Published online Feb 15, 2000. doi: 10.3748/wjg.v6.i1.136
Revised: August 26, 1999
Accepted: September 19, 1999
Published online: February 15, 2000
- Citation: Dai YM, Shou ZP, Ni CR, Wang NJ, Zhang SP. Localization of HCV RNA and capsid protein in human hepatocellular carcinoma. World J Gastroenterol 2000; 6(1): 136-137
- URL: https://www.wjgnet.com/1007-9327/full/v6/i1/136.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i1.136
The relation of HCV to hepatocytic carcinoma (HCC) has been emphasized recently in low HBV infection countries and regions. The distribution patterns of HCV in liver tissues are not well understood although studies on HCV infection in blood and hepatocytes have been conducted by PCR. In this study, 42 liver cancers and surrounding liver tissues were detected for HCV RNA and HCAg using photosensitive biotin-labeled HCV cDNA probe and Immuno-gold-silver stain (IGSS) method.
Fourty-two samples including cancers and their surrounding tissues were collect ed and processed into formalin-fixed, paraffin-embedded blocks.
NS5 cDNA probe PCR product from NS5 region was inserted into charomid 9-42. This resulted in recombinant HCV plasmid which could be spliced into specific cDNA fragment with DNA length 534 bp[1].
HCV cDNA probe HCVC cDNA probe was the PCR product of C region.
The preparation of photosensitive biotin-labelled probe was as described in literature[2].
5 μm sections were dewaxed, rehydrated in buffered solutions, then digested in proteinase K, and fixed in polyformaldehyte. After dehydration, prehybridization was performed and then the sections were denatured (steaming bath at 100 °C, 10 min, then rapidly cooling down in ice bath), and hybridized again (42 °C, overnight). After rinse, the sections were conjugated with gold-labeled strepto-biotin, stained with AgNO3 buffered solution, then restained in hemoxylin and eosin or eosin. Positive stain should be present as intracytoplasmic or intranuclear black granules.
Immunohistochemical detection of HCAg PAP method was used. The specific human anti-HCV antibody was from Laboratory of Immunology at Tongji Medical University.
Negative control Sections were treated with RNase (0.1 g/L), 1 h, at 37 °C.
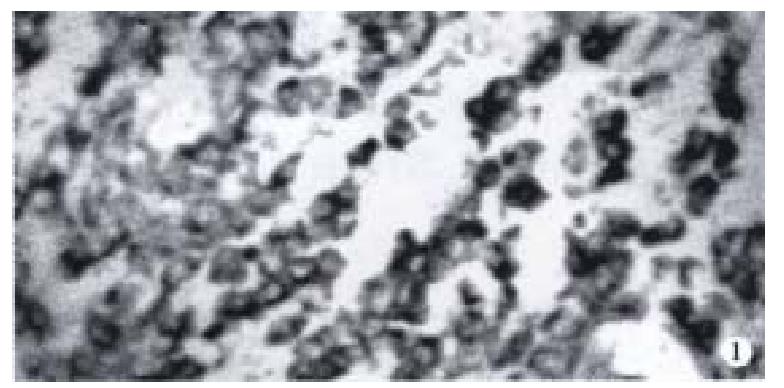
HCV RNA positivity was detected in 8 cancers and their surrounding tissues including 2 positivity in cancers, 1 in both cancer and their surrounding tissue, 5 in surrounding liver tissues. Positive cells appeared diffuse, clustered, or discrete in cancer and its surrounding tissues. Three types of positive granules were noted: ① cytoplasmic, diffuse granules in cytoplasm with prominent perinuclear staining (Figure 1); ② nuclear, evenly diffuse granules in nucleus; ③ nuclear-cytoplasmic, positive granules present in both cytoplasm and nucleus. Detection by NS5 cDNA probe revealed that 7 out of 42 were positive; detection by C cDNA probe showed one nuclear positive in 2 cases.
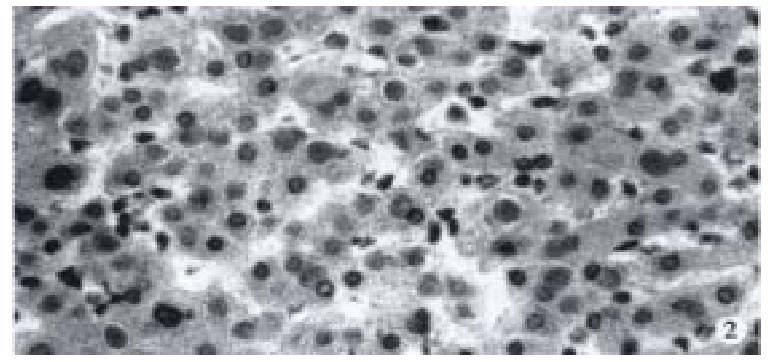
Ten out of 42 stained positive for HCAg and positive cells were distributed in cancers and its surrounding tissues. HCAg positive cells appeared diffuse, clustered or discrete. Two distributing patterns of intracellular positive granules: ① cytoplasmic type: diffuse positive granules in cytoplasm; ② inclusion type: positive granules present as inclusions in cytoplasm (Figure 2).
HCV is a single-stranded RNA virus which reproduces itself on the template of negative strand and its copies are rarely present in tissues. It has been established that replication of HCV RNA occurs mainly in liver tissues. How it infects liver cells and at which point it starts duplication remains unknown.
Since 1992, there have been some reports on detection of HCV RNA in hepatic tissues using ISH, and HCV RNA was present in cytoplasm or nucleus[3-8]. However, it is still unclear whether HCV RNA is present in hepatocytic carcinoma. Using ISH, we found that HCV RNA did infected cancers and their surrounding tissues, which was in good agreement with other authrs’ reports. HCV infect ed cells appeared diffuse, clustered or discrete in cancers and their surrounding tissues.
There is a viewpoint that the reverse transcriptive form of HCV RNA does not exist at all during its replication. Therefore, it is disputable on the significance of its presence in nucleus. Kayhan[8] reported that HCV RNA was present in nucleus and nucleoli, and suggested that HCV duplication might use its host’s genes or intranuclear apparatus, or HCV granules accumulated in infected cells like HBV. Our results were in accordance with the above findings. The studies on yellow fever virus also supported the above viewpoints: NS5 protein could be detected in nucleus and nucleoli, indicating that viral replication could occur in nucleus[9].
In 42 liver cancers, 10 were positive for HCAg, while only 8 expressed HCV RNA by ISH, indicating that positivity of viral genes was lower than that of viral antigen. It is suggested that this phenomenon could be accounted by the variations of viral genes, and the fixing time by formalin. HCV is a variable virus, it may differ individually and may vary at different stages of infection. Therefore, the variation of gene sequence may result in less sensitivity of ISH detection. In addition, longer fixation may cause loss of viral genes or false negativity. The above defects could be overcome by using probes of different fragments of the same gene. Two cases with HCAg(+), NS5 cDNA(-) were hybridized again and revealed positive signals of NS5 cDNA in one case. This agreed well with Haruna’s[7] findings. Although ISH is not as sensitive as PCR, it is a promising technique in the study of relation of HCV RNA and infected cells[10-12].
The significance and mechanism of continual variation of HCV gene in vitro remain to be resolved in recent years. It was reported that HCVs continual variation in vivo may be related to virus escape from host immune deletion and virus configurative change brought about by immune injury introduced by the host[13-15].
Project supperted by the National Natural Science Foundation of China, No.39370294.
Edited by Wang XL
| 1. | Qi ZT, Pan WD, Du P. cDNA cloning and sequencing of hepatitis C virus in Chinese people. Dier Junyi Daxue Xuebao. 1992;13:301-306. |
| 2. | Zhang HQ. Application of photosensitive biotin-labeling, Hantan virus cDNA probe and gold-strepto-biotin in dot blot hybridization. Junshi Yixue Kexueyuan Yuankan. 1990;14:309-311. |
| 3. | Yamada S, Koji T, Nozawa M, Kiyosawa K, Nakane PK. Detection of hepatitis C virus (HCV) RNA in paraffin embedded tissue sections of human liver of non-A, non-B hepatitis patients by in situ hybridization. J Clin Lab Anal. 1992;6:40-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Yamada G, Nishimoto H, Endou H, Doi T, Takahashi M, Tsuji T, Yoshizawa H, Nozawa M, Koji T, Nakane PK. Localization of hepatitis C viral RNA and capsid protein in human liver. Dig Dis Sci. 1993;38:882-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Negro F, Pacchioni D, Shimizu Y, Miller RH, Bussolati G, Purcell RH, Bonino F. Detection of intrahepatic replication of hepatitis C virus RNA by in situ hybridization and comparison with histopathology. Proc Natl Acad Sci USA. 1992;89:2247-2251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 100] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Tsutsumi M, Urashima S, Takada A, Date T, Tanaka Y. Detection of antigens related to hepatitis C virus RNA encoding the NS5 region in the livers of patients with chronic type C hepatitis. Hepatology. 1994;19:265-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Haruna Y, Hayashi N, Hiramatsu N, Takehara T, Hagiwara H, Sasaki Y, Kasahara A, Fusamoto H, Kamada T. Detection of hepatitis C virus RNA in liver tissues by an in situ hybridization technique. J Hepatol. 1993;18:96-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Nouri Aria KT, Sallie R, Sangar D, Alexander GJ, Smith H, Byrne J, Portmann B, Eddleston AL, Williams R. Detection of genomic and intermediate replicative strands of hepatitis C virus in liver tissue by in situ hybridization. J Clin Invest. 1993;91:2226-2234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 78] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Buckley A, Gaidamovich S, Turchinskaya A, Gould EA. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J Gen Virol. 1992;73:1125-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Yamada G, Nishimoto H, Endou H, Doi T, Takahashi M, Tsuji T, Yoshizawa H, Nozawa M, Koji T, Nakane PK. Localization of hepatitis C viral RNA and capsid protein in human liver. Dig Dis Sci. 1993;38:882-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Haruna Y, Hayashi N, Kamada T, Hytiroglou P, Thung SN, Gerber MA. Expression of hepatitis C virus in hepatocellular carcinoma. Cancer. 1994;73:2253-2258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Sansonno D, Cornacchiulo V, Racanelli V, Dammacco F. In situ simultaneous detection of hepatitis C virus RNA and hepatitis C virus-related antigens in hepatocellular carcinoma. Cancer. 1997;80:22-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | McGuinness PH, Bishop GA, Painter DM, Chan R, McCaughan GW. Intrahepatic hepatitis C RNA levels do not correlate with degree of liver injury in patients with chronic hepatitis C. Hepatology. 1996;23:676-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Kojima S, Tanaka Y, Enomoto N, Marumo F, Sato C. Distribution of hepatitis C virus RNA in the liver and its relation to histopathological changes. Liver. 1996;16:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Zhao XP, Shen HX, Tian DY, Zhang DS, Peng ZH, Yang DL, Hao LJ. Express ion and significance of HCV RNA NS5 antigen in liver tissues of patients with he patitis C. Shijie Huaren Xiaohua Zazhi. 1999;7:516-518. |