-hCaM III; 4. Induced DH5α/pBV220-hCaM III; 5. Purified rhCaM by Phenyl-sepharose CL-4B column; 6. Standard Human brain CaM (Sigma); 7. Protein molecular weight marker.
Amino acid composition analysis
The amino acid compositions of purified rhCaM were identical to those of the pre viously reported bovine brain CaM. The acidic amino acid (such as Asp and Glu) composition was about 30% or more.
NAD kinase assay
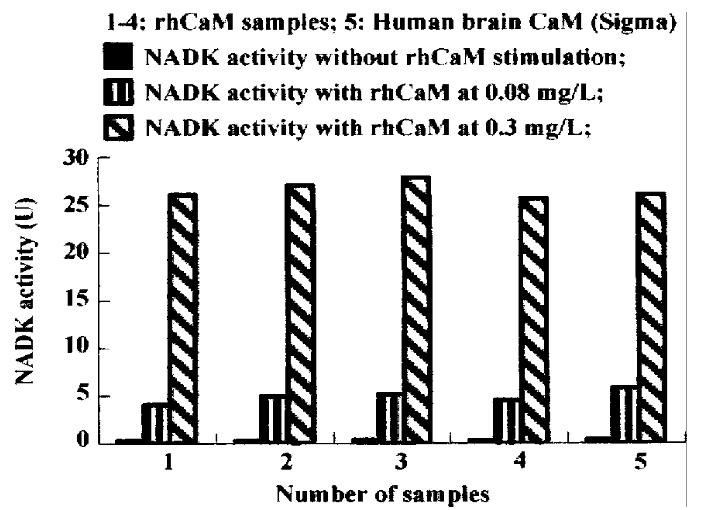
The results of NAD kinase (NADK) assay showed that purified rhCaM was able to activate CaM-dependent NADK activity to the same extent as the standard human brain CaM (Sigma, Figure 3).
Figure 3 Activation of NADK by rhCaM.
Effect of extracellular rhCaM on cell proliferation
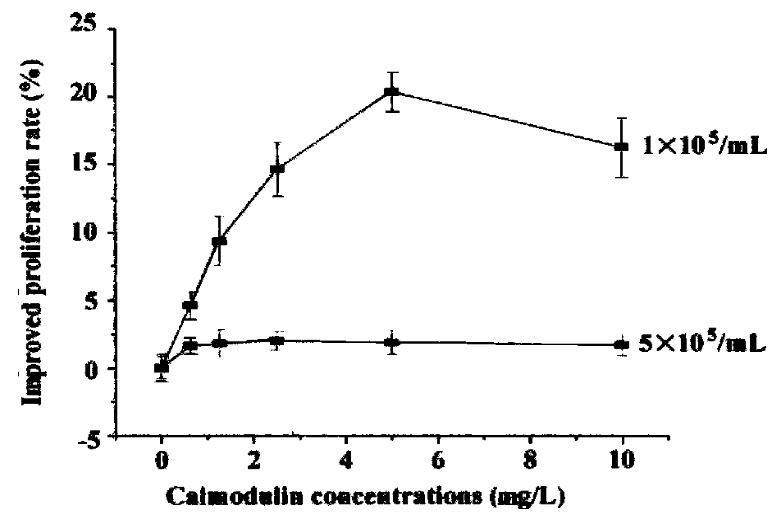
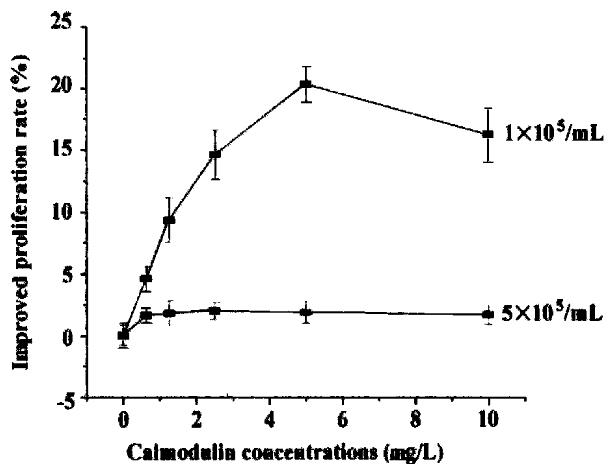
Cells were diluted with RPMI 1640 (containing 0. 5% NCS) to 1-5 × 105/mL and se eded in 96-well plates. rhCaM was added to the final concentration of 0.1-10 μg/mL and each concentration was triplicated. Cells were incubated with different concentrations of rhCaM under 37 °C, 5% CO2 conditions for 48 h prior to MTT assay. The effect of extracellular rhCaM on cell proliferation was investigated (Figure 4). Evidence indicated that rhCaM within a certain concentration (0.1-7.5 mg/L) could stimulate cell proliferation. The stimulatory effect declined when the rhCaM concentration was higher than 10 mg/L. Effect of a ddition of pu re rhCaM was also dependent in some degree upon the cell density of the initial culture. For a certain CaM concentration, the lower the cell density of the init ial culture, the higher the promoting effect. Moreover, the effect of exogenous rhCaM was also influenced by the amount of NCS added to the culture medium. When NCS in medium accounted for 0.5%-1%, a significant stimulatory effect could be observed. However, when NCS in medium was increased to 2%-10%, no stimulatory effect was observed for rhCaM (data not supplied).
Figure 4 The effect of extracellular rhCaM on cultured SP2/0 cells.
Inhibitory effect of TFP on cell proliferation
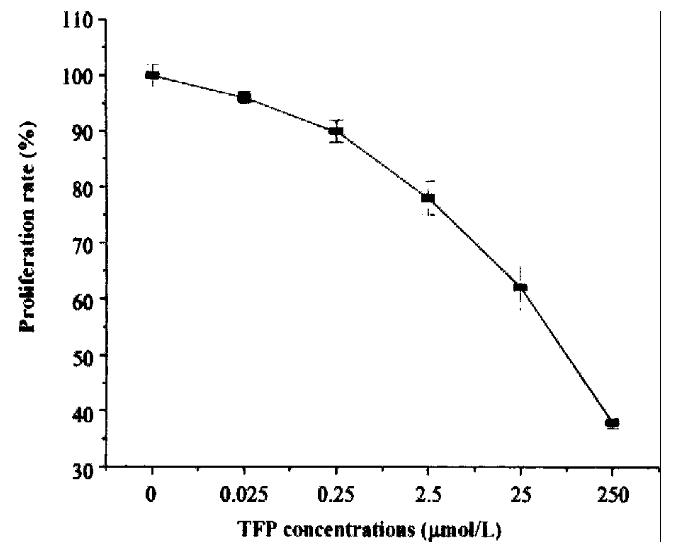
Cells were seeded in 96-well plates as above. CaM-antagonist TFP (Sigma) was added to the final concentration of 0.025-250 μmol/L. Each concentra tion was do ne in triplicate. Cells were incubated with TFP under 37 °C, 5% CO2 conditions for 48 h and determined by MTT assay. Results indicated that addition of various concentrations of TFP to cell culture medium could significantly inhibit cell p roliferation rate and the inhibitory effect strengthened with increase in the co ncentrations of TFP (Figure 5).
Figure 5 The inhibitory effect of CaM-antagonist TFP on cell proliferation.
Effect of extracellular rhCaM on TFP-inhibited cells
TFP was added to the SP2/0 cell (1 × 105/mL) culture medium until the final concentrations were 25 μmol/L. Cells were cultured in 5% CO2 incuba ted under 37 °C for 6 h, washed twice with RPMI 1640, then seeded into 96-well plate at the density of 1 × 105/mL. Various concentrations of rhCaM were added and cells were cultured for 48 h. Results demonstrated that addition of rhCaM could alleviat e the inhibitory effect of TFP. rhCaM 5 mg/L could offset the inh ibition almost to normal (Figure 6).
Figure 6 The effect of extracellular rhCaM on TFP inhibited cells.
DISCUSSION
CaM, a heat-stable, acidic and multifunctional calcium-binding protein, exists in almost all eukaryotic cells. On the gene level, a multigene family of th ree maximally divergent members is responsible-at least in mammals-for the gene ration of the single CaM protein with the same amino acid sequence. Three human CaM cDNAs called hCaM I, hCaM II, and hCaM III have been cloned[14,18-23]. In this paper we described the constructi on of expression plasmid for human CaM III gene and expression of hCa M protein in E. coli. About 3-4 mg of purified rhCaM was obtained from 1 L of E.coli culture. This level of production is higher than that achieved by the classic purification method. If the genetic engeneering techniques are used for a large scale expression, the product can be much higher.
It is well known that intracellular CaM plays an important role in the regulatio n of cell proliferation. The levels of intracellular CaM and CaM mRNA vary during cell cycle and accelerate cell proliferation. Moreover, CaM antagonists TFP, W7, W13 and anti-CaM antibodies can block the effect of CaM and arrest cell cycle at G1/S boundary[24-27]. In the last few years, extracellular CaM was detected and evidence accumulated that extracellular CaM could also affect cell proliferation.
In our study, effects of purified rhCaM and CaM antagonist TFP on SP2/0 prolife ration rate were examined. The results revealed that certain concentrations of extracellular rhCaM (0.1-7.5 mg/L) could stimulate cell divergent members is responsible-at least in mammals-for the gene ration of the single CaM protein with the same amino acid sequence. Three human CaM cDNAs called hCaM I, hCaM II, and hCaM III have been cloned[14,18-23]. In this paper we described the constructi on of expression plasmid for human CaM III gene and expression of hCa M protein in E. coli. About 3-4 mg of purified rhCaM was obtained from 1 L of E.coli culture. This level of production is higher than that achieved by the classic purification method. If the genetic engeneering techniques are used for a large scale expression, the product can be much higher.
It is well known that intracellular CaM plays an important role in the regulatio n of cell proliferation. The levels of intracellular CaM and CaM mRNA vary during cell cycle and accelerate cell proliferation. Moreover, CaM antagonists TFP, W7, W13 and anti-CaM antibodies can block the effect of CaM and arrest cell cycle at G1/S boundary[24-27]. In the last few years, extracellular CaM was detected and evidence accumulated that extracellular CaM could also affect cell proliferation.
In our study, effects of purified rhCaM and CaM antagonist TFP on SP2/0 prolife ration rate were examined. The results revealed that certain concentrations of extracellular rhCaM (0.1-7.5 mg/L) could stimulate cell proliferation in a dose-dependent manner and the stimulatory effect was dependent upon the cell density of the initial culture. The lower the cell density, the higher the stimulatory effect.The results are consistent with previous literature[4-7]. TFP inhibited cell proliferation and the inhibition could be alleviated by addition of extracellular rhCaM, which further supports that extracellular CaM could acce lerate cell proliferation. CaM was also detected in many extracellular body flui ds such as breast milk, saliva and serum. Serum, which is necessary for the succ essful culture of many cell types, contains high levels of CaM which could amoun t to 0.9-8 mg/L[28]. This might explain why the effect of CaM was inf luenced by NCS added to culture media. While CaM could facilitate cell prolifera tion, compared with NCS, the effect of CaM was less important.
So far, how extracellular calmodulin achieves its effects is yet unclear. Evidence indicates that the mechanisms may be as follow: (1) CaM can interact with so me cell proliferation-related factors such as epidermal growth factor (EGF), transforming growth factor (TGF) and platelet derived growth factor (PDGF). It is possible that extracellular CaM exerts its effects by strengthening these factors’ binding to their membrane receptors. It was reported that EGF receptor conta ned a CaM-binding domain[29] and the binding of EGF to cells could be inhibited by CaM antagonist[30]. MacNeil et al[28] showed that there was a significant positive correlation between CaM level and EGF concentrations in normal body fluids, indicating that CaM together with EGF, TGF, PDGF etc may be members of a functionally co-ordinated group of mitogens. (2) Ca M may directly interact with CaM-binding proteins (CaMBPs) on cell membrane. CaMBPs were found to exist on cell wall of wheat coleoptiles[31]. Extrac ellular CaMBPs were also detected in the suspension-cultured cells of Angelica Dahurica and carrot[32]. CaMBP is a kind of glycoprotein which can bind with CaM and may function as a bridge between CaM and intracellular metabolic processes.