Published online Jun 15, 2000. doi: 10.3748/wjg.v6.i3.377
Revised: February 11, 2000
Accepted: February 18, 2000
Published online: June 15, 2000
AIM: To construct the recombinant of HDV cDNA and HBV-specific ribozyme gene by recombinant PCR in order to use HDV as a transporting vector carrying HBV-specific ribozyme into liver cells for inhibiting the replication of HBV.
METHODS: We separately cloned the ribozyme (RZ) gene and recombinant DVRZ (comprising HDV cDNA and HBV-specific ribozyme gene) into the downstream of T7 promoter of pTAdv-T vector and studied the in vitro cleavage activity of their transcripts (rRZ, rDVRZ) on target RNA (rBVCF) from in vitro transcription of HBV C gene fragment(BVCF).
RESULTS: Both the simple (rRZ) and the recombinant ribozyme rDVRZ could efficiently catalyze the cleavage of target RNA (rBVCF) under different temperatures (37 °C, 42 °C and 55 °C) and Mg2+ concentrations (10 mmol/L, 15 mmol/L and 20 mmol/L) and their catalytic activity tended to increase as the temperature was rising. But the activity of rRZ was evidently higher than that of rDVRZ.
CONCLUSION: The recombinant of HDV cDNA and ribozyme gene had the potential of being further explored and used in gene therapy of HBV infection.
- Citation: Wen SJ, Xiang KJ, Huang ZH, Zhou R, Qi XZ. Construction of HBV-specific ribozyme and its recombinant with HDV and their cleavage activity in vitro. World J Gastroenterol 2000; 6(3): 377-380
- URL: https://www.wjgnet.com/1007-9327/full/v6/i3/377.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i3.377
Hepatitis B virus (HBV) can cause acute and chronic B-type hepatitis in man. The conventional ways available for curing this disease have not been very efficient. This promotes people to explore novel genetic therapeutical ways. Hammerhead ribozyme is a kind of antisense RNA which can specifically cleave the target RNA[1,2]. In the light of this, people have developed many effective genetic vectors containing ribozyme genes, the transcripts of which showed catalytic activity in vitro and in vivo[3-5]. But how to improve the stability and efficiency of ribozyme and specifically carry ribozyme gene into only target cells or tissues has been a tackling problem. HDV, a human hepatitis agent, is a defective RNA virus, the replication cycle of which relies on the infection of HBV[6]. So HDV can be developed as a specific transporting and replicating vector in vivo for ribozyme to reach liver[7]. In this study, we constructed HBV-specific hammerhead ribozyme gene (RZ) and the recombinant (DVRZ) of HDV cDNA (DV) and ribozyme gene (RZ) and made a careful investigation of their in vitro catalytic activity under various conditions. The positive results encouraged us to further explore the feasibility of- using HDV as a vector carrying ribozyme for inhibiting the replication of HBV in vivo.
The plasmid pSVC-D3 (containing HDV cDNA) as one of two templates of recombinant PCR was kindly offered by Prof. Taylor of American. pTAdV-T vector used for cloning was purchased from Clontech Corporation.
RiboMax transcription kit, acrylamide, bisactylamide, dNTP, rNTP and Taq polymer ase were purchased from Promega. [α-32P]UTP from Beijing Yahui Corporation. Advantage-TM PCR pure kit (gel purification kit) from Clontech. X-film from Kodak. DNA polymerase I klenow from Biolabs. T7/Sp6 sequencing kit from Pharmacia.
Primers P1 and P2 covered the whole sequence of designed HBV-specific hammerhea d ribozyme gene(RZ) and were used to amplify ribozyme gene(53 bp) because of 9 nt base-pairing of their 3'ends. The sequence of HBV-specific hammerhead ribozyme was designed according to the requirement of domains of ribozyme[1] and the sequence of HBV C gene fragment.
The ribozyme gene (RZ: 53 bp) was also used as one of two templates for recombinant PCR to construct the recombinant (DVRZ) of HDV cDNA and ribozyme gene (mentioned below).
P3, P4 were both recombinant primers, each of which was composed of partial sequences of ribozyme and HDV in order to replace the sequence (17-67) of near 5'-end of HDV with ribozyme by recombinant PCR. P5 was the 3'-end sequence of HDV cDNA.
P3, P4, P5 were used to construct the recombinant (DVRZ) of HDV cDNA (DV) and HBV-specific ribozyme gene (RZ) by one-tube recombinant PCR[8].
P6, P7 were used to amplify HBV C gene fragment (BVCF), which was the transcription template of target RNA (rBVCF). T7 was partial sequence of T7 promoter region of pTAdV-T vector and was used for sequencing (with Sp6) and identifying(with P2, P5, P7) whether 5'-end of foreign fragment forwardly inserted the downstream of T7 promoter.
The sequencing of all gene fragments was performed on ABI391 automatic sequencer (Pharmacia). The sequences and positions of these primers are listed in Table 1.
| Primer name | Sequences | Positions |
| P1 | 5'-AACATTGACATAGCTCTGATGAGTCCGTGAG-3' | RZ: 1-31 |
| P2 | 5'-TCCAGGGAATTAGTAGTTTGTCCTCACGGAC-3' | RZ: 53-23 |
| P3 | 5'-AGCAAGCTTGAGCCAAAACATTGACATAGCTCT-3' | HDV: 1-16, RZ: 53-37 |
| P4 | 5'-CTCCGACGTTCCAATGCTCCAGGGAATTAGTAGT-3' | HDV: 84-68, RZ: 53-37 |
| P5 | 5'-GTCGAATTCGGGCTCGGGCGGCGATCCAGCAGTC-3' | HDV: 1680-1647 |
| P6 | 5'-GATAAGCTTTTACATAGAGGACTCTTGG-3' | HBV: 1650-1677 |
| P7 | 5'-CTGGAATTCGGCGAGGGAGTTCTTCTTCTAG-3' | HBV: 2480-2450 |
Because of 9 nt base-pairing between 3'ends of P1 and P2, direct PCR could produce complete 53 bp ribozyme gene (Figure 1). After gel-purification (according to Advantage-TM PCR pure kit, Clontech Corporation), the PCR product (RZ) was directly cloned into the downstream of T7 promoter of pTAdV-T vector and so the resulting recombinant plasmid pTA-RZ was obtained. PCR with T7/P2 as primers and pTA-RZ as template could produce about 100 bp DNA fragment if 5'-end of ribozyme gene was forwardly inserted in to the downstream of T7 promoter of pTAdV-T vector and so could be used to identify the recombinant plasmid pTA-RZ of forward insert. The preparation of competent DH5 α cells and transformation of plasmids were performed according to the reference[9]. The cloned ribozyme gene was finally verified by sequencing with T7/Sp6 primers.
Thirty μL PCR reaction system was established[8]: 20 mmol/L Tris-HCl (pH8.3), 50 mmol/L KCl, 2 mmol/L MgCl2, each dNTP 200 μmol/L; Primers P3, P4, P5 were separately 0.05 μmol/L, 0.005 μmol/L, 0.05 μmol/L. The two templates were recombinant plasmids pTA-RZ (containing ribozyme gene) and pSVC-D3 (containing HDV cDNA), each 10 ng. Taq polymerase 2 μL, denaturation at 92 °C-50 s; annealing at 53 °C-50 s; elongation at 70 °C-120 s; 33 cycles; the final elongation at 70 °C lasted 5 min. PCR product (DVRZ, 1.7 kb) was identified by agrose-gel (1.5%) electro phoresis and then purified with Advantage-TM PCR pure kit. The obtained DNA fragment (DVRZ) was directly cloned into the downsteam of T7 promoter of pTAdV-T vector. The resulting recombinant plasmid pTA-DVRZ (positive clones on Ampr & white-blue plate) was verified by PCR with primers T7/P5 and sequencing with Sp6/T7 primers. PCR with primers T7/P5 could produce about 1.7-1.8 kb DNA fragment and was used to identify the forward insertion of 5'-end of DVRZ. Sequencing was performed on ABI 391 automatic sequencer (Pharmacia).
PCR with primers P6, P7 was performed to amplify HBV C gene fragment from serum of HBV. Extraction of sample DNA and PCR reaction were performed according to the reference[10]. PCR product (BVCF) was directly cloned into the downstream of T7 promoter of pTAdV-T vector and the resulting recombinant plasmid pTA-BVCF (positive clones on Ampr & blue-white plate) was verified by PCR with T7/P7 and bidirectional sequencing with T7/Sp 6 primers. PCR with primers T7/P7 was utilized to identify the forward-direction insertion of 5'-end of BVCF.
These plasmids were seperately digested with BamH I and then filled with Klenow fragment for in vitro transcription of inserted gene fragments (RZ, DVRZ, BVCF).
32P-UTP-labeled transcription of the two plasmids was first carried out in order to test the effect of transcription. But the plasmids' transcripts (rRZ, rDVRZ) used for catalyzing the cleavage of target RNA (rBVCF-transcript of plasmid pTA-BVCF containing HBV C gene fragment-BVCF) were not 32P-labeled. In vitro transcription was performed with T7 RNA polymerase according to RiboMax Transcripti on kit and the reference[11]. The transcripts (rRZ, rDVRZ) would additionally contain 100 nt partial sequence of pTAdV-T vector at their both ends. rRZ and rDVRZ were extracted by phenol/chloroform/iso-propyl alcohol (25:24:1), precipitated with ethanol and resuspensed with H2O (RNase-free). The latter was the recombinant of HDV and ribozyme.
Linearized plasmid pTA-BVCF (containing HBV C gene fragment) was in vitro transcripted and the target RNA (rBVCF) was produced. 20 μL transcription system was so established: ATP, CTP, GTP each 2.5 mmol/L and UTP 0.1 μmol/L, 0.5 μCi/μL [α-32P]UTP, 2 μg linearized plasmid pTA-BVCF, T7 RNA polymerase 20 U, RNasin 10 U, 37 °C-60 min. The product was loaded for 5% PAGE-7M urea autoradiographed electrophoresis. Gel-band of 1 cm lengt h and 0.5 cm breadth was cutted off from the position of target RNA and soaked overnight with NES buffer (0.5 mol/L NH4Ac, 1 mmol/L EDTA, 0.1% SDS) and extracted with phenol/chloro form/iso-propyl alcohol (25:24:1). The supernate was precipitated with ethanol and then the pellet was harvested and resuspensed with H2O (RNase-free). rBVCF (831 + 100 nt) contained also 100 nt partial sequence of pTAdV-T vector at its two ends.
Ten μL reaction system was established: 0.1 mol/L Tris-HCl (pH8.0), 20 mmol/L MgCl2, rRZ or r-DVRZ and their target RNA (rBVCF) each 2 μL, mixed and incubated under different temperatures (37 °C, 42 °C, 55 °C) for 1 h. Negative control with 32P-labeled rBVCF incubated at 55 °C without rRZ & rDVRZ was performed. In the meantime, rRZ and rDVRZ were separately incubated with target RNA (rBVCF) at 37 °C under different Mg2+ concentrations (10 mmol/L, 15 mmol/L). In the end, 10 μL ion-free formamide and 1 μL loading buffer (50% glycerol, 1 mmol/L EDTA, 0.04% bromphenol blue) were added to terminate the reaction. Then after incubated at 65 °C for 10 min, 5 μL reaction sample was loaded for 5% PAGE-7M urea electrophoresis.
53 bp DNA fragment was obtained by PCR with primers P1/P2 (Figure 1). This was in accordance with the size of anticipated ribozyme gene. PCR using T7/P2 as primers and plasmid pTA-RZ as the template amplified a DNA fragment of about 100 bp, which identified forward-direction insertion of ribozyme gene into the downstream of T7 promoter of pTAdV-T vector. Sequencing verified the HBV-specific ribozyme gene. The sequence of ribozyme gene was as follows: 5'-AACATTGACATAGCTCTGATGAGTCCGTGAGGACAAACTACTAATTCCCTGGA3'
About 1.7 kb DNA was amplified by recombinant PCR, which indicated the anticip ated recombinant DNA molecule DVRZ (Figure 1). After DVRZ's cloning into pTAdV-T vector and transforming into DH5α on Ampr & white-blue plate, PCR with primers T7/P5 produced about 1.8 kb-DNA fragment and identified the positive recombinant plasmid pTA-DVRZ of correct DVRZ's insertion direction. Sequencing of DVRZ with primers T7/Sp6 (pTA-DVRZ as template) confirmed the construct DVRZ.
Anticipated 831 bp target DNA fragment was amplified by PCR from the serum of HBV (Figure 1). After its cloning into pTAdV-T vector and then transformed into DH5α, PCR with primers T7/P7 produced an about 0.9 kb DNA fragment and so identified the positive recombinant plasmid pTA-BVCF of BVCF's correct insertion direction. Finally, sequencing with primers T7/Sp6 confirmed the inserted BVCF.
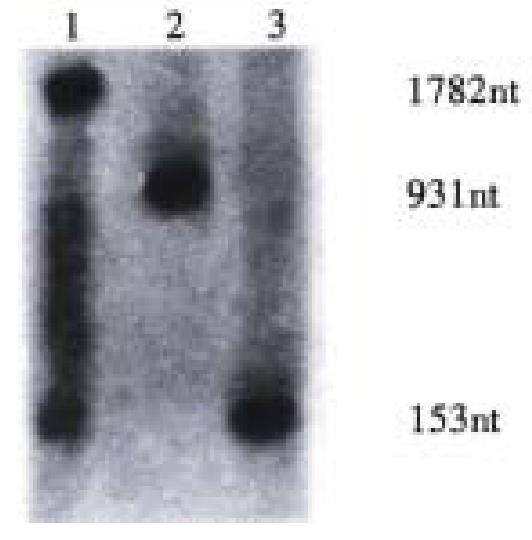
Target RNA (rBVCF) of 931 nt was transcripted from pTA-BVCF. HBV-specific ribo zyme (rRZ) of 153 nt was transcripted from pTA-RZ.
HBV-specific recombinant ribozyme (rDVRZ, containing HDV) of 1782 nt was transcripted from pTA-DVRZ (Figure 2). All these transcripts (rBVCF, rRZ, rDVRZ) contained the same partial sequence of pTAdV-T vector at their both ends after transcription with T7 RNA polymerase.
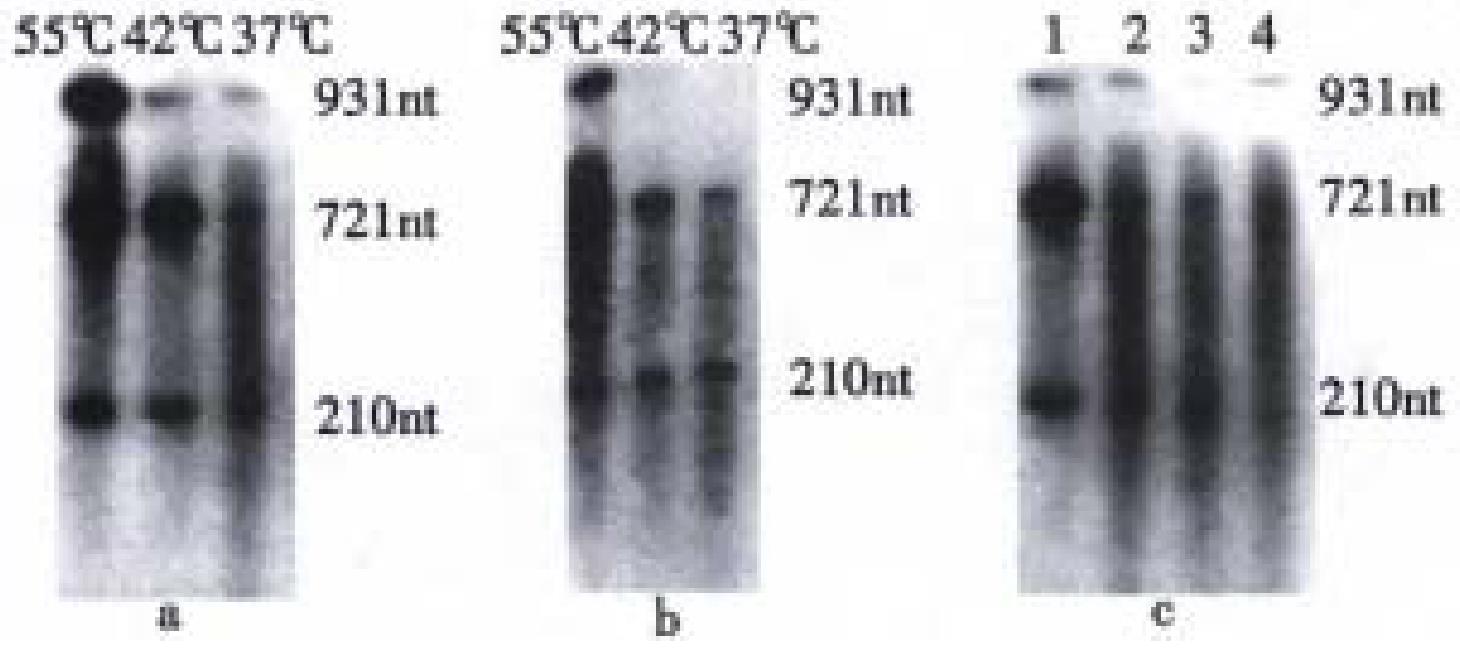
Ribozymes rRZ and rDVRZ without 32P-label were separately incubated with target RNA (32P-labeled rBVCF) under different temperatures(37 °C, 42 °C, 55 °C) and Mg2+ concentrations. The results from autoradiog raphed electrophoresis showed that under these temperatures and Mg2+ concentrations, both rRZ and rDVRZ could catalyze the cleavage of target rBVCF into two RNA fragments (721 nt, 210 nt) and their catalytic activity tended to increas e with the rising of temperature. Comparatively, the catalytic activity of rRZ was higher than that of rDVRZ. But it seemed that Mg2+ from 10 mmol/L to 20 mmol/L had no obvious effect on their cleavage activity (Figure 3).
Hammerhead ribozyme is a kind of antisense RNA with specific catalytic activity, which can catalyze the specific cleavage of target RNA[1-3]. So far, many specific ribozyme constructs have demonstrated their catalytic activity both in vitro and in vivo[12-15]. HDV, is a defective RNA virus, the replication cycle of which must be dependent on the infection of HBV[6]. In view of the characteristics of hammerhead ribozyme and HDV, we constructed the recombinant (rDVRZ) of HDV and ribozyme by one-tube recombinant PCR with 3 primers and intended to have HBV-specific ribozyme carried into liver cells by using HDV as a transporting vector. As the first step, we studied the in vitro cleavage activity of HBV-specific ribozymes-rRZ and rDVRZ. The results showed that both rRZ and rDVRZ had the a bility of catalyzing the specific cleavage of target RNA (rBVCF-in vitro transcript of HBV C gene fragment) into two RNA fragments (721 nt, 210 nt). The activity of ribozyme rRZ (without containing HDV) was much higher than that of recombinant rDVRZ, which in part cleaved the target RNA. One possible reason is that rDVRZ has much longer two arms outside its base-pairing region and can easily form a complex secondary or tertiary structure, an obstacle to subsequent base-pairing with target RNA (rBVCF). Its three dimensional structure simulated by computer (not presented here) supports this conclusion. However, we could conclude that the post-transcripted hammerhead ribozyme should not be too long though we are unsure how long ribozyme and its target RNA are optimal to their interaction.
We found that with the temperature rising, the ribozyme activity increased, possibly because higher temperature helped transit the complex tertiary structure into comparatively extended state and so partially delete the structural obstacle to base- pairing. We also found that the requirement of ribozyme for Mg2+ was not strict. It appeared that different Mg2+ concentrations from 10 mmol/L to 20 mmol/L could meet the ribozymes' activity. But overhigh Mg2+ seemed to cause the non-specific cleavage of target RNA (not presented). The reason for this was unclear. Presently, we are studying the activity of the recombinant ribozyme in vivo and its repressive effect on HBV replication.
Shu-Juan Wen, graduated from First Military Medical University in 1985, major in gene diagnosis, having 6 papers published.
Edited by You DY
proofread by Sun SM
| 1. | Cech TR. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987;236:1532-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 343] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 2. | Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627-3640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 401] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 3. | von Weizsäcker F, Blum HE, Wands JR. Cleavage of hepatitis B virus RNA by three ribozymes transcribed from a single DNA template. Biochem Biophys Res Commun. 1992;189:743-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 4. | Funato T, Ishii T, Kanbe M, Scanlon KJ, Sasaki T. Reversal of cisplatin resistance in vivo by an anti-fos ribozyme. In Vivo. 1997;11:217-220. [PubMed] |
| 5. | Kim YK, Junn E, Park I, Lee Y, Kang C, Ahn JK. Repression of hepatitis B virus X gene expression by hammerhead ribozymes. Biochem Biophys Res Commun. 1999;257:759-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Karayiannis P. Hepatitis D virus. Rev Med Virol. 1998;8:13-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Hsieh SY, Taylor J. Delta virus as a vector for the delivery of biologically-active RNAs: possibly a ribozyme specific for chronic hepatitis B virus infection. Adv Exp Med Biol. 1992;312:125-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Marini F, Naeem A, Lapeyre JN. An efficient 1-tube PCR method for internal site-directed mutagenesis of large amplified molecules. Nucleic Acids Res. 1993;21:2277-2278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning laboratory mannual, 2nd. New York: Cold Spring Harbor Laboratory Press 1989; . |
| 10. | Bruce AW. PCR Protocols: current methods and applications. New Jersey: Human Press 1993; . |
| 11. | Xu ZK, Anzola JV, Nalin CM, Nuss DL. The 3'-terminal sequence of a wound tumor virus transcript can influence conformational and functional properties associated with the 5'-terminus. Virology. 1989;170:511-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Beck J, Nassal M. Efficient hammerhead ribozyme-mediated cleavage of the structured hepatitis B virus encapsidation signal in vitro and in cell extracts, but not in intact cells. Nucleic Acids Res. 1995;23:4954-4962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Zern MA, Ozaki I, Duan L, Pomerantz R, Liu SL, Strayer DS. A novel SV40-based vector successfully transduces and expresses an alpha 1-antitrypsin ribozyme in a human hepatoma-derived cell line. Gene Ther. 1999;6:114-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Wands JR, Geissler M, Putlitz JZ, Blum H, von Weizsäcker F, Mohr L, Yoon SK, Melegari M, Scaglioni PP. Nucleic acid-based antiviral and gene therapy of chronic hepatitis B infection. J Gastroenterol Hepatol. 1997;12:S354-S369. [PubMed] |
| 15. | Albuquerque-Silva J, Milican F, Bollen A, Houard S. Ribozyme-mediated decrease in mumps virus nucleocapsid mRNA level and progeny in infected vero cells. Antisense Nucleic Acid Drug Dev. 1999;9:279-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |