Published online Jun 15, 2000. doi: 10.3748/wjg.v6.i3.395
Revised: February 3, 2000
Accepted: March 3, 2000
Published online: June 15, 2000
- Citation: Wang Y, Liu H, Zhou Q, Li X. Analysis of point mutation in site 1896 of HBV precore and its detection in the tissues and serum of HCC patients. World J Gastroenterol 2000; 6(3): 395-397
- URL: https://www.wjgnet.com/1007-9327/full/v6/i3/395.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i3.395
Hepatitis B is one of the common infectious diseases, which severely impairs the health of the people in our country and has close relationship to the initiation and progression of chronic hepatitis, cirrhosis, and liver cancer. The recent researches indicate that the mutation of HBV precore exists in the patients with these diseases as stated above[1-5]. According to the recent publications, the mutation of HBV attracts great interests of investigators. The major mutation points in HBV precore are the point in sites 1896 (A1893) and 83 (A83), which are both of G→A point mutations[6]. Based on the DNA sequence of precore region of HBV, the method of 3'base specific polymerase chain reaction (3'-BS-PCR) is applied to analyze the 1896 site mutation of HBV[7] in 126 clinical serum samples and 23 patients' tissues and ser a whose tumors have been surgically excised and pathologically diagnosed.
Primers were ordered from Life Technology Company, USA; Proteinase K, Taq Polymerase, MgCl2, PCR reaction buffer ordered from Hua Mei Biotechnology Company; other reagents were of analytical purity and made in China. Hema Thermocycler was the product from Zhuhai Hema Medical Instruments Company.
The 126 serum samples were collected from outpatients and inpatients of Department of Infectious Disease, the First Affiliated Hospital of Anhui Medical University from November 1996 to August 1997. Seventy-two patients were infected by HBV, and 14 by HAV. Forty were HBsAg negative diagnosed by the method of RPHA. The 23 samples of liver cancer tissues were the surgical specimens from HCC patients in Department of Surgery, the First Affiliated Hospital of Anhui Medical University who were corroborated by pathological diagnosis. The corresponding serum was obtained before operation. These patients are diagnosed according to the diagnostic criteria stipulated by the Sixth National Meeting of Hepatitis, 1990.
Into 125 μL of the serum to be examined, add 125 μL HBV DNA extraction buffer (200 mmol·L-1 NaCl, 2 mmol·L-1-EDTA, 1% SDS, 100 mmol·L-1 Tris-HCl, pH8.0) and 6.25 μL Proteinase K (2 g·L-1).Incubate in 37 °C water bath for 6 h. Extract with phenol and chloroform, precipitate the HBV DNA with ethanol, dissolve in 20 μL sterile re-distilled water and store at -20 °C.
Into the 200 μg HCC tissue, add 500 μL of DNA extract buffer (100 mmol·L-1 NaCl, 1 mmol·L-1 EDTA, 0.5% SDS, 50 mmol·L-1 Tris-HCl, pH8.0). Homogenize on ice. Add 50 μL of Proteinase K(2 g·L-1). Incubate in 37 °C water bath for 6 h Extract with phenol and chloroform, precipitate the HBV DNA with ethanol, dissolve in 20 μL sterile re-distilled water and store at -20 °C.
The primers are designed according to the recorded HBV DNA sequence, the principle of 3'-BS-PCR, and the papers published by Goergen[6] i.e. 3' primer (GW-1-1d) TCCACACTCCAAAAGACA (2287-2270), wild type 5' primer (GW-1-1a) GTGCCTTGGGTGGCTTTG (1879-1896) and mutant type 5' primer (GW-1-1b) GTGCCTTGGGTGGCTTTA (1879-1896). Into the 10 μL DNA extracted from serum or tissue diluted into 5 ng·μL-1, add PCR reaction buffer [10 mmol·L-1, Tris, 50 mmol·L-1 KCl, 2 mmol·L-1 MgCl2, 0.001% Gelatin, 200 μmol·L-1 dNTPs, 0.5 μmol·L-1 primers (wild type: GW-1-1a + GW-1-1d; mutant type: GW-1-1a + GW-1-1b) and 0.5 μL Taq DNA polymerase (3 U·μL-1)], covered with mineral oil, run PCR: 93 °C 1 min, 64 °C 1 min, 72 °C 1 min, 30 cycles, 72 °C 5 min. Aspirate 8 μL of PCR products, check the results with 1.5% agarose gel electrophoresis. The positive band is of 408 bp.
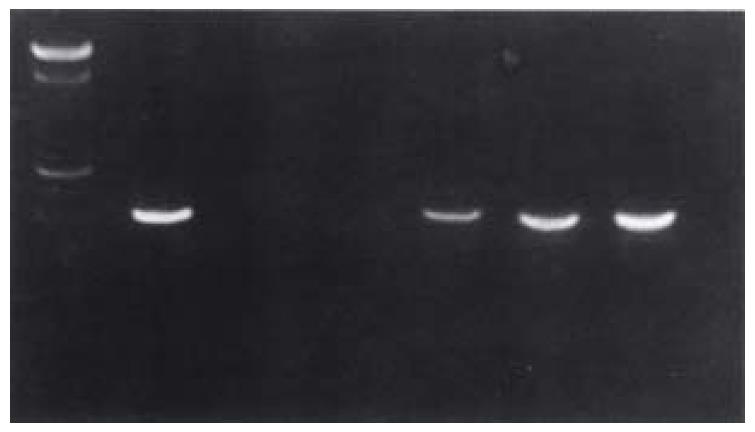
Using the specific oligonucleotides designed according to the DNA sequences of mutant and wild types the results showed that the amplified specific DNA of wild and mutant types of HBV in precode C were both of 408 bp. Figure 1 shows the PCR results of the DNA extracted from 126 patients' serum obtained at different stages, which demonstrates that constant results can be achieved when repeated. The PCR results of the 14 patients infected by HAV were all negative.
Of the 72 patients with chronic hepatitis examined with the 3'-BS-PCR of HBV DNA, 58 were positive in wild type, 13 were positive in mutant type only, 21 were positive in both types. The positive rate of mutant type was 47.2% (34/72); the positive rate of mutant type only was 18.1%, the positive rate of both types was 29.1% (21/72). Neither of these types were detected in the 40 patients with negative HBsAg.
| Total | HBV precore positive | HBV precore negative | HBV precore positive | |||
| Mutant type | Wild type | Mixed type | ||||
| Tissue | 23 (100%) | 12 (52.2) | 6 (26.1) | 5 (21.7) | 12 (52.2) | 5 (21.7) |
| Serum | 23 (100%) | 7 (30.4) | 16 (69.6) | 6 (26.1) | 4 (17.4) | 3 (13.0) |
The positive rate of the 23 HCC patients' surgically excised tissues and pre-operational serum was 52.2% (12/23) and 30.4% (7/23) respectively. In order to exclude the possibility of contamination, negative controls and positive controls were performed each time.
The mutation of HBV may have a close relationship to the continuous HBV infection and the deterioration of liver after the infection. The research on the mutation of HBV will greatly promote the clinical analysis of HBV infection and its subsequent diseases. The common methods to detect point mutation are DNA sequencing and SSCP, which are expensive, time-consuming and unsaitable for clinical practice. In order to fulfill the requirements of further researches on HBV mutation and its application to the clinic, we established the 3'-BS-PCR method for the analysis of point mutation in site 1896 of HBV precore.
According to the principle of primer design that only when the last base of primer's 3' end must strictly match its corresponding template could PCR be accomplished, the reports of Georgen[6] and the phenomenon that the nucleotide of site 1896 of HBV precore mutates from G to A, the corresponding 5' primers of wild and mutant types whose 3' terminal base are G and A respectively were designed to amplify the wild or mutant type DNA of the HBV precore region. Georgen[6] has successfully applied this kind of method to detect the point mutation of site 1896 in the HBV precore in the patients with positive anti-HBe results, which have been confirmed by DNA sequencing. The positive rates of this site's point mutation were slightly higher than the other reports[9]. It may be because most patients investigated were inpatients from the Department of Infectious Diseases whose liver damages were more severe than those from out patients. This explanation is supported by the recorded reports that the positive rates of mutant type are unanimous with the conditions of the liver damages. The establishment of this method provides a new approach to study the HBV infection and analyze the liver damage.
Many researches have shown that the infection of HBV has a close relationship with the carcinogenesis of liver cancer and is one of important factors inducing the liver cancer. With the research progresses in molecular oncology, two kinds of genes, oncogenes and anti-oncogenes, that have close relationship to carcinogenesis have been discovered. In the development of HCC, the activation of oncogene and the inactivation of anti-oncogene are frequent events, which cause the changes of the qualities or quantities of the important proteins that they are encoded and eventually lead to the carcinogenesis of normal cells. The integration of HBV DNA in the genome of the cells infected by the HBV is an important factor that brings the instability of chromosomes in liver cells[8].
Therefore, it is meaningful to investigate the HBV-DNA fragments' integration and its effect on the activation of oncogene and inactivation of anti-oncogene. Our research shows that 12 of 23 primary HCC patients were found to have the HBV precore in the cancer tissues whose positive rate was 52.2%. Among these patients, the HBV precore DNA were detected in 7 patients' sera whose positive rate was 30.4%. The results indicate that the HBV precore gene widely exists in liver cancer tissues and the replication of HBV is accompanied by the development of HCC. HCC patients (21.8%, 5/23) had negative results in serum but positive results in cancer tissues, which indicates that these patients had HBV precore DNA fragments integrated in their liver cells and there was no virus replication. We plan to study whether there is an integration in the genome of liver cells using Southern Blot. Among these HBV precore positive patients, the patients with mutant type detected in the tissues were all wild type. Only half of the patients with mutant type detected in the serum were also wild type. A hypothesis could be made that the HBV precore may mutate during long host in the liver cell and may play an important role in the transformation of normal liver cells.
Dr. Yuan Wang, graduated from Anhui Medical University (1982) and Shanghai Medical University (1987) postdoctor in Vanderbilt University and Northwester n University, USA from 1993 to 1995, professor and director, majoring in molecular biology and biochemistry, having 42 papers published.
Project supported by the Natural Science Foundation of Anhui Province, NO. 9741006 and Natural Science Foundation of Anhui Educational Commission, NO. JL-97-077.
Edited by You DY
proofread by Sun SM
| 1. | Ulrich PP, Bhat RA, Kelly I, Brunetto MR, Bonino F, Vyas GN. A precore-defective mutant of hepatitis B virus associated with e antigen-negative chronic liver disease. J Med Virol. 1990;32:109-118. [PubMed] |
| 2. | Fattovich G, McIntyre G, Thursz M, Colman K, Giuliano G, Alberti A, Thomas HC, Carman WF. Hepatitis B virus precore/core variation and interferon therapy. Hepatology. 1995;22:1355-1362. [PubMed] |
| 3. | Guptan RC, Thakur V, Sarin SK, Banerjee K, Khandekar P. Frequency and clinical profile of precore and surface hepatitis B mutants in Asian-Indian patients with chronic liver disease. Am J Gastroenterol. 1996;91:1312-1317. [PubMed] |
| 4. | Luber B, Arnold N, Stürzl M, Höhne M, Schirmacher P, Lauer U, Wienberg J, Hofschneider PH, Kekulé AS. Hepatoma-derived integrated HBV DNA causes multi-stage transformation in vitro. Oncogene. 1996;12:1597-1608. [PubMed] |
| 5. | Robinson WS, Klote L, Aoki N. Hepadnaviruses in cirrhotic liver and hepatocellular carcinoma. J Med Virol. 1990;31:18-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Georgen B. Comparison of mutation specific PCR and direct sequencing of PCR-products for the detection of the HBV pre-C stop codon. Hepatology. 1992;16:232. |
| 7. | Wang Y, Li X, Zhou Q. Analysis of precore site mutation of hepatitis B virus by polymerase chain reaction. Anhui Yike Daxue Xuebao. 1998;33:251-253. |
| 8. | Robinson WS. The role of hepatitis B virus in the development of primary hepatocellular carcinoma: Part I. J Gastroenterol Hepatol. 1992;7:622-638. [PubMed] |
| 9. | Wang XF, Liu HD, Wang JR, Zhou F, Lei PJ. Studies of HBV precore region 1896 site mutation of patients with hepatitis B and hepatoma. Zhonghua Chuanranbingxue Zazhi. 1996;14:11-13. |